Abstract
COFECO is a web-based tool for a composite annotation of protein complexes, KEGG pathways and Gene Ontology (GO) terms within a class of genes and their orthologs under study. Widely used functional enrichment tools using GO and KEGG pathways create large list of annotations that make it difficult to derive consolidated information and often include over-generalized terms. The interrelationship of annotation terms can be more clearly delineated by integrating the information of physically interacting proteins with biological pathways and GO terms. COFECO has the following advanced characteristics: (i) The composite annotation sets of correlated functions and cellular processes for a given gene set can be identified in a more comprehensive and specified way by the employment of protein complex data together with GO and KEGG pathways as annotation resources. (ii) Orthology based integrative annotations among different species complement the defective annotations in an individual genome and provide the information of evolutionary conserved correlations. (iii) A term filtering feature enables users to collect the specified annotations enriched with selected function terms. (iv) A cross-comparison of annotation results between two different datasets is possible. In addition, COFECO provides a web-based GO hierarchical viewer and KEGG pathway viewer where the enrichment results can be summarized and further explored. COFECO is freely accessible at http://piech.kaist.ac.kr/cofeco.
INTRODUCTION
High-throughput experiments such as microarrays, serial analysis of gene expression (SAGE), chromatin immunoprecipitation (ChIP)-on-CHIP and proteomics generate a number of interesting gene sets that are functionally correlated within a certain biological condition. For an interpretation of the functions and biological processes for gene sets under study, enrichment based functional annotation is a common and suitable method. Various enrichment tools have been developed and are widely used but some challenging issues still remain unresolved (1–12). Annotation databases need to be extended for the comprehensive identification of biological processes for a gene set of interest. In addition, the interrelationship of heterogeneous annotations should be integrated in order to make functional annotations more interpretable within a network context. Currently, enrichment tools contain various biological annotation resources such as Gene Ontology (GO), Pfam domains, InterPro motifs, KEGG pathways and so on. However, protein complex information has not been used extensively for enrichment resources. The physical interactions of co-complexed proteins support a solid basis for assigning correlated proteins working together for specific functions. Other various functional annotations can be associated in a protein complex as correlated functions. Protein complex data show many similar variants that may reflect the dynamic changes of functional modules under various cellular conditions. Therefore, the interrelationship of cellular functions can be comprehensively and specifically delineated by integrating the information of a protein complex with other functional annotation resources. To obtain the integrated functional annotations from heterogeneous resources, composite annotation methods can be applied (8,9,10). All annotation terms including concurrent genes are composited and evaluated in order to provide the best composite annotations for the given gene set. The interrelating feature of a composite annotation algorithm can be combined synergistically with protein complex annotations. For the same purpose, a protein interaction network, alone or together with complexes, can be suggested but the functional boundary of an interaction network is ambiguous and still suffers from a high false-positive rate mainly due to the wrong interpretation of co-complex data (13). COFECO is a web-based tool that improves on the aforementioned issues in current enrichment tools by using a composite function annotation with protein complex data, KEGG pathways (14) and GO terms (15) for a given set of genes and their orthologs. COFECO enables comparative analyses between different gene sets with cross comparison tool. The correlated functions for an annotated complex can be conveniently explored at the GO and KEGG pathways using graphical viewers. We combined wide-spread protein complex datasets (15–22) so that it covers a large enough number of proteins and annotations so as to be comparable to other annotation resources (Table 1).
Table 1.
