Abstract
Metagenomic sequencing projects yield numerous sequencing reads of a diverse range of uncultivated and mostly yet unknown microorganisms. In many cases, these sequencing reads cannot be assembled into longer contigs. Thus, gene prediction tools that were originally developed for whole-genome analysis are not suitable for processing metagenomes. Orphelia is a program for predicting genes in short DNA sequences that is available through a web server application (http://orphelia.gobics.de). Orphelia utilizes prediction models that were created with machine learning techniques on the basis of a wide range of annotated genomes. In contrast to other methods for metagenomic gene prediction, Orphelia has fragment length-specific prediction models for the two most popular sequencing techniques in metagenomics, chain termination sequencing and pyrosequencing. These models ensure highly specific gene predictions.
INTRODUCTION
Metagenomics is an approach to the characterization of microbial genomes without the cultivation of individual species under laboratory conditions. In metagenomic sequencing projects, DNA is directly isolated from the environment and sequenced. Currently, the most common sequencing methods utilized in this field are chain termination sequencing (also named Sanger sequencing) (1), which yields an average read length of ∼700 bp, and the more cost efficient pyrosequencing (2), which results in reads of average length ∼300 bp. Regardless of the read length, it is in many cases impossible to reliably assemble metagenomic sequencing reads into longer contigs because diversity in metagenomic samples is often too large to provide a high sequencing coverage of single species. To answer one of the major questions of metagenomic sequencing projects, which parts of the sequencing reads encode for proteins, methods are required that can identify genes directly in short and anonymous DNA fragments.
In principle, metagenomic gene prediction is accomplished by two approaches. One is the identification of genes through homology-based methods, for instance by BLAST search of the input sequence against a database of known proteins (3). This approach is limited to the prediction of genes that are highly similar to already known genes. By the clustering of open reading frames (ORFs), homology-based methods can also find novel genes which are conserved within the metagenomic sample (4,5). However, these methods become computationally expensive for large samples. A different approach is gene prediction by means of statistical models. Model-based gene prediction methods have the advantage that they can discover novel genes at lower computational cost and without the prerequisite of a high conservation of these genes within the sample. On the other hand, most model-based methods are sensitive to sequencing errors in form of frame shifts. Up to now, three model-based gene prediction tools for metagenomic DNA fragments are available, namely MetaGene (6), its successor, the MetaGeneAnnotator (7), and GeneMark with a heuristic model (8). All three tools are available as web server applications. In contrast to the MetaGene and MetaGeneAnnotator web servers, the GeneMark web server was not designed to treat single entries of a multiple fasta file separately, which limits its applicability to metagenomic data. Nevertheless, all tools achieve a good performance on fragments of Sanger read length. Prediction accuracies on 300 bp DNA fragments are lower.
Here, we introduce the ab initio gene prediction web server application ‘Orphelia’, which is based on our previously published machine learning approach to metagenomic gene prediction (9). While the other three tools utilize the same prediction model for all read lengths, Orphelia currently supports two separate models for the most common sequencing techniques in metagenomics, thereby also providing highly specific gene predictions in fragments <300 bp. A high gene prediction specificity can be very important for high-throughput metagenome analysis, because the large number of sequences usually makes a manual curation of the predictions impossible.
METHODS
In a first step, Orphelia identifies all ORFs in the input sequence. By our definition, ORFs begin with a start codon (ATG, CTG, GTG, or TTG), are followed by at least 18 subsequent triplets, and end with a stop codon (TGA, TAG, or TAA). Due to the short input sequence length, we also consider incomplete ORFs of at least 60 bp input length that lack a start and/or stop codon. After extraction, all ORFs are scored by a gene prediction model that is based on machine learning techniques. Finally, a greedy method with a maximal overlap constraint selects a combination of highly probable genes.
