Abstract
Pomelo II (http://pomelo2.bioinfo.cnio.es) is an open-source, web-based, freely available tool for the analysis of gene (and protein) expression and tissue array data. Pomelo II implements: permutation-based tests for class comparisons (t-test, ANOVA) and regression; survival analysis using Cox model; contingency table analysis with Fisher's exact test; linear models (of which t-test and ANOVA are especial cases) that allow additional covariates for complex experimental designs and use empirical Bayes moderated statistics. Permutation-based and Cox model analysis use parallel computing, which permits taking advantage of multicore CPUs and computing clusters. Access to, and further analysis of, additional biological information and annotations (PubMed references, Gene Ontology terms, KEGG and Reactome pathways) are available either for individual genes (from clickable links in tables and figures) or sets of genes. The source code is available, allowing for extending and reusing the software. A comprehensive test suite is also available, and covers both the user interface and the numerical results. The possibility of including additional covariates, parallelization of computation, open-source availability of the code and comprehensive testing suite make Pomelo II a unique tool.
INTRODUCTION
There is a continuous demand for web-based applications for the analysis of genomic and proteomic data. For end-users, a key feature of web-based applications is that they make few demands on users' software and hardware, since only a web browser is needed (1). Moreover, and of particular relevance, when dealing with large datasets, computational capabilities are not limited by the user's hardware (only by the servers'). In this context, web-based applications allow developers to take advantage of the increased availability of multicore processors and clusters built with off-the-shelf components. These are probably the major opportunities for significant performance gains in the near future (2–5). When deployed in a computing cluster, parallelization [such as provided by MPI (6)], harvests computational resources that are rarely available to individual researchers and can deliver significant decreases in waiting time, while being completely transparent to the end-user. Moreover, web-based applications can offer a user interface and experience very similar to that of desktop applications [e.g. by usage of Javascript (7)]. Finally, web-based tools offer the opportunity to quickly bring new methodological developments to many potential users. Interpretation of results (8,9) can also be easily provided by web-based tools, by linking to additional sources of information [e.g. PubMed references, Gene Ontology terms, etc.), which also permits further analysis with this additional information, such as identifying features [e.g. pathways, GO terms, etc.] which might be characteristic of the set of differentially expressed genes.
In addition to the above general features of ‘omics’ web-based applications, when searching for differentially expressed genes (and similarly for protein and tissue array analysis) it is of course imperative to incorporate the best statistical practices in the field. Depending on the type of response data, different tests should be applied. The most common type of data (gene expression data for different types of patients) are often analyzed to search for differentially expressed genes using ANOVA, t-tests and related approaches that compare two or more classes. Tissue array data, however, are of a categorical or presence/absence nature, and require contingency table methods. Survival data, in contrast, require methods (such as Cox model) that can explicitly deal with right-censored observations. Thus, a tool for the search of differentially expressed genes (proteins) should incorporate the above methods to cover some of the more common needs of wet-lab researchers. In all these cases, and regardless of the type of test, it is by now been well recognized (10,11) that multiple testing problems should be taken into account. In addition, and since many microarray studies are really observational studies with human patients, it is often necessary to include additional clinical covariates to minimize confounding problems (12,13). Finally, in some cases the statistical methodology exists that will allow us to borrow information from all genes in the array when carrying out the test for each gene, using moderated statistics and Empirical Bayes approaches [e.g. (14)].
Finally, availability of source code, under an open-source license, is well recognized as an important feature of bioinformatics applications (13,15): it allows for fast methodological development based upon previous work by permiting other researchers to extend the methods and provide improvements and bug fixes, it makes it possible to verify claims made by method developers, and ensures that the international research community remains the owner of the tools it needs to carry out its work. The impact of code availability is further enhanced when standard best practices in software development [see review and references in (16)] and the usual open-source development mode (17), are followed. Of particular importance, especially with applications that perform complex analysis, is to provide testing suites that allow to verify the results of the analysis performed by the web-based application.