Statistics of annotations and proteins in annotation resources of COFECO
| Organisms | Proteins |
Annotations |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Complex | KEGG | GO BP | GO MF | GO CC | Complexa | KEGG | GO BP | GO MF | GO CC | |
| M. musculus | 5670 | 5313 | 33 714 | 39 440 | 34 231 | 2118 | 199 | 3584 | 2320 | 614 |
| S. cerevisiae | 5246 | 1237 | 5147 | 5020 | 5761 | 8049 | 110 | 1556 | 1484 | 511 |
| H. sapiens | 4123 | 5507 | 36 780 | 40 115 | 36 594 | 2858 | 205 | 3082 | 2612 | 684 |
| R. norvegicus | 2957 | 2674 | 9407 | 10 729 | 9837 | 1850 | 197 | 2059 | 1858 | 463 |
| G. gallus | 1820 | 604 | 4379 | 5058 | 4345 | 1287 | 121 | 894 | 984 | 289 |
| A. thaliana | 1606 | 1947 | 19 694 | 25 962 | 13 380 | 619 | 132 | 1043 | 1240 | 269 |
| D. melanogaster | 1583 | 2216 | 13 862 | 17 892 | 10 666 | 865 | 135 | 2196 | 1545 | 439 |
| S. pombe | 899 | 966 | 3553 | 3429 | 4790 | 526 | 106 | 1153 | 1082 | 370 |
| C. elegans | 806 | 1012 | 9891 | 11651 | 6456 | 600 | 122 | 1163 | 969 | 237 |
aThe numbers of annotations for complexes are the number of complexes for each genome. Among the whole 19 064 complexes, 63% have their own function annotations or complex names and 37%, which are from high-throughput co-complex data in general, do not have complex name. In this system, complex itself is used as an annotation unit representing functionally correlated group.
MATERIALS AND METHODS
Inputs
A list of gene sets can be the input into COFECO. Gene (or protein) identifiers from various databases including UniProtKB, iProClass, Entrez Gene, UniGene, RefSeq, EMBL, ENSEMBL, SGD, RGD, MGI, HGNC and IPI are allowed. COFECO also accepts microarray probe identifiers of Affymetrix and Agilent.
Data resources
The protein complex datasets employed in COFECO are as follows: complex terms specified within the GO cellular component category (15), CORUM (16), Reactome (17), MPact (18), PINdb (19) and three high-throughput TAP/Mass datasets (20–22). The statistics of protein complexes completely included in three GO categories and KEGG pathways is as follows: 7737 (40%) in GO biological processes, 8767 (46%) in GO cellular components, 8693 (46%) in GO molecular functions, and 7860 (41%) in KEGG pathways, respectively. Those complexes create new complex-subclasses in GO or KEGG pathways, which more specify or cross-correlate the classes in both resources. 6431 (34%) complexes have proteins both included and not-included in GO or KEGG. Especially, 1473 (8%) complexes has completely new members that are not included in GO or KEGG. The complex datasets support the following 21 organisms: Homo sapiens, Mus musculus, Rattus norvegicus, Arabidopsis thaliana, Drosophila melanogaster, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Dictyostelium discoideum, Gallus gallus, Bos taurus, Mycobacterium tuberculosis, Danio rerio, Xenopus laevis, Sulfolobus solfataricus, Sus scrofa, Canis lupus familiaris, Xenopus tropicalis, Methanocaldococcus jannasc, Synechocystis sp. PCC 6803 and Pan troglodytes. KEGG pathways, GO terms and other annotation files were downloaded from public ftp servers. The orthologs of query genes in user-specified organism are acquired from the eukaryotic ortholog database, InParanoid (23) that is generated by using orthologs and in-paralogs detection algorithm. Ortholog set is analyzed separately from original query gene set. More details on the data collection can be found in supplementary material.
Composite enrichment algorithm
COFECO uses a modified form of the a priori algorithm (24) for composite enrichment that generates sets of associated annotations which co-occur significantly in a set of genes. Composite enrichment algorithm consists of two processes: generation of composite annotation terms and statistical evaluation of them. In the generation of composite annotations, the a priori algorithm begins by selecting the set of all single annotation terms that occur in at least k concurrent genes. In the next step, two terms that occur in at least k concurrent genes are merged to a new associated term. The process continues until the longest associated terms are found. In the statistical evaluation process, composite annotations which are significantly enriched in a given gene set are evaluated. As the number of annotation resources increases and k decreases, the computational complexity might drastically grow to enumerate all possible compositions of annotation terms. In addition, the protein redundancy among complexes may also lead to huge computation in COFECO analysis. To solve this problem, we developed the greedy algorithm that select the top ranked K terms determined by P-value calculations at each step of composite annotation. The greedy algorithm is optionally applicable to the composite annotation depending on user's preference. More details on the greedy algorithm can be found in supplementary material. A statistical significance test is applied to all single and associated terms found in the above process. A hypergeometric distribution, binomial test, Fisher's exact test, or chi-squared test can be applied in COFECO. The multiple testing correction of P-value can be conducted using Bonferroni correction, the Holm–Bonferroni method, or a false discovery rate (FDR) method (25). In COFECO, these processes are implemented as both single and composite enrichment of various combination of annotation resources can be performed simultaneously or selectively. COFECO implements special composite annotation processes. First, annotation resource combination can be performed by four types: mandatorily including protein complex and at least one different resource, mandatorily including protein complex, mandatorily including at least two different resources, and all possible combinations. These operations support term-term associations by considering annotation resources and specify biological annotations. In addition, COFECO provides term filtering function that enables the enrichment of selected annotation terms with user-specified keywords.