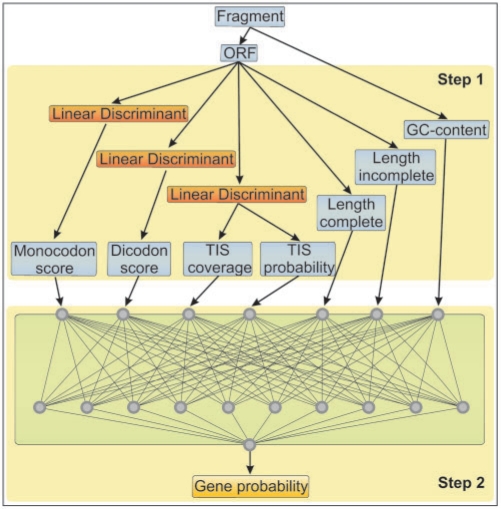
The gene prediction model is sketched in Figure 1. At first, features for monocodon usage, dicodon usage and translation initiation sites are extracted from the ORF sequence using linear discriminants. The discriminants were trained on 131 fully sequenced prokaryotic genomes (9). After feature extraction, an artificial neural network combines the sequence features with ORF length and fragment GC-content, and computes a posterior probability of an ORF to encode a protein. The neural network was trained on randomly excised DNA fragments of a specified length from the genomes that were used for linear discriminant training. In our previous publication, we provided a prediction model in which the neural network was trained on 700 bp fragments for predicting genes in Sanger read length fragments. We showed that this model is robust with respect to varying sequence length (above ∼300 bp). On fragments as short as ∼300 bp, we observed a drastic decrease in performance. Therefore, the Orphelia web server also provides an additional prediction model that was trained on 300 bp fragments, which corresponds to the average read length of pyrosequencing.
Figure 1.
Orphelia's ORF scoring model. In Step 1, 7 ORF/fragment features are computed. Step 2 calculates a final gene probability, combining the features by means of a neural network.
Besides the discriminant-based translation initiation site (TIS) probability as inferred from a 60 bp TIS region around the potential start codon, we now use the ‘TIS coverage’ as an additional feature. The TIS coverage is the fraction of the TIS region, which is actually contained in the sequence fragment. This feature accounts for incomplete TIS regions and completely missing start codons, which imply a zero coverage.
WEB SERVER
Input
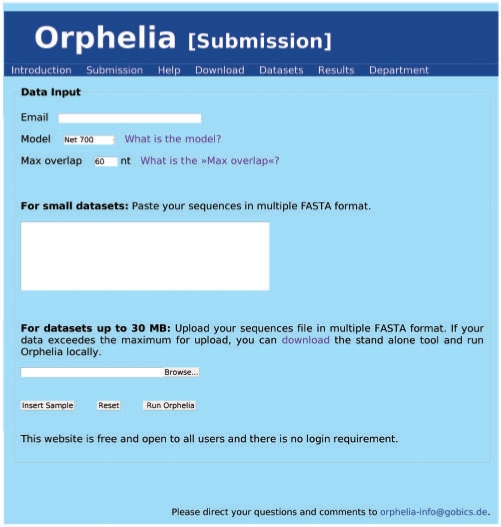
The Orphelia submission page is shown in Figure 2. Orphelia requires as input data a set of DNA sequences in standard multiple FASTA format. Small data sets can be pasted into the sequence window, larger data sets should be uploaded via the ‘Browse’ button. Currently, the upload is limited to 30 MB. If a data set exceeds this size, we recommend either the splitting into smaller files, or the usage of our standalone command-line tool for 64-bit architecture Linux systems.
Figure 2.
Screenshot of the Orphelia web server application submission page.
Further, the prediction model to be utilized can be specified: Net700 should be selected for Sanger reads, Net300 for reads shorter than 300 bp. For calculating the final combination of predicted genes per fragment, Orphelia by default allows a maximal overlap of 60 bp between genes. The maximal overlap can be varied through the web interface. Finally, the user must provide a valid e-mail address to which an URL with a link to results will be sent.
