Abstract
Profile–profile methods are well suited to detect remote evolutionary relationships between protein families. Profile Comparer (PRC) is an existing stand-alone program for scoring and aligning hidden Markov models (HMMs), which are based on multiple sequence alignments. Since PRC compares profile HMMs instead of sequences, it can be used to find distant homologues. For this purpose, PRC is used by, for example, the CATH and Pfam-domain databases. As PRC is a profile comparer, it only reports profile HMM alignments and does not produce multiple sequence alignments. We have developed webPRC server, which makes it straightforward to search for distant homologues or similar alignments in a number of domain databases. In addition, it provides the results both as multiple sequence alignments and aligned HMMs. Furthermore, the user can view the domain annotation, evaluate the PRC hits with the Jalview multiple alignment editor and generate logos from the aligned HMMs or the aligned multiple alignments. Thus, this server assists in detecting distant homologues with PRC as well as in evaluating and using the results. The webPRC interface is available at http://www.ibi.vu.nl/programs/prcwww/.
INTRODUCTION
Sequence-alignment techniques are essential in providing predictions of protein function and evolution. The introduction of sequence–profile methods, such as hmmpfam, hmmsearch (1) and PSI-BLAST (2,3), increased the detection of homologous sequences considerably compared to sequence-sequence methods [e.g. (4)], such as BLAST (3). A profile numerically encodes a multiple sequence alignment and its amino acid diversity by counting the amino acids in each column. Profile hidden Markov models (HMMs), or (profile) HMMs, are statistically more advanced than numerical profiles and allow for variable gap penalties (1). Clearly, profiles, based on an alignment, contain more information than a single sequence. Indeed, including distant but true homologues in the alignment, further increases the chance of detecting of similar families (5). We here use the word ‘profiles’ to refer to both numerical profiles and profile HMMs.
The last decade the sequence–profile methods have been advanced to profile–profile methods. Profile–profile methods provide a more sensitive (6–9) way to find distant homologies between proteins. Using profiles for both query and subject (domain database), has been shown to lead to more sensitive detection of evolutionary remote relationships [e.g. (9,10)]. Different profile–profile methods have been developed, including prof_sim (9) and FFAS (11). We here focus on three widely used state-of-the-art profile–profile programs: Profile Comparer [first released in 2002 (12)], COMPASS [COmparison of Multiple Protein sequence Alignments with assessment of Statistical Significance (6,13)] and HHsearch (7,14). Profile Comparer [PRC, (12)] is a stand-alone program for scoring and aligning HMM and is routinely used by, for example, the CATH (15) and Pfam (16,17) domain databases. The CATH pipeline uses PRC to detect extremely remote homologues and group them in superfamilies [http://www.cathdb.info/wiki/doku.php?id=about:intro, (15)]. Initially, Pfam used only PRC to detect similar domains (16), but now also uses HHsearch (14) [and SCOOP (18)] to establish Pfam clans (17). In addition, internal links from one Pfam family to another are generated with PRC and SCOOP.
In contrast to HHsearch and COMPASS (7,13), PRC did not have a web interface available yet. We therefore have implemented webPRC, a server for searching several public domain databases with additional functionality, including HMM-to-alignment translation, as compared to stand-alone PRC.
METHODS
Database construction
Several major domain databases are provided: Pfam (17), NCBI's Conserved Domain Database (19), KOG (20), TIGRFAMs (21), CATH (22) and SUPERFAMILY (23). We briefly indicate how the profile HMMs and their seed alignments were obtained.
Pfam-A: The Pfam-A (16,17) profile HMMs have been rebuilt locally using the seed alignments downloaded from the Pfam FTP site (http://pfam.sanger.ac.uk) and the hmmbuild options provided therein. When building the HMMs the starting alignment, also for CDD/KOG and TIGRFAMs, was re-saved by hmmbuild (HMMER v2.3.2; http://hmmer.janelia.org/). This re-saved alignment includes an ‘RF’ line that indicates which alignment columns are absent from the HMM. This line is used to translate the HMM coordinates of the PRC results back to the alignment coordinates.