Outputs
COFECO reports a summary of annotation result, composite annotation, single annotation and details of enriched protein complex for requested genes and their orthologs. A summary table provides the frequency of enriched annotation terms and associated gene sets. A composite annotation table provides a list of annotations, their associated genes, P-value and public website links for the annotation resources. A single annotation table displays typical enrichment results of individual resources without term association process. An enriched protein complex table shows all available information of the complex including KEGG pathways and GO terms. KEGG pathways and GO terms in an enriched annotation table and a protein complex table are summarized by web-based viewers. COFECO outputs are accessible at specified URL addresses that are notified by an Email and can be used as an input file of cross-comparison analysis in COFECO.
Implementation
COFECO was implemented on Linux and runs on Apache Web Server combined by a Tomcat servlet engine. A composite annotation algorithm was implemented in Java to take advantage of serialization, reusability of data objects and platform independence. Java serialization supports much faster and simpler manipulation of output objects in different processes such as cross comparisons or output reporting. All preprocessed data used within our system were stored in Oracle 9i DBMS. COFECO web pages were developed with Java Server Page and tested with most available web browsers. A GO hierarchical viewer was implemented in a Java Applet and JUNG library at http://jung.sourceforge.net. KEGG pathway viewer was developed using the open web services of KEGG at http://soap.genome.jp/KEGG.wsdl. More organisms, annotation resources and identifiers will be systematically updated regularly.
Functionalities and characteristics
The details of functionalities and characteristics of COFECO are as follows.
Employment of protein complexes as an annotation resource
The addition of protein complex within annotation resources means more than simple expansion of annotation space. Protein complexes show the precise composition of collaborative proteins for specific functions under various cellular conditions in different time and locations. The conservation and variation of protein members and corresponding functions in complex give the information of dynamic cross correlation among cellular functions and processes. In these senses, the composite annotation of protein complexes with other annotation resources such as GO terms and KEGG pathways provides more specified protein groups with comprehensively correlated functional contexts of GO and KEGG within complex unit.
Comparative analysis with orthologs
A unique feature of COFECO is orthologs-based annotation analysis. COFECO performs composite or single annotations of ortholog set in a user-specified organism and reports the result separately from original query genes. User can acquire putative complementary annotations from different organisms with the insight of evolutionary conserved or differentiated functional groups in a given gene set by comparing the annotations for queried genes and orthologs.
