Abstract
Genome-wide data enables us to clarify the underlying molecular mechanisms of complex phenotypes. The Online Mendelian Inheritance in Man (OMIM) is a widely employed knowledge base of human genes and genetic disorders for biological researchers. However, OMIM has not been fully exploited for omics analysis because its bibliographic data structure is not suitable for computer automation. Here, we characterized diseases and genes by generating feature profiles of associated drugs, biological phenomena and anatomy with the MeSH (Medical Subject Headings) vocabulary. We obtained 1 760 054 pairs of OMIM entries and MeSH terms by utilizing the full set of MEDLINE articles. We developed a web-based application called Gendoo (gene, disease features ontology-based overview system) to visualize these profiles. By comparing feature profiles of types 1 and 2 diabetes, we clearly illustrated their differences: type 1 diabetes is an autoimmune disease (P-value = 4.55 × 10−5) and type 2 diabetes is related to obesity (P-value = 1.18 × 10−15). Gendoo and the developed feature profiles should be useful for omics analysis from molecular and clinical viewpoints. Gendoo is available at http://gendoo.dbcls.jp/.
INTRODUCTION
The major aims of omics analysis are to identify disease-relevant genes and to understand their mechanisms. Genome sequences and transcriptomics provide large amounts of data, and researchers have attempted to interpret these genetic data in conjunction with clinical phenotypes (1–3). To analyze these data, we can easily obtain gene information such as gene names and genomic location, and their features in the form of Gene Ontology (GO) terms (4) from Entrez Gene (5,6) and Ensembl (7). Additionally, as a disease database, we generally refer to the Online Mendelian Inheritance in Man (OMIM: http://www.ncbi.nlm.nih.gov/omim/) (8,9).
OMIM contains nearly 18 000 detailed entries for human genes and genetic disorders. OMIM is a useful resource for obtaining information about diseases. However, it is difficult to utilize OMIM's data for omics analysis because almost all of its sections are written in natural language, namely English sentences (10). To enable computers to handle OMIM data, certain studies (11–15) have organized OMIM by selecting terms referred to in the Clinical Synopsis (CS) section as keywords. The CS section describes clinical features of disorders and their mode of inheritance such as ‘autosomal dominant’. Some of the terms in the CS section for Prader–Willi syndrome (OMIM ID: #176270) are shown in Table 1 as an example. Previous studies (12,14) characterized diseases according to corresponding tissue and etiology with CS terms. By using these terms, researchers do not have to use text mining techniques to automatically extract disease information from OMIM for omics analysis. However, even though OMIM includes detailed biological and genetic descriptions, CS terms are mainly clinical and diagnostic terms so that it is difficult to decipher disease information in conjunction with biological process data such as gene expression data. In addition, CS terms, such as ‘Cardiac’ and ‘Cardiovascular’, are ambiguous because the assigned terms are often defined by the author's original description of the cited articles (8).
Table 1.
Symptoms referred to in OMIM Clinical Synopsis section for Prader–Willi syndrome (partial)
| Inheritance: |
| Isolated cases |
| Growth: |
| Height |
| Mean adult male height, 155 cm |
| Mean adult female height, 147 cm |
| Steady childhood growth |
| Weight |
| Onset of obesity from 6 months to 6 years |
| Central obesity |
| Respiratory: |
| Hypoventilation |
| Hypoxia |
| Skeletal: |
| Osteoporosis |
| Osteopenia |
| Endocrine features: |
| Hyperinsulinemia |
| Growth hormone deficiency |
| Hypogonadotropic hypogonadism |
| Miscellaneous: |
| Food related behavioral problems include excessive appetite and obsession with eating |
| Temperature instability |
| High pain threshold |
| Molecular basis: |
| Microdeletion of 15q11 in 70% of patients confirmed by fluorescent in situ hybridization |
| Remainder of cases secondary to maternal disomy |
| Rare cases secondary to chromosome translocation |
Clinical features of a disorder are listed in the Clinical Synopsis (CS) section of the OMIM database. The CS section mainly describes morphologies and events in clinical and diagnostic fields. Each feature is itemized, but a controlled vocabulary is not used.