Output
A typical run of Orphelia takes several minutes (for 10 MB input). Upon completion of the job, Orphelia sends an e-mail with two files to the user: the original input sequences, seq.fna, and the predicted genes, gene.pred. Predicted genes are given in a one-line-per-gene format:
>FragNo, GeneNo, Coord1_Coord2_Str_Fr_C_FH
FragNo is the fragment number in the input file, GeneNo is a numerical identifier of a Gene within the fragment. Coord1 and Coord2 indicate the positions of a predicted gene in the fragment, starting with position 1 at the beginning of the fragment. Str is the strand on which a predicted gene is encoded. The input sequence from 5′ to 3′ direction is assigned the ‘+’ strand. Fr gives the reading frame of a gene counted from the 5′-end of the sequence. Reading frame 1 begins at the first nucleotide position of the input sequence, frame 2 at the second position and frame 3 at the third position. C is a label which indicates whether a candidate is complete (C) or incomplete (I). FH stands for the FASTA header of the input sequence. The first three entries are separated by a comma (,), all subsequent entries are separated by an underscore (_).
EXPERIMENTAL RESULTS
Evaluation on simulated data
We evaluated the accuracy of Orphelia's prediction models on DNA fragments of 300 and 700 bp, respectively. The fragments were randomly excised to a 10-fold genome coverage from 12 annotated test genomes that were not contained in the training set of Orphelia and that were first proposed by Noguchi et al. in 2006 (6). We measured sensitivity, which reflects how many of the existing genes were detected, and specificity, which shows how many of the predicted genes are annotated. In addition, the harmonic mean, which combines sensitivity and specificity within a single measure was used according to: 2 × (Sensitivity × Specificity)/(Sensitivity + Specificity). All predicted genes that match at least 60 bp in the same reading frame on the same strand with the annotation were counted as true positives. Table 1 shows the mean and standard deviation of performance over all species. Orphelia (Net700) has a prediction sensitivity of 88%, a specificity of 93% and a harmonic mean of 90.5% on 700 bp fragments. On 300 bp fragments (Net300), sensitivity (82%), specificity (92%) and the harmonic mean (86.6%) are lower than on 700 bp fragments, but the specificity is still very good.
Table 1.
Mean and standard deviation of sensitivity, specificity and harmonic mean on 300 and 700 bp DNA fragments that were randomly excised from 12 test species
| 300 bp fragments |
700 bp fragments |
|||||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Harmonic mean | Sensitivity | Specificity | Harmonic mean | |
| Orphelia Net300 | 82.1 ± 3.6 | 91.7 ± 3.8 | 86.6 ± 2.7 | 49.5 ± 13.8 | 79.3 ± 6.9 | 59.4 ± 10.2 |
| Orphelia Net700 | 83.8 ± 3.4 | 88.1 ± 4.9 | 85.8 ± 3.9 | 88.4 ± 3.1 | 92.9 ± 3.2 | 90.6 ± 2.9 |
| MetaGene | 89.3 ± 3.3 | 84.2 ± 6.0 | 86.6 ± 4.3 | 92.6 ± 3.1 | 88.6 ± 5.9 | 90.4 ± 4.0 |
| MetaGeneAnnotator | 90.1 ± 2.8 | 86.2 ± 5.7 | 89.1 ± 3.1 | 92.9 ± 3.0 | 90.0 ± 6.0 | 91.5 ± 3.3 |
| GeneMark | 87.4 ± 2.8 | 91.0 ± 4.2 | 89.1 ± 3.1 | 90.9 ± 2.7 | 92.2 ± 5.1 | 91.5 ± 3.1 |
Orphelia Net300 represents Orphelia with the 300 bp prediction model, Orphelia Net700 represents the 700 bp prediction model. In addition, the performance of MetaGene, MetaGeneAnnotator and GeneMark is shown.