CDD/KOG: NCBI's Conserved Domain Database [CDD (19)] and KOG (20) HMMs have been built from the seed alignments downloaded via the CDD site (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). As there can be multiple identical sequence identifiers in CDD alignments, the sequence identifiers in the re-saved alignments were made unique by prepending a number to the entire identifier for reoccurring identifiers only.
TIGRFAMs: The TIGRFAMs (21) HMMs have been rebuilt locally from the seed alignments and the hmmbuild options provided in the TIGRFAMs HMM files.
CATH: The CATH (15,22) HMMs have been obtained from the CATH web site (http://www.cathdb.info). These models are not based on Pfam-like seed alignments, but are produced iteratively starting from a single sequence (24). This can result in huge alignments with high gap content (up to about 80 000 sequences, >50 000 columns, or 680 Mb for a single alignment). For this reason, the CATH models are used directly. Their underlying alignments have been processed to include an ‘RF’ line and a maximum of the first 200 sequences are included in the alignment output.
SUPERFAMILY: The SUPERFAMILY (23) models were retrieved from http://supfam.org.
User input
The user can provide a single protein sequence or multiple sequence alignment via the paste or upload field. A variety of alignment formats is accepted (ClustalW, FASTA, GCG MSF, Stockholm and SELEX). The user may configure the following search parameters: the domain database, PSI-BLAST options, several PRC options, the number of unique hits to be visualized in the hit graphic, and the use of the hmmbuild ‘–hand’ option. This option can be used to mark regions of the alignment that should be absent from the HMM produced by hmmbuild, which is useful for searching with discontinuous domains. The ‘RF’ annotation line, required for the optional ‘–hand’ option, is supported for the SELEX (#= RF) and Stockholm (#=GC RF) formats. Finally, the user may choose to generate logos from the HMM alignments or from the aligned multiple sequence alignments [with LogoMat-P (25) and Two Sample Logo (26), respectively] to visualize the alignments. Example input and output are provided, including the possibility to regenerate the example output (‘rerun the example’).
Alignment calculation
The webPRC searches run on a 64-CPU computer cluster. The processing scripts are coded in Perl, Bioperl [Bio::Graphics and Bio::SimpleAlign; (27)], PHP and Javascript. PRC is run with the selected domain library and domain descriptions of the hits are parsed from the chosen domain database. Since PRC results are reported in profile HMM space, both the PRC alignment output and the re-saved alignment files, produced by hmmbuild, are processed to provide a mapping of PRC results to the query and hit multiple sequence alignments. Then, these alignments are sliced according to the calculated alignment coordinates and joined in one alignment. The IDs of the hit alignment in this combined alignment file are prepended with ‘Hit:’. In addition, an ‘aligned alignments’ view is constructed which contains the first sequence and the consensus sequence from each alignment. For viewing the alignment interactively, an extended version of Jalview (28) is used that supports regular expressions to parse sequence identifiers for its linkUrl parameters.
Logo generation
The logos are generated with local installations of LogoMat-P (25) and Two Sample Logo (26). LogoMat-P was adapted such that the generated logos correspond exactly to the HMM alignments reported by PRC. Thus, LogoMat-P is not executing a new pair-wise PRC search to find an HMM alignment between the query and the single subject HMM, but now directly uses the alignment produced by the PRC library run against the domain databases.
RESULTS AND DISCUSSION
The webPRC server facilitates the use of PRC for finding domains related to a query alignment. Besides the possibility to run PRC against different domain databases, webPRC offers additional functionality not available with a PRC stand-alone run.