Here, to organize the disease features referred to in OMIM, we attempted to use the MeSH (Medical Subject Headings) controlled vocabulary (16). MeSH contains >20 000 keywords and hierarchically categorized into 15 concepts including ‘disease’, ‘chemicals and drugs’ and ‘anatomy’. It is originally curated for indexing MEDLINE articles by National Library of Medicine (NLM). In our previous study (17), to annotate genes from biological viewpoint excluded by GO such as disease and drug fields, we assigned MeSH to each gene by using Entrez Gene as gene data. In this article, we therefore generated feature profiles of diseases by applying MeSH to OMIM data with the method previously described (17). By comparing these feature profiles of genes developed (17) and diseases derived from this work, we aim to assist to interpret omics data from the molecular and clinical aspects.
METHODS
Data collection
We retrieved OMIM data available in February 2008 by downloading from the National Center for Biotechnology Information (NCBI) FTP site (ftp://ftp.ncbi.nih.gov/repository/OMIM/) and by using the web service with Entrez Programming Utilities (http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html). We obtained MeSH terms (2008 release) from the NLM web site (http://www.nlm.nih.gov/mesh/meshhome.html).
Articles extraction related to each OMIM entry
To generate OMIM–MeSH associations, we need to retrieve articles referred to in each OMIM entry because MeSH terms are not assigned to OMIM entries directly, but to MEDLINE. A schematic view of the pipeline for generating OMIM–MeSH associations is shown in Supplementary Figure S1. We retrieved PubMed IDs (PMIDs) cited in the reference section of OMIM (Supplementary Figure S1a) and extracted OMIM IDs described in the abstracts in MEDLINE (Supplementary Figure S1b). We also retrieved PMIDs by searching PubMed by inputting disease names (Supplementary Figure S1c). One of the problems is that one disease often has many names (18), e.g. ‘type 2 diabetes’, ‘non-insulin dependent diabetes’ and ‘NIDDM’. Another problem is that the same abbreviation may refer to several diseases, genes and drugs (19); for example, ‘EVA’ refers to ‘enlarged vestibular aqueduct’ (disease), ‘epithelial V-like antigen’ (gene) and ‘ethylene vinyl acetate’ (chemical). We therefore created abbreviation/long-form pairs for disease names such as ‘PWS’ and ‘Prader–Willi syndrome’ and searched MEDLINE for articles co-occurring with both names. Accordingly, we retrieved 426 141 unique OMIM ID and PMID pairs and generated 1 760 054 OMIM–MeSH pairs.
Scoring of associations between OMIM entries and MeSH terms
OMIM contains gene entries as molecular mechanisms and disease entries as their phenotypes (8). These types are indicated by symbols prefixed to the OMIM ID. We divided the OMIM entries into three groups according to these types: sequence known (*, +), locus known (%) and phenotype (#, none). We then calculated P-values as a score of OMIM–MeSH pairs in each group. The P-value is the probability of the actual or a more extreme outcome under the null-hypothesis. The lower P-value means the larger significance of association. We also calculated information gain to rank the associations of the OMIM–MeSH pairs as described in (17). Briefly, information gain refers to the frequency of co-occurrence of a disease name and a MeSH term and also refers to the specificity of the MeSH term.
Data visualization
We updated the web-based software application called Gendoo (gene, disease features ontology-based overview system) to visualize associations between OMIM entries and relevant MeSH terms. It was originally developed to visualize gene–MeSH associations (17). Gendoo accepts OMIM IDs, OMIM titles, Entrez Gene IDs, gene names and MeSH terms as input queries. For disease names, Gendoo currently uses descriptions of ‘title’ and ‘alternative titles; symbols’ sections of OMIM, so that not all synonyms are included in the disease name dictionary. We will increase the synonyms by involving the canonical name and synonyms (entry terms) of corresponding MeSH terms, and extracting disease names from MEDLINE and OMIM resources with text mining approach. Gendoo generates high-scoring lists that display relevant MeSH terms for diseases, drugs, biological phenomena and anatomy together with their scores (Supplementary Figure. S2a). These MeSH terms are sorted according to their information gain, and the background color of each association indicates its P-value. Gendoo also gives a hierarchical-tree view of MeSH terms associated with diseases of interest by using JavaScript and cascading style sheet (CSS) resources from the Yahoo! User Interface (YUI) library (http://developer.yahoo.com/yui/) (Supplementary Figure S2b).