In comparison to MetaGene, Orphelia has a lower sensitivity but shows a higher specificity, while the harmonic mean of both methods differs by <1%. The MetaGeneAnnotator shows a slightly higher harmonic mean than Orphelia and MetaGene, particularly on 300 bp fragments. Orphelia still has a higher specificity than the MetaGeneAnnotator. A direct comparison of GeneMark and Orphelia on the test setup shown here seems unfair if one considers that the model used by GeneMark was built using some of the test species. Keeping this in mind, GeneMark has a harmonic mean that is similar to MetaGeneAnnotator but has a specificity that is comparable with Orphelia.
In order to determine input sequence length-specific optimal models, we evaluated gene prediction accuracy of both models on fragments ranging in length from 200 bp to 500 bp in 20 bp intervals. The fragments were randomly excised to a 1-fold genome coverage from the test species mentioned above. While Net700 shows a softly decreasing sensitivity and specificity on shorter fragments, and a good performance on fragments as long as 60 000 bp (previously demonstrated in supplementary materials, Figure 4 of (9)), Net300 drastically drops in accuracy for fragments >300 bp. We therefore recommend the usage of Net300 for fragments ranging from 200 bp to 300 bp length, and Net700 for all longer fragments. More details can be seen in Supplementary Data, Figures 1 and 2.
In order to determine the effect of sequencing errors on gene prediction accuracy, several scenarios were simulated using the MetaSim software (10) and the same test species as before. We simulated error-free Sanger reads (with a mean length of 700 bp), and Sanger reads with error rates of 1 × 10−2, 1 × 10−3, 1 × 10−4 and 1 × 10−5 at the beginning of the read and error rates of 2 × 10−2, 2 × 10−3, 2 × 10−4 and 2 × 10−5 at the end of the read, respectively (more details are given in the Methods section of Supplementary Data).
For a comprehensive evaluation of the effect of sequencing errors on gene prediction performance, predicted nucleotide sequences were translated to amino acid sequences using the standard translation table for prokaryotes. Predicted sequences were then aligned to annotated protein sequences using BLAT (11) with standard parameters. Matching amino acids were counted as true positives, amino acids that occur only in the annotation were counted as false negatives and amino acids that occur only in the prediction were counted as false positives. Based on these counts, we observe a decrease of sensitivity and specificity for Orphelia Net700 on Sanger reads with increasing error rates (see Supplementary Data, Table 1). For an error rate of ∼10−4, which was suggested by (6) as a realistic error rate, Orphelia shows a drop in accuracy of <1%.
Application to real data
The hypersaline microbial mat metagenome consists of samples from 10 spatially successive layers of Guerrero Negro (12). Each sample was Sanger sequenced and contains ∼13 000 reads. The original gene annotation of those reads was created with the commercial program FGENESB (http://www.softberry.com). Note that FGENESB integrates model-based gene prediction with homology-based annotation. In contrast to Orphelia and MetaGene, FGENESB also annotates rRNA and tRNA genes. For the following comparison of gene predictions, all RNA genes were removed from the FGENESB annotation.
