Abstract
Protein contact map prediction is useful for protein folding rate prediction, model selection and 3D structure prediction. Here we describe NNcon, a fast and reliable contact map prediction server and software. NNcon was ranked among the most accurate residue contact predictors in the Eighth Critical Assessment of Techniques for Protein Structure Prediction (CASP8), 2008. Both NNcon server and software are available at http://casp.rnet.missouri.edu/nncon.html.
INTRODUCTION
Predicting residue contacts is an important problem in protein structure prediction. Contact maps, a matrix representation of protein residue–residue contacts within a distance threshold, provide an avenue for predicting protein 3D structure (1,2). There have been several algorithms developed to reconstruct protein 3D structure from an accurate contact map using distance-based algorithms developed for protein structure prediction and nuclear magnetic resonance (NMR) structure determination (3–7).
Even though contact prediction is presumably as hard as ab initio 3D structure prediction, it can be readily formulated as a classification problem, which can be tackled by knowledge-based reasoning methods, such as correlated mutation (8–14) and machine learning (15–27).
As more and more evidence shows that sequence-based contact predictions can be used to infer protein folding rates, evaluate protein models (28), and improve 3D structure prediction (29), contact map prediction is becoming increasingly important and useful. To date, however, only a few contact prediction servers [e.g. SCRATCH, Distill, SVMcon, SAM, RECON (30–34)] and a software package (SVMcon) are publicly available. To fill the gap, we describe a fast, state-of-the-art neural network-based contact map predictor NNcon that was ranked among the best methods in the Eighth Critical Assessment of Techniques for Protein Structure Prediction (CASP8), 2008 (35).
HYBRID CONTACT PREDICTION METHODS
We used 2D-Recursive Neural Network (2D-RNN) models to predict both general residue-residue contacts and specific beta contacts (i.e. beta-residue pairs in beta sheets).
General contact prediction
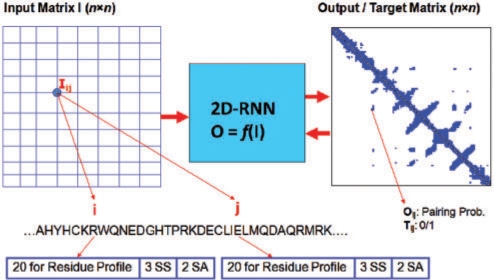
2D-RNN is a 2D machine learning method designed to map 2D input information into 2D output targets (36). The basic architecture of 2D-RNN contact predictions is illustrated in Figure 1.
Figure 1.
The 2D-RNN architecture for contact prediction. For a protein sequence with length n, the input to a 2D-RNN is an n × n input matrix and the output is an n × n probability matrix residue contacts.
The 2D-RNNs in NNcon are trained on a large data set consisting of 482 proteins and validated on a data set of 48 proteins. The real contacts were calculated as those residue pairs with C-α atoms within a set distance threshold. Ten 2D-RNN models were trained in order to create an ensemble of models that predict contacts. We trained two sets of 2D-RNN to predict contacts at an 8 Å and 12 Å threshold, respectively.
Beta-contact prediction
The general residue–residue contacts are defined based on a standard distance threshold of 8 and 12 Å. To take advantage of physiochemical constraints (i.e. hydrogen bonds) in beta sheets, we use 2D-RNN to directly predict beta-residue pairings within beta sheets (37). NNcon treated the prediction of inter-strand residue pairings as an additional binary classification problem, and refined these regions locally. The 2D-RNNs were trained and validated on the data set using 10-fold cross-validation on a large data set consisting of 916 chains and 2533 beta sheets (37). The ensemble of these 10 models is used to make predictions.
Combination of general and specific contact maps
Since the specific beta-contact predictor models predict beta contacts more accurately than the general contact map predictor models, we combined the predictions from these two methods for those proteins containing beta sheets. If the probability of a beta-residue pairing from the general contact model is less than that predicted by the beta-specific contact predictor, the general prediction is replaced by the beta-specific prediction value. The revised general contact map predictions are then finalized as the final contact map.