POMELO II: UNIQUE FEATURES
There are several other web-based applications that can be used to identify differentially expressed genes (18–31). However, all of these fail one or more of the requirements mentioned in the Introduction section. Many of them incorporate some of the same procedures as Pomelo II, but few offer as comprehensive a set of analysis as Pomelo does. Most tools are limited to two-class comparisons. Multi-class comparisons are only available from EMAAS (18), EzArray (24), GenePublisher (26), WebArray (27) and GEPAS (32). Survival analysis and regression are only available in GEPAS (32), but GEPAS is not open source. Contingency tables, however, are not available in other tools except Pomelo II. Several other tools make it explicit that they run in clusters (20,33), which allows for load balancing and swapping jobs to idle nodes. However, with the exception of EMAAS (18), parallel computing seems not to be used by any other tools.
More importantly in microarray data analysis, a unique characteristic of Pomelo II is that it allows to incorporate additional covariates, such as age or sex, a much needed feature in many microarray studies with human subjects, where these variables can have an effect in gene expression (12,13). As part of our emphasis on this feature, when using additional covariates, the user is alerted to possible aliasing and confounding and to the available degrees of freedom available (Input and output section).
FUNCTIONALITY, INPUT, OUTPUT
Available statistical methods
Pomelo II incorporates a range of validated, well-know, statistical methods for identifying differentially expressed genes (or proteins). Fisher's exact test is available for contingency tables (this test is useful specially with tissue array data). Linear regression, with a P-value obtained by a permutation test, is of interest when we try to model the values of an interval-scaled variable using gene expression data. Cox model is a widely used method for censored data, such as when we want to find the relationship between patient survival and gene expression. Two-class comparisons are available as a permutation-based t-test, as a parametric t-test using moderated statistics with an empirical Bayes approach (14) as implemented in Limma (34), and as a paired t-test (also using Limma). Class comparisons for two or more classes are available as ANOVA, using permutation for significance, or as linear models, using Limma. If using linear models, we can adjust for the possible effects of ‘additional covariates’ (e.g. sex, age, etc.). For all the tests implemented, we return unadjusted P-values as well as FDR-adjusted P-values, using the approach of Benjamini and Hochberg [see details and discussion in (10,11)].
Input and output
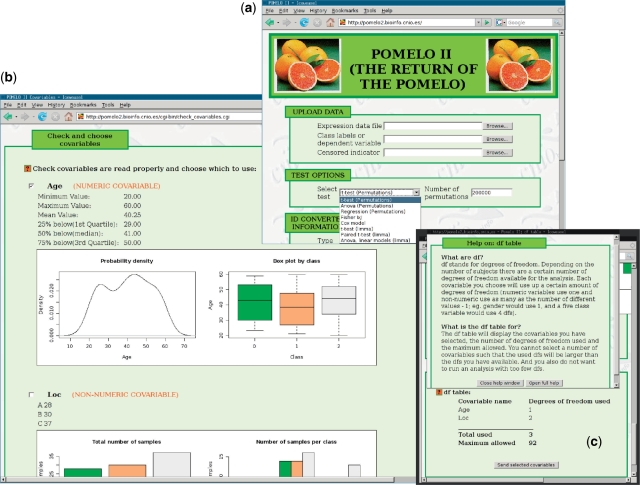
Input are plain text files. For all analysis (except survival analysis), two files are needed: the expression data, and the class labels data. In addition, for linear models, and if additional covariates are used, a file with the additional covariables will be required. For survival analysis, three files are needed: expression data, survival times and censored indicators. A screenshot of the main input screens showing the methods available is shown in Figure 1a.
Figure 1.
Three input screens from Pomelo II. (a) Initial input, showing available statistical methods. (b) Additional covariates check page, with figures showing distributions at different levels of the class variable. (c) Additional covariates check page, showing degrees of freedom available and help.
When using linear models, the user can use additional covariables. These are other subject attributes (e.g. subject age, gender, weight, etc.), often readily available from the clinical history. This information can allow Pomelo II to check if gene expression differences or similarities may be due to these factors instead of due to belonging to a certain class.