RESULTS
Table 2 lists top-three keywords related to Prader–Willi syndrome for the features of the ‘Disease’, ‘Chemicals and Drugs’, ‘Biological Phenomena’ and ‘Anatomy’ fields. Prader–Willi syndrome results from deletion of paternal copies of the imprinted SNRPN (small nuclear ribonucleoprotein polypeptide N) and necdin genes within chromosome 15 (20). Gendoo shows the keyword phrases clearly reflecting the features of Prader–Willi syndrome, including ‘Chromosomes, Human, Pair 15’, ‘Genomic Imprinting’ and ‘Ribonucleoproteins, Small Nuclear’. Gendoo illustrates the disease features from not only a clinical perspective, but also a biological one, unlike the symptoms referred to in the CS section shown in Table 1. To retrieve more clinical and diagnostic features with MeSH, we can increase the number of novel associations by using terms from the ‘Analytical, Diagnostic and Therapeutic Techniques and Equipment’ category of MeSH.
Table 2.
Lists of top-three keywords related to Prader–Willi syndrome
| MeSH terms | P-value |
|---|---|
| Diseases | |
| Prader–Willi syndrome | 0 |
| Angelman syndrome | 4.05 × 10−140 |
| Obesity | 6.94 × 10−128 |
| Chemicals and Drugs | |
| Human growth hormone | 5.86 × 10−68 |
| Ribonucleoproteins, small nuclear | 4.29 × 10−62 |
| Ghrelin | 1.58 × 10−50 |
| Biological Phenomena | |
| Chromosomes, human, pair 15 | 0 |
| Genomic imprinting | 2.47 × 10−131 |
| Obesity | 1.69 × 10−121 |
| Anatomy | |
| Chromosomes, human, pair 15 | 0 |
| Chromosomes, human, 13–15 | 1.25 × 10−30 |
| Adipose tissue | 3.93 × 10−13 |
We generated feature profiles by using the MeSH vocabulary. Unlike the symptoms referred to in the CS section of OMIM (Table 1), these profiles give not only clinical, but also biological information about the disease.
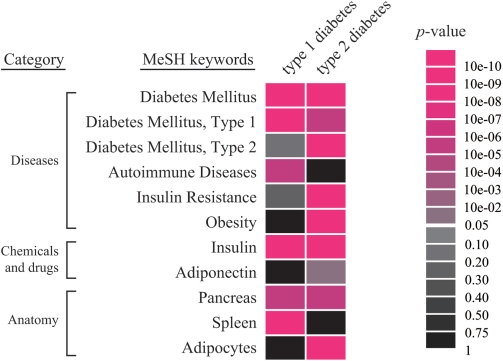
We applied this analysis to types 1 and 2 diabetes (OMIM IDs are %222100 and #125853, respectively). Figure 1 summarizes the feature profiles; type 1 diabetes is closely related to ‘Autoimmune Diseases’ and ‘Spleen’ (their P-values are 4.55 × 10−5 and 5.53 × 10−7, respectively), whereas type 2 diabetes is associated with ‘Obesity’ (P-value = 1.18 × 10−15) and ‘Adipocytes’ (P-value = 5.17 × 10−5). Type 1 diabetes is involved in immune systems, and type 2 diabetes is a metabolic disorder (21). This result suggests that the MeSH profiles produced by Gendoo can clarify the differences and similarities in features between OMIM entries.
Figure 1.
Differences and similarities between feature profiles of types 1 and 2 diabetes. Typical features and scores of types 1 and 2 diabetes are shown. The background colors of each association reflect the P-value. Type 1 diabetes is an autoimmune disorder, whereas type 2 diabetes is a metabolic disorder. These profiles clarify the differences between the features of these diseases.
We provide more practical results shown in Supplementary Table S1.