Summarization and exploration of annotated functions via intuitive graphical views
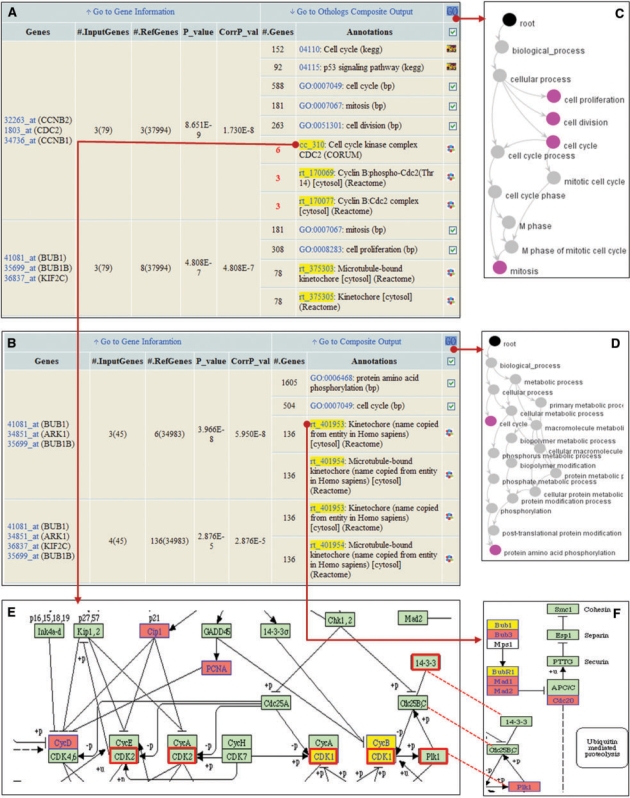
The hierarchical relationship of enriched GO terms can be summarized efficiently by a web-based GO viewer, that GO terms are color-marked by the enrichment types or your selection (Figure 1C and D). KEGG viewer has color-marking function for a significant set of genes which are co-annotated with protein complex and KEGG pathways. The enriched members with the other co-complexed proteins are marked in the KEGG pathway (Figure 1E and F).
Figure 1.
Screenshot depicting the results of the analysis of 85 differentially expressed genes (DEGs) in human testis tissue. (A) Composite annotation results from the function annotation of protein complexes, KEGG pathways and GO biological processes. (B) Composite annotation results of an ortholog list of 85 DEGs. (C) and (D) Graphical GO views of A and B, respectively. (E) Inset of KEGG cell-cycle pathway, which involves co-complexed proteins of cell cycle kinase complex. (F) Inset of KEGG cell-cycle pathway, which involves co-complexed proteins of kinetochore. Red outlined rectangles in Figure 1E indicate co-complexed proteins of the protein complex named as ‘centrosome containing phosphorylated Nlp’, which is actually ranked the third annotation, not displayed in Figure 1B. Yellow boxes indicate statistically significant genes in enriched composite annotations and pink boxes indicate co-complexed proteins except for statistically significant genes. Dotted lines indicate same genes.
Specified composite annotation via term filtering
User can optionally specify the annotation terms to be included or excluded in the enrichment by setting the keywords for the annotation terms.
Cross comparison between the annotation outputs
A cross-comparison is used to identify changes/trends between the annotation results of two different datasets. This functionality is useful to compare various types of outputs, for examples, enriched annotations of input list of genes with those of ortholog list of input genes, and annotation outputs from different input sets.
An example of COFECO analysis
Figure 1 summarizes the functionality of COFECO with an example of 85 human testis-specific genes (26) that has been used previously for the test of other enrichment tools (3,8). The previous composite annotation analysis (8) could successfully point out new explicit connection between the terms of ‘protein amino-acid phophorylation’ and ‘cell cycle’ out of large categories annotated by single annotation analysis (3). The new connection was interpreted with five best composite annotations from GO biological process and InterPro motifs. In a single and composite annotation with typical analysis setting with GO and KEGG pathway, COFECO showed basically same results as previous studies. However, composite annotation based on complex and orthology summarized all previous conclusions and more specific annotations from the first ranked composite annotations (Figure 1A and B). From a few composite annotations with the significant P-value, more specified annotations and gene sets could be summarized on complex units with comprehensively correlated functional contexts of GO and KEGG. The relevance of enriched annotations could be confirmed by the known protein complex information. For example, cell cycle related kinase complexes of the first annotation shown Figure 1A perform the control of the cell cycle at the G2/M (mitosis) transition through cyclin-dependent kinase activity (27). Kinetochore related complexes of the second annotation have kinase activity and interact with centromere and spindle during cell division (28). The importance of orthology-based composite annotation is shown in Figure 1B. The GO and KEGG annotations in human and mouse were complementary and informative, especially with specific terms like ‘p53-signaling pathway’ and ‘protein amino-acid phosphorylation’. The annotation results of mouse also suggest an additive gene of interest such as, ARK1, which is serine/threonine protein kinase 6 and is involved in microtubule formation/stabilization. There were interesting observations related to human protein complex, ‘cell-cycle kinase complex CDC2 complex’, which consists of six proteins: CCNB1 (CycB), CCNB2 (CycB), CDC2, CCND1 (CycD), CDKN1A (Cip1) and PCNA. KEGG cell cycle pathway completely contains these six co-complexed proteins (Figure 1E). CCND1, CDKN1A and PCNA which were not involved in the enriched genes could be inferred as importantly correlated genes by this integrated analysis. Figure 1E and F provide insight into the associated relationships among three annotated protein complexes: ‘cell cycle kinase complex’, ‘kinetochore’ and ‘centrosome containing phosphorylated N1p’ (third annotation in mouse which is not shown at Figure1B) with KEGG viewer summary. Fourteen proteins [CycB, CycD, CDK1, CDK2, Cip1, PCNA, Pik1, 14-3-3, Bub1, Bub3, BubR1 (BUB1B), Mad1 (MAD1L1), Mad2 (MAD2L1) and Cdc20] of the three protein complexes above were annotated in KEGG cell cycle pathway. These proteins can be highlighted as significant gene sets that narrows down the scope of KEGG cell cycle correlated with a human testis-specific expression. Another example showing the unique functionalities and features of COFECO can be found in supplementary material.