IMPLEMENTATION OF WEB SERVER
Both the NNcon web server and executable are freely available to all users at http://casp.rnet.missouri.edu/nncon.html and there is no login requirement. The input for the web server includes an e-mail address where the results will be sent, a target name, and an unformatted protein sequence. The e-mail output includes both a main message and several attachments. The main message includes the selected residue–residue contacts, in CASP format, at an 8 Å threshold, with sequence separation ≥6 and a predicted probability ≥0.1; and the average contact order and the average contact number derived from the predicted contact probability matrix at 8 Å. The attachments include a contact map image file, the full-contact probably matrix at 8 Å, and the full contact probability matrix at 12 Å. Users can select contacts from these probability matrices according to any probability threshold.
The server can accept multiple submissions concurrently through a task queue. NNcon predictions are much faster than support vector machine contact map predictors, such as SVMcon, which contain hundreds of thousands of support vectors. NNcon can make a prediction for a protein of average size (250 residues) in just a few minutes. The server can also make predictions for large proteins with up to 1000 residues in under an hour.
A Linux version of the contact prediction software is also available for download at the web site and the readme file contains the necessary installation instructions. This version of NNcon requires two input parameters at the command prompt: the name of a FASTA file and an output directory. The prediction results in the output directory include name.cm8a and name.cm12a, which are the predicted contact probability matrices for 8 and 12 Å, respectively.
EVALUATION OF WEB SERVER
NNcon was blindly tested in the CASP8 data set. We first evaluated NNcon against SVMcon, one of the top ranked contact map predictors in CASP7, on 116 CASP8 protein targets (Table 1). Both NNcon and SVMcon use pure ab initio methods to predict contacts within a protein. Next, we compared NNcon with all the CASP8 contact predictors on the 11 ab initio CASP8 domains, as shown in Table 2. All the contact predictions for these predictors and the 3D structures of the protein targets were downloaded from http://predictioncenter.org/casp8/.
Table 1.
Results of NNcon and SVMcon on 116 CASP8 targets
| Method | Acc6 | Cov6 | Acc12 | Cov12 | Acc24 | Cov24 |
|---|---|---|---|---|---|---|
| NNcon (L/5) | 0.58 | 0.07 | 0.51 | 0.06 | 0.31 | 0.05 |
| SVMcon (L/5) | 0.5 | 0.06 | 0.42 | 0.06 | 0.27 | 0.05 |
Acc6, Acc12, Acc24 denote prediction accuracy (specificity) at sequence separation ≥6, 12, 24 residues, respectively. Cov6, Cov12, Cov24 denote prediction coverage (sensitivity) at sequence separation ≥6, 12, 24 residues, respectively.
Table 2.
Multiple contact map predictors evaluated on 11 CASP8 ab initio domains
| Method | Acc6 | Cov6 | Acc12 | Cov12 | Acc24 | Cov24 |
|---|---|---|---|---|---|---|
| NNcon | 0.68 | 0.11 | 0.51 | 0.09 | 0.18 | 0.05 |
| SVMcon | 0.68 | 0.09 | 0.39 | 0.09 | 0.18 | 0.05 |
| SAM08_2stage | 0.28 | 0.05 | 0.26 | 0.06 | 0.17 | 0.05 |
| SAM06 | 0.26 | 0.04 | 0.24 | 0.05 | 0.16 | 0.06 |
| Fang | 0.44 | 0.07 | 0.31 | 0.06 | 0.16 | 0.05 |
| MUprot | 0.59 | 0.09 | 0.37 | 0.08 | 0.15 | 0.05 |
| Distill | 0.32 | 0.05 | 0.16 | 0.03 | 0.14 | 0.05 |
| 3Dpro | 0.05 | 0.01 | 0.33 | 0.07 | 0.14 | 0.05 |
| SAM08_server | 0.24 | 0.04 | 0.21 | 0.05 | 0.13 | 0.05 |
| SVMSEQ | 0.56 | 0.09 | 0.34 | 0.07 | 0.13 | 0.05 |
| Hamilton | 0.08 | 0.01 | 0.12 | 0.02 | 0.12 | 0.02 |
| Spine | 0.09 | 0.01 | 0.09 | 0.02 | 0.07 | 0.02 |
| Lee | 0.1 | 0.01 | 0.09 | 0.02 | 0.07 | 0.02 |
| Pairings | 0.36 | 0.05 | 0.35 | 0.06 | 0.05 | 0.01 |
For each domain, select top L/5 predicted contacts ranked by contact probabilities. Acc6, Acc12, Acc24 denote prediction accuracy (specificity) at sequence separation ≥6, 12, 24 residues, respectively. Cov6, Cov12, Cov24 denote prediction coverage (sensitivity) at sequence separation ≥6, 12, 24 residues, respectively.