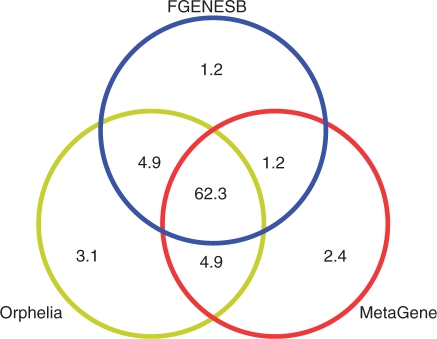
We applied Orphelia (Net700) and MetaGene to the hypersaline microbial mat metagenome (all samples). The number of nucleotides that were predicted as protein encoding was counted and all possible intersections of nucleotides that were predicted as protein coding by Orphelia MetaGene, and FGENESB were calculated. The results are shown in Figure 3. All three methods predict ∼62.3 × 106 nt as protein coding. FGENESB predicts ∼1.2 × 106 nt, MetaGene predicts ∼2.4 × 106 nt and Orphelia predicts ∼3.1 × 106 nt as protein coding that were not predicted by any other method. FGENESB has an intersection of ∼4.9 × 106 nt with Orphelia, and an intersection of ∼1.2 × 106 nt with MetaGene. Both Orphelia and MetaGene predict about ∼4.9 × 106 nt as protein coding that were not predicted by FGENESB. Mavromatis et al. (13) reported FGENESB to overlook ∼20% of the genes on single sequencing reads from annotated genomes, and that FGENESB ‘newly predicted’ ∼10% genes in the same reads. We think that the intersection of nucleotides that were predicted by all methods contains highly reliable genes, and that at least the nucleotides commonly predicted by Orphelia and MetaGene, but not by FGENESB, are worth further investigation because they are likely to contain genes that were overlooked by FGENESB. Gene predictions of Orphelia and MetaGene on this dataset are available through the Orphelia web site.
Figure 3.
Venn diagram of the number of million nucleotides predicted as protein encoding by FGENESB, Orphelia (Net700) and MetaGene in the hypersaline microbial mat metagenome samples.
IMPLEMENTATION
Orphelia's ORF finder is implemented in Java, while the ORF scoring routine and the greedy strategy for calculating the final gene combination are implemented in MATLAB using fast C (‘mex’) code for time critical subroutines. MATLAB routines are integrated as a MATLAB compiler generated program. The web server is based on Java Servlet technology. Submitted jobs are scheduled via a batch queuing system which allows simultaneous processing of several requests.
CONCLUSION
The evaluation on simulated data sets demonstrates that Orphelia shows high gene prediction accuracy on short DNA fragments and has—compared with the other web servers for metagenomic gene prediction—a particularly high gene prediction specificity. We showed that realistic sequencing error rates influence prediction performance only mildly. Therefore, the Orphelia web server application can be a valuable tool for predicting genes in metagenomic sequencing reads.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Georg-Christoph-Lichtenberg stipend granted by the state of Lower Saxony (to K.J.H.); fellowship within the Postdoc-program of the German Academic Exchange Service (DAAD to T.L.). Funding for open access charge: Department for Bioinformatics, Institute for Microbiology and Genetics, Georg-August-Universität Göttingen.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rasmus Steinkamp for helping us with the web server at GOBICS. We thank Dr Mario Stanke for discussions about the evaluation of gene predictions on the hypersaline microbial mat data set.
REFERENCES
- 1.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronaghi M, Uhlén M, Nyreén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Krause L, Diaz NN, Bartels D, Edwards RA, Pühler A, Rohwer F, Meyer F, Stoye J. Finding novel genes in bacterial communities isolated from the environment. Bioinformatics. 2006;22:e281–e289. doi: 10.1093/bioinformatics/btl247. [DOI] [PubMed] [Google Scholar]
- 5.Yooseph S, Li W, Sutton G. Gene identification and protein classification in microbial metagenomic sequence data via incremental clustering. BMC Bioinformatics. 2008;9:182. doi: 10.1186/1471-2105-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoff KJ, Tech M, Lingner T, Daniel R, Morgenstern M, Meinicke P. Gene prediction in metagenomic fragments: a large scale machine learning approach. BMC Bioinformatics. 2008;9:217. doi: 10.1186/1471-2105-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim – a sequencing simulator for genomics and metagenomics. PLoS ONE. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent WJ. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, von Mering C, Bebout BM, Pace NR, Bork P, et al. Millimeter-scale genetic gradients and community level molecular convergence in a hypersaline microbial mat. Mol. Syst. Biol. 2008;4:198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, Rigoutsos I, Salamov A, Korzeniewski F, Land M, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods. 2007;4:1548–7091. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.