When entering additional covariates for the linear model, the user can choose which of the covariates to use. In addition, we show plots of each of the covariates at the different levels of the class variable (Figure 1b). This allows the user to check that the program has correctly interpreted the variables showing which are numerical and which are categorical. In addition, it alerts the user of the possible existence of confounding and aliasing (35). Suppose that in a study comparing expression profiles of breast cancer patients with non-breast cancer control subjects, most breast cancer patients were females and the non-breast cancer subjects were males. This situation would be readily detectable with the plots provided by Pomelo II. In addition, some studies have small numbers of subjects but try to correct for too many covariates; when entering additional covariates, the user is informed about the available degrees of freedom, as well as the degrees of freedom used by any of the covariates included; help is available immediately, explaining the meaning of the table (Figure 1c).
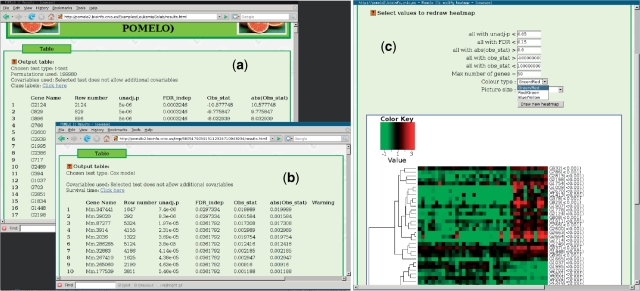
The main output from the program is a table with the results of the analysis and a heatmap. The results table contains a header indicating the test you have used, number of permutations and which covariables were used (if any); see Figure 2a and b, for two examples, corresponding to a permutation t-test and a Cox model. The table shows an index corresponding to the original ordering in the data file, gene names, P-values (unadjusted), FDR-adjusted P-values and statistics (and the absolute value of the statistic); in the case of Cox models, an additional column, ‘Warnings’, might show warnings from the fit (e.g. lack of convergence). At the bottom of the output, there is a figure with a heatmap (Figure 2c) where you can filter how and which genes to plot, and allows you to choose the color scale. Both tables and heatmap are clickable and will take you to a page with additional information [our IDConverter Light (36)] and will allow you to send selected genes (based on user-specified selection criteria) to PaLS (37) to examine PubMed references, Gene Ontology terms, KEGG pathways or Reactome pathways that are common to that set of genes.
Figure 2.
Output. Output table from a permutation t-test (a) and a Cox model (b), and a heatmap with dendrogram, showing available options for heatmap redrawing (c).
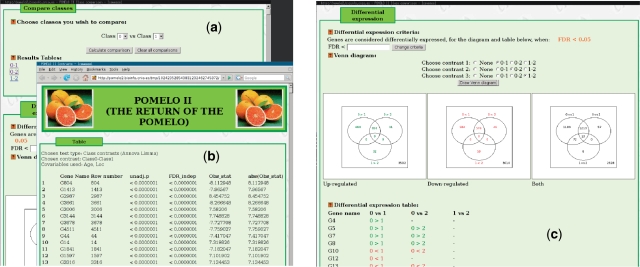
If you have run an ‘Anova, linear models (limma)’ test, the output will also contain a Class compare section containing a button. By clicking on the button, we will be taken to a class compare page (Figure 3a), where we will be able to compare specific pairs of classes. For each comparison, a table will appear (e.g. Figure 3b), showing a table with (moderated) t-statistics (and associated P-values and FDR-corrected P-values), similar to the one in Figure 2. The Class compare page is provided because in linear models (ANOVAs) with three or more classes, we might be interested in comparing particular pairs of classes in addition to the overall F-test (if our linear model had only two classes originally, this option is not really necessary, since of course the overall F-test is equivalent to the t-test for the two-class comparison). Note that a particular two-class comparison in a, say, three-class analysis is not necessarily identical to conducting just a two-class comparison with a t-test: in linear models, we use all available data to estimate the error term and, moreover, the empirical Bayes method implemented in Limma (14) borrows information from gene expression data across all classes. Thus, in experiments that comprise more than two classes, it is always preferable to carry out specific contrasts after a full, global model, is fitted to all the data, rather than conducting many two-group analyses that discard information from the other groups.
Figure 3.