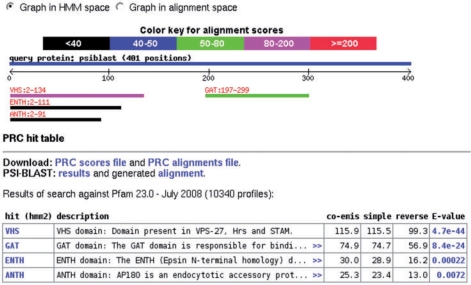
After completion of a PRC search, the raw PRC output is reformatted into a BLAST-like report, which includes a domain hit distribution graphic and a hit table (Figure 1). This makes interpreting PRC output as straightforward as reading a BLAST report. The reformatted PRC alignments now include the match, insert, and delete percentages (Figure 2). In addition, several other features aiding the evaluation of the hits are included in the report: hits in the table are linked to the source domain database and include a description from the selected domain database. The alignments section contains links to the optionally produced logos. These logos are graphical representations of the aligned HMMs or the aligned alignments and can help in the evaluation of the found domains. LogoMat-P (25) produces pair-wise HMM logos based on the reported PRC alignment. These HMM logos are related to the HMM logos (29) used to visualize the HMMs of protein families in Pfam (17). In addition, Two Sample Logos are produced. These logos are based on two multiple sequence alignments and show the positions that are significantly different between the alignments (26). Furthermore, the alignments section contains an ‘aligned alignments’ presentation. Specifically, this translation of ‘raw’ PRC results to query and hit alignments facilitates the identification of conserved residues. The combined multiple sequence alignments can be viewed in Jalview (28). The sequence labels in the Jalview applet are linked to several sequence databases, including UniProt and Entrez Protein, to facilitate the retrieval of sequence annotations. Finally, the alignments can be downloaded for additional analyses. For example, Sequence Harmony can be used to predict specificity-determining residues from these alignments (30).
Figure 1.
An example of the webPRC domain graphic and hit table section for GGA1_HUMAN run against Pfam (after running PSI-BLAST). The graph can be viewed in HMM or alignment space and the hits are hyperlinked to the alignments. The PRC hit table provides links to the original PRC and PSI-BLAST output and shows a table with annotated hits, including the name and, after clicking on ‘>>’, the description from the domain database. The hits are hyperlinked to the source database and E-values are hyperlinked to the alignments. Co-emission, simple and reverse scores are calculated by PRC [cf. (12)]. The E-value is calculated from the reverse score.
Figure 2.
An example alignment showing hit number (#1), links, PRC alignment and aligned alignments (truncated). The original PRC HMM alignment is formatted in a BLAST-like style and now includes the counts and percentages of the Match, Insert and Delete states (M–M, M–I, D–∼ pairs, respectively). The aligned alignments view shows the PRC result in multiple sequence alignment space and includes the first sequence of the query and hit alignment as well as their consensus sequences. The alignments are separated by a mid-line that indicates the PRC match states (M) with a ‘+’. Gaps present in the seed alignments are indicated by ‘–’, gaps introduced by PRC by ‘∼’ and positions corresponding to columns missing from the HMM by ‘:’. The entire (aligned) alignments can be viewed with Jalview or downloaded by clicking on ‘View alignment’ or ‘Download’, respectively.
The translation from HMM alignments to sequence alignments is provided for most databases. However, the sequence alignments resulting from searches against CATH generally include a large number of gaps (indicated with ‘:’ in the web output). Many alignment columns are indeed absent from their corresponding HMMs due to the high gap content of the seed alignments: for the entire CATH database only 15% of all alignment columns are represented in the HMMs as opposed to 91% for Pfam-A.
Figures 1 and 2 illustrate the webPRC output of a search with ADP-ribosylation factor-binding protein GGA1 (UniProt: GGA1_HUMAN) against Pfam and explain the aligned alignments view. A search with the single sequence indeed finds the known domains: VHS, GAT, and GAE (cf. UniProt). PSI-BLAST was run on this sequence to build an alignment (three iterations, E-value 0.0005, NCBI's NR database). Now, not only the VHS, but also the ENTH and ANTH domains are detected, while the GAE domain is not detected anymore. Indeed, the VHS, ENTH and ANTH domains are related, though in general, especially an E-value like that for the ANTH match (0.007) would require further data to state a homologous relationship. In addition to further profile–profile based searching, it is worthwhile to check the Pfam and CDD databases for information on the retrieved hits: CDD contains superfamilies and Pfam groups related families into clans and also provides ‘internal database links’. Pfam and CDD provide information on this VHS/ENTH/ANTH cluster. Hence, webPRC can be used to easily find such clusters and links for any query alignment.
E-values can be used to judge the significance of the hits returned by PRC. However, they are accurate only if the library contains more than 1000 profile HMMs (12). The author of PRC indicated that ‘for libraries of sufficient size, E < 0.003 can be taken as indicative of homology and E < 10−5 as a strong match’ (12). For profile–profile comparisons, Pfam uses an E < 0.001 as an indication of a significant match and E-values between 0.1 and 0.001 as an indication of a true relationship (16).