The Mendelian Inheritance in Man (MIM) is an excellent knowledge bank that has been annotated by Dr McKusick and his colleagues for >40 years, and its online version, OMIM, is accessible through the internet from NCBI (22). However, its bibliographic data structure has prevented OMIM from being fully exploited for omics analysis. To alleviate this problem, we comprehensively characterized human genes and genetic disorders referred to in OMIM with the MeSH vocabulary, and this will enable researchers to decipher their genome-wide data in conjunction with clinical phenotypes by using Gendoo. For example, the developed feature profiles can be applied to analyses of disease-relevant genes by comparing the similarities among profiles of OMIM entries and groups of genes such as those found in the clustering results of gene expression data. Researchers can also make overviews of features of unfamiliar diseases with Gendoo (Supplementary Table S1c and d).
AVAILABILITY
Gendoo can be openly accessed at http://gendoo.dbcls.jp/. Every association file including Entrez Gene/OMIM IDs, MeSH and their scores is available from the web site. Dictionary files including gene/disease names, synonyms and IDs are also downloadable. These web service and files are freely available under a Creative Commons Attribution 2.1 Japan license (http://creativecommons.org/licenses/by/2.1/jp/deed.en).
CONCLUSIONS
We characterized diseases and genes by generating feature profiles of associated drugs, biological phenomena and anatomy with the MeSH vocabulary and developed a web-based application called Gendoo to visualize these associations. MeSH profiles illustrate the features of genes and diseases. Comparing profiles emphasizes the differences and similarities between the features of genes and diseases. Gendoo will accelerate the analysis of omics data from biological and clinical perspectives.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Integrated Database Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Integrated Database Project.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Shoko Kawamoto and Prof. Kousaku Okubo for their helpful discussions.
REFERENCES
- 1.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat. Biotechnol. 2006;24:55–62. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez-Iratxeta C, Wjst M, Bork P, Andrade MA. G2D: a tool for mining genes associated with disease. BMC Genet. 2005;6:45. doi: 10.1186/1471-2156-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez-Iratxeta C, Bork P, Andrade MA. Association of genes to genetically inherited diseases using data mining. Nat. Genet. 2002;31:316–319. doi: 10.1038/ng895. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's online mendelian inheritance in man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bajdik CD, Kuo B, Rusaw S, Jones S, Brooks-Wilson A. CGMIM: automated text-mining of Online Mendelian Inheritance in Man (OMIM) to identify genetically-associated cancers and candidate genes. BMC Bioinformatics. 2005;6:78. doi: 10.1186/1471-2105-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masseroli M, Galati O, Manzotti M, Gibert K, Pinciroli F. Inherited disorder phenotypes: controlled annotation and statistical analysis for knowledge mining from gene lists. BMC Bioinformatics. 2005;6(Suppl. 4):S18. doi: 10.1186/1471-2105-6-S4-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hishiki T, Ogasawara O, Tsuruoka Y, Okubo K. Indexing anatomical concepts to OMIM Clinical Synopsis using the UMLS Metathesaurus. In Silico Biol. 2004;4:31–54. [PubMed] [Google Scholar]
- 13.Cantor MN, Lussier YA. Mining OMIM for insight into complex diseases. Medinfo. 2004;11:753–757. [PMC free article] [PubMed] [Google Scholar]
- 14.Freudenberg J, Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18(Suppl. 2):S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- 15.van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA. A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 16.Nelson SJ, Schopen M, Savage AG, Schulman JL, Arluk N. The MeSH translation maintenance system: structure, interface design, and implementation. Stud. Health Technol. Inform. 2004;107:67–69. [PubMed] [Google Scholar]
- 17.Nakazato T, Takinaka T, Mizuguchi H, Matsuda H, Bono H, Asogawa M. BioCompass: a novel functional inference tool that utilizes MeSH hierarchy to analyze groups of genes. In Silico Biol. 2008;8:53–61. [PubMed] [Google Scholar]
- 18.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 19.Gaudan S, Kirsch H, Rebholz-Schuhmann D. Resolving abbreviations to their senses in Medline. Bioinformatics. 2005;21:3658–3664. doi: 10.1093/bioinformatics/bti586. [DOI] [PubMed] [Google Scholar]
- 20.Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am. J. Med. Genet. A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- 21.Rother KI. Diabetes treatment—bridging the divide. N. Engl. J. Med. 2007;356:1499–1501. doi: 10.1056/NEJMp078030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.