Output from linear model. (a) Class comparison page. (b) Output table from one of the two-class comparisons. (c) Details of Class comparison, showing Venn diagram and table of up- and down regulated genes for each two-class comparison.
From the Class compare page, we can also obtain differential expression tables which, again, are particularly useful with more than two classes. They are also useful with two classes since the F-statistic, which is always of positive sign, gives no indication of whether the mean of the first group is larger or smaller than the mean of the second group. As shown in Figure 3c, for the user-selected group comparisons we obtain Venn diagrams that provide a quick visual information about the number of up- and down regulated genes in each two-class comparison and their intersection (e.g. the number of genes that are up regulated in both the contrast between classes 0 and 1 and the contrast between classes 0 and 2 are 656 in the figure). We also obtain a table showing which genes are differentially expressed in each two-class comparison; we use color codes (green and red) and the ‘<’ and ‘>’ signs to allow for fast differentiation between up- and down regulated genes. The FDR threshold below which genes are considered differentially expressed can be changed by the user, and the Venn diagram and table will be regenerated automatically.
Documentation, help, tutorials
Online help, including full documentation, pre-run examples, sample files and loading of sample data sets is available from the main page of Pomelo II. We also provide video tutorials (see http://pomelo2.bioinfo.cnio.es/help/flash_tutorials/video_tutorials.html) of some of the most common or most involved analysis. In most screens, there is help available to options specific of that step, accessible by clicking on the ‘?’ symbol (e.g. see Figure 1c). The help files are licensed under a Creative Commons license (http://www.creativecommons.org), allowing for redistribution and classroom use.
IMPLEMENTATION, AVAILABILITY, MATURITY AND TESTING
Most of the statistical functionality is written in R (38) or in C/C++, with extensive use of parallelization using MPI (6) and R interfaces to MPI [via the R-packages Rmpi (39), by H. Yu, and papply (40) by D. Currie]. Parallelization is used in all permutation-based tests and the Cox model computations. Cox model fitting uses the survival package, by T. Therneau (41). For linear models, we use Limma (34), by G. K. Smyth and collaborators. The web interface is written in Python and Javascript. Control of the application, fault-tolerance and booting and halting the LAM/MPI universes is accomplished by a combination of Python and shell scripts. We create a new LAM/MPI universe for each run of each application, and the actual nodes/CPUs that are used in a LAM/MPI universe are determined at run time (thus excluding nodes that are down).
Our publicly accessible installation, available at http://pomelo2.bioinfo.cnio.es, runs on a cluster with 31 two dual-core AMD Opteron 2.2 GHz CPUs and six GB RAM, running Debian GNU/Linux. Shared storage space uses RAID 50, which provides protection against hard disk failure, as well as access to results and data from nodes different from the one where computations started. Redundancy and load-balancing of the web service is achieved with Linux Virtual Server with heartbeat and mon, which ensures balancing of the master nodes for MPI and of the non-parallelized executions.
All of the code (including repository history) is available under open source licenses (GNU GPL and Affero GPL) from the Launchpad at http://launchpad.net/pomelo2.
Testing, maturity and number of accesses
Pomelo II includes a comprehensive test suite that uses FunkLoad (http://funkload.nuxeo.org). These tests cover the user interface, handling of error conditions and incorrectly formated files and the numerical output, and can be run on demand, and wherever new changes are introduced in the software, thus ensuring appropriate quality control and regression testing. The complete code is also available, under the GNU GPL and Affero GPL licenses, from http://launchpad.net/functional-testing (go to the Pomelo II directory in the source code). An additional test using Selenium (http://www.openqa.org/selenium/) is available (http://pomelo2.bioinfo.cnio.es/tests.html); these tests verify that the AJAX component of the application runs correctly under different operating systems and browsers.
Pomelo II is a mature application. The server has been running for more than four years. In the last 2 years, over 6000 experimental datasets have been analyzed. Usage and testing includes four groups at the developers institution (CNIO), and users world wide.