We here describe our PRC web interface and refrain from including another PRC validation. We would like to refer the reader to several benchmarking studies that report on the performance of PRC [(8,12,14,18), http://toolkit.tuebingen.mpg.de/hhpred/help_ov]. Reid et al. (24) benchmarked profile–profile and profile-sequence methods, including PRC, COMPASS, HHsearch, and concluded that PRC is the best method for distinguishing homologous from non-homologous domains. Depending on the specific benchmarking study, PRC performs better or worse than HHsearch, but generally better than COMPASS. We encourage prospective webPRC users to have a look at these benchmarking studies as well as the COMPASS (13) and HHsearch web servers (7).
CONCLUSION
The webPRC server provides a web-based front end to PRC, one of the state-of the-art methods for detecting remote homology, to carry out similarity searches against well-established domain databases. Since the input is a single sequence or an alignment, users need not build an HMM themselves. In addition to the domain hit distribution graphic and logo visualizations, webPRC features the translation of the PRC HMM alignments to multiple sequence alignments. This supports evaluation of a hit based on multiple sequence alignments. To this end, the Jalview applet is implemented. Furthermore, the hit, query and combined alignments can be downloaded for additional analyses.
FUNDING
ENFIN, a Network of Excellence funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LSHG-CT-2005-518254. Funding for open access charge: ENFIN.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr James Procter for extending the Jalview alignment editor with regular expression based link parsing.
REFERENCES
- 1.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 2.Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J, Karplus K, Barrett C, Hughey R, Haussler D, Hubbard T, Chothia C. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- 5.Sadreyev RI, Grishin NV. Quality of alignment comparison by COMPASS improves with inclusion of diverse confident homologs. Bioinformatics. 2004;20:818–828. doi: 10.1093/bioinformatics/btg485. [DOI] [PubMed] [Google Scholar]
- 6.Sadreyev R, Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326:317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 7.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadreyev RI, Grishin NV. Accurate statistical model of comparison between multiple sequence alignments. Nucleic Acids Res. 2008;36:2240–2248. doi: 10.1093/nar/gkn065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yona G, Levitt M. Within the twilight zone: a sensitive profile-profile comparison tool based on information theory. J. Mol. Biol. 2002;315:1257–1275. doi: 10.1006/jmbi.2001.5293. [DOI] [PubMed] [Google Scholar]
- 10.Madera M, Gough J. A comparison of profile hidden Markov model procedures for remote homology detection. Nucleic Acids Res. 2002;30:4321–4328. doi: 10.1093/nar/gkf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rychlewski L, Jaroszewski L, Li W, Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madera M. Profile Comparer: a program for scoring and aligning profile hidden Markov models. Bioinformatics. 2008;24:2630–2631. doi: 10.1093/bioinformatics/btn504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadreyev RI, Tang M, Kim BH, Grishin NV. COMPASS server for remote homology inference. Nucleic Acids Res. 2007;35:W653–W658. doi: 10.1093/nar/gkm293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 15.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bateman A, Finn RD. SCOOP: a simple method for identification of novel protein superfamily relationships. Bioinformatics. 2007;23:809–814. doi: 10.1093/bioinformatics/btm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited—architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 24.Reid AJ, Yeats C, Orengo CA. Methods of remote homology detection can be combined to increase coverage by 10% in the midnight zone. Bioinformatics. 2007;23:2353–2360. doi: 10.1093/bioinformatics/btm355. [DOI] [PubMed] [Google Scholar]
- 25.Schuster-Böckler B, Bateman A. Visualizing profile-profile alignment: pairwise HMM logos. Bioinformatics. 2005;21:2912–2913. doi: 10.1093/bioinformatics/bti434. [DOI] [PubMed] [Google Scholar]
- 26.Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 27.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 29.Schuster-Böckler B, Schultz J, Rahmann S. HMM Logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feenstra KA, Pirovano W, Krab K, Heringa J. Sequence harmony: detecting functional specificity from alignments. Nucleic Acids Res. 2007;35:W495–W498. doi: 10.1093/nar/gkm406. [DOI] [PMC free article] [PubMed] [Google Scholar]