CONCLUSION
A large and linear list of enriched annotation terms in the outputs of functional enrichment tools is often incomprehensive or irrelevant toward understanding the biological functions of a gene set under study. Here, we present COFECO, a web-based tool for the composite annotation of protein complexes, KEGG pathways and GO terms within a class of genes and their orthologs under study. As has been illustrated in an example, the composite annotation of protein complexes with other annotation resources such as GO and KEGG pathway provides more specified annotations and gene groups with comprehensively correlated functional contexts of GO and KEGG within complex unit. Our tool can also furnish additional proteins of interest among co-complexed proteins. In addition, comparative analysis with orthology and a cross comparison between annotation outputs provide interesting phenomena that are not addressed by the annotation outputs of a single dataset. The aforementioned features make COFECO a useful tool for users discovering the biological functions of experimental data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Development of Next-Generation New Technology program (10024715-2008-21).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank anonymous reviews for constructive criticisms and fruitful discussions. This research was partially supported by the MKE (Ministry of Knowledge Economy), Korea, under the ITRC (Information Technology Research Center) support program supervised by the IITA (Institute of Information Technology Advancement), IITA-2009-C1090-0902-0014 and by “Development of Intelligent Robot Technologies for Laboratory Medicine by Applying Biotechnology” under the Development of Next-Generation New Technology program (10024715-2008-21) of the Ministry of Knowledge Economy (MKE), Korea.
REFERENCES
- 1.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 2.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vencio RZ, Koide T, Gomes SL, Pereira CA. BayGO: Bayesian analysis of ontology term enrichment in microarray data. BMC Bioinformatics. 2006;7:86. doi: 10.1186/1471-2105-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Shahrour F, Minguez P, Tarraga J, Medina I, Alloza E, Montaner D, Dopazo J. FatiGO+: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0-a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;22:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nam D, Kim SB, Kim SK, Yang S, Kim SY, Chu IS. ADGO: analysis of differentially expressed gene sets using composite GO annotation. Bioinformatics. 2006;24:2249–2253. doi: 10.1093/bioinformatics/btl378. [DOI] [PubMed] [Google Scholar]
- 9.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonov AV, Schmidt T, Wang Y, Mewes HW. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart GT, Ramani AK, Marcotte EM. How complete are current yeast and human protein-interaction networks. Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stumpflen V, et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vastrik I, D'Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S, Matthews L, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luc PV, Tempst P. PINdb: a database of nuclear protein complexes from human and yeast. Bioinformatics. 2004;20:1413–1415. doi: 10.1093/bioinformatics/bth114. [DOI] [PubMed] [Google Scholar]
- 20.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 21.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 22.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 23.Berglund AC, Sjolund E, Ostlund G, Sonnhammer EL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal R, Imielinski T, Swami A. Mining association rules between sets of items in large databases. Proc. ACM SIGMOD Int. Conf. Manage. Data. 1993:207–216. [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 26.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol. Biol. Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.