FUNDING
Fundacion de Investigacion Medica Mutua Madrileña and Project [TIC2003-09331-C02-02 of the Spanish Ministry of Education and Science (MEC); Red Tematica de Investigacion Cooperativa COMBIOMED. Funding for open access charge: Red Tematica de Investigacion Cooperativa COMBIOMED.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We want to thank many testers at CNIO and elsewhere for feedback on the applications and bug reports. Bionformatics.org and Launchpad provide hosting for the repositories. Our applications would not have been possible without the excellent and free R language and its many freely available packages. A. Valencia for covering publication costs. Two reviewers for comments on the article.
Footnotes
Present address: Systems Biology DTC, University of Warwick, Coventry House, Gibbet Hill Road, Coventry, CV4 7AL, UK
REFERENCES
- 1.Graham P. The other road ahead. In: Graham P, editor. Hackers and Painters. Sebastopol: O'Reilly Media; 2004. pp. 56–86. [Google Scholar]
- 2.Sutter H. The free lunch is over: a fundamental turn toward concurrency in software. Dr. Dobb's J. 2005;30:202–210. [Google Scholar]
- 3.Kontoghiorghes EJ. Handbook of Parallel Computing and Statistics. CRC, Boca Raton, FL: Chapman & Hall; 2006. [Google Scholar]
- 4.Dongarra J, Gannon D, Fox G, Kenned K. The impact of multicore on computational science software. CTWatch Quarterly. 2007;3:3–10. [Google Scholar]
- 5.Turek D. High performance computing and the implications of multi-core architectures. CTWatch Q. 2007;3:31–33. [Google Scholar]
- 6.Pacheco P. Parallel programming with MPI. San Francisco: Morgan Kufman; 1997. [Google Scholar]
- 7.Geraci F, Pellegrini M, Renda ME. Amic@: all microarray clusterings @ once. Nucleic Acids Res. 2008;36(Web Server issue):W315–W319. doi: 10.1093/nar/gkn265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hyatt G, Melamed R, Park R, Seguritan R, Laplace C, Poirot L, Zucchelli S, Obst R, Matos M, Venanzi E, et al. Gene expression microarrays: glimpses of the immunological genome. Nat. Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- 9.Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat. Genet. 2005;37(Suppl):S31–S37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 10.Ge Y, Dudoit S, Speed T. Resampling-based multiple testing for microarray data analysis (with discussion) TEST. 2003;12:1–77. [Google Scholar]
- 11.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 12.Potter JD. Epidemiology, cancer genetics and microarrays: making correct inferences, using appropriate designs. Trends Genet. 2003;19:690–695. doi: 10.1016/j.tig.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Díaz-Uriarte R. Supervised methods with genomic data: a review and cautionary view. In: Azuaje F, Dopazo J, editors. Data Analysis and Visualization in Genomics and Proteomics. New York: Wiley; 2005. pp. 193–214. [Google Scholar]
- 14.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 15.Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- 16.Baxter SM, Day SW, Fetrow JS, Reisinger SJ. Scientific software development is not an oxymoron. PLoS Comput. Biol. 2006;2:e87. doi: 10.1371/journal.pcbi.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fogel KF. Producing Open Source Software. Sebastopol, CA: O'Reilly; 2005. [Google Scholar]
- 18.Barton G, Abbott J, Chiba N, Huang DW, Huang Y, Krznaric M, Smith JM, Saleem A, Sherman B, Tiwari B, et al. Emaas: an extensible grid-based rich internet application for microarray data analysis and management. BMC Bioinformatics. 2008;9:493. doi: 10.1186/1471-2105-9-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weniger M, Engelmann JC, Schultz J. Genome expression pathway analysis tool – analysis and visualization of microarray gene expression data under genomic, proteomic and metabolic context. BMC Bioinformatics. 2007;8:179. doi: 10.1186/1471-2105-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Argraves G, Jani S, Barth J, Argraves SW. Arrayquest: a web resource for the analysis of dna microarray data. BMC Bioinformatics. 2005;6:287. doi: 10.1186/1471-2105-6-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehrauer H, Zoller S, Schlapbach R. Magma: analysis of two-channel microarrays made easy. Nucleic Acids Res. 2007;35(Web Server issue):W86–W90. doi: 10.1093/nar/gkm302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapushesky M, Kemmeren P, Culhane AC, Durinck S, Ihmels J, Körner C, Kull M, Torrente A, Sarkans U, Vilo J, et al. Expression profiler: next generation–an online platform for analysis of microarray data. Nucleic Acids Res. 2004;32(Web Server issue):W465–W470. doi: 10.1093/nar/gkh470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Psarros M, Heber S, Sick M, Thoppae G, Harshman K, Sick B. Race: remote analysis computation for gene expression data. Nucleic Acids Res. 2005;33(Web Server issue):W638–W643. doi: 10.1093/nar/gki490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Zhu Y, Xu W. Ezarray: a web-based highly automated affymetrix expression array data management and analysis system. BMC Bioinformatics. 2008;9:46. doi: 10.1186/1471-2105-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. Carmaweb: comprehensive r- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudsen S, Workman C, Sicheritz-Ponten T, Friis C. Genepublisher: automated analysis of dna microarray data. Nucleic Acids Res. 2003;31:3471–3476. doi: 10.1093/nar/gkg629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia X, McClelland M, Wang Y. Webarray: an online platform for microarray data analysis. BMC Bioinformatics. 2005;6:306. doi: 10.1186/1471-2105-6-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luscombe NM, Royce TE, Bertone P, Echols N, Horak CE, Chang JT, Snyder M, Gerstein M. Expressyourself: a modular platform for processing and visualizing microarray data. Nucleic Acids Res. 2003;31:3477–3482. doi: 10.1093/nar/gkg628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romualdi C, Vitulo N, Favero MD, Lanfranchi G. MIDAW: a web tool for statistical analysis of microarray data. Nucleic Acids Res. 2005;33:W644–W649. doi: 10.1093/nar/gki497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel S, Lyons-Weiler J. cageda: a web application for the integrated analysis of global gene expression patterns in cancer. Appl Bioinformatics. 2004;3:49–62. doi: 10.2165/00822942-200403010-00007. [DOI] [PubMed] [Google Scholar]
- 31.Tarraga J, Medina I, Carbonell J, Huerta-Cepas J, Minguez P, Alloza E, Al-Shahrour F, Vegas-Azcarate S, Goetz S, Escobar P, et al. Gepas, a web-based tool for microarray data analysis and interpretation. Nucleic Acids Res. 2008;36(Web Server issue):W308–W314. doi: 10.1093/nar/gkn303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montaner D, Tarraga J, Huerta-Cepas J, Burguet J, Vaquerizas JM, Conde L, Minguez P, Vera J, Mukherjee S, Valls J, et al. Next station in microarray data analysis: Gepas. Nucleic Acids Res. 2006;34:W486–W491. doi: 10.1093/nar/gkl197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hokamp K, Roche FM, Acab M, Rousseau M.-E, Kuo B, Goode D, Aeschliman D, Bryan J, Babiuk LA, Hancock RE, et al. Arraypipe: a exible processing pipeline for microarray data. Nucleic Acids Res. 2004;32(Suppl 2):W457–W459. doi: 10.1093/nar/gkh446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, R. Irizarry WH, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 35.Woodward M. Study Design and Data Analysis. London: Chapmand & Hall, CRC; 2005. Epidemiology. [Google Scholar]
- 36.Alibès A, Yankilevich P, Cañada A, Diaz-Uriarte R. Idconverter and idclight: conversion and annotation of gene and protein ids. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alibes A, Canada A, Diaz-Uriarte R. Pals: filtering common literature, biological terms and pathway information. Nucleic Acids Res. 2008;36(Web Server issue):W364–W367. doi: 10.1093/nar/gkn251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R: a language and Environment for Statistical Computing R Foundation for Statistical Computing. Vienna, Austria: 2004. R Development Core Team. [Google Scholar]
- 39.Yu H. Technical report. Department of Statistics. University of Western Ontario; 2004. Rmpi: interface (wrapper) to mpi (message-passing interface) [Google Scholar]
- 40.Currie D. papply: parallel apply function using MPI [Google Scholar]
- 41.Therneau T. survival: survival analysis, including penalised likelihood. (Ported to R by Thomas Lumley); 2008. [Google Scholar]