COMPARISON OF NNcon AND SVMcon
Both NNcon and SVMcon were evaluated on 116 CASP8 targets. For each target, the top L/5 predicted contacts were selected, where L is the residue length of the protein. Then we calculated prediction coverage (sensitivity [TP/(TP + FN)]) and accuracy (specificity [TP/(TP + FP)]) for sequence separation of at least 6 residues, 12 residues and 24 residues, respectively, where TP, FP, TN and FN, are true positive, false positive, true negative and false negative predictions, respectively. NNcon had higher performance statistics than SVMcon in both coverage and accuracy for all sequence separation distances (Table 1). The sensitivities, overall, are lower than specificities in all predictions because only a small number of predicted contacts (L/5) are selected.
Comparison with other predictors on CASP8 ab initio domains
NNcon, as well as other CASP8 predictors, were evaluated on 11 CASP ab initio domains and then compared. The top L/5 predicted contacts were again used in the calculations. As Table 2 shows, NNcon performed favorably when compared with other predictors, especially at sequence separations ≥12 residues.
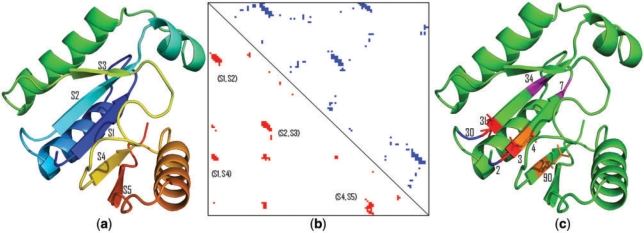
A good CASP8 contact prediction example
Figure 2 shows the predictions from the NNcon server for the target T0507. NNcon correctly identified key contacts in the beta sheets which can be very useful for predicting the final structure of the protein.
Figure 2.
(a) The 3D structure of CASP8 target T0507. The protein has five strands (S1–S5) that forms a parallel beta sheet. (b) The true contact map (upper triangle, blue) and predicted contact map (lower triangle, red). Each dot denotes a contact. It shows that some key contacts in four strand pairs (S1–S2, S2–S3, S1–S4, S4–S5) are correctly predicted. (c) Selective visualization of four residue–residue contacts correctly predicted (2–30, 3–31, 4–90, 7–34).
INFERENCE OF CONTACT ORDER AND CONTACT NUMBER
For each of the 48 proteins in the test data set, the average contact number and the average contact order of all the residues were calculated, and then correlated with the actual values. The actual (resp. predicted) contact number for each residue at 8 Å threshold was calculated as the total number of actual (resp. predicted) contacts with sequence separation greater than five residues.
The actual (resp. predicted) contact order for each residue is the sum of sequence separations of actual (resp. predicted) contacts with sequence separation greater than five residues, and then normalized by the protein sequence length. The Pearson correlations between the average actual and predicted contact number (0.85) and contact order (0.65) were strong, indicating that NNcon can successfully infer the actual average contact number and contact order of each protein from the predicted contact map. In the web server, the average contact number and order for the entire query protein are reported.
CONCLUSION
We have described NNcon—a fast and reliable web server and software for protein contact map prediction. NNcon was ranked among the most accurate methods in the CASP8 experiment, 2008. The contact map predicted by NNcon can be used to estimate the contact order and contact number of a protein. On average, a contact map prediction can be made in under a few minutes on one single-processor PC, making the method a valuable tool in large-scale contact map predictions.
FUNDING
MU Bioinformatics consortium, a MU research board grant and a MU research council grant to J.C. and a NLM fellowship to A.N.T. Funding for open access charge: MU faculty startup grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bonneau R, Ruczinski I, Tsai J, Baker D. Contact order and ab initio protein structure prediction. Protein Sci. 2002;11:1937–1944. doi: 10.1110/ps.3790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartoli L, Capriotti E, Fariselli P, Martelli PL, Casadio R. The pros and cons of predicting protein contact maps. In: Zaki MJ, Bystroff C, editors. Protein Structure Prediction. Totowa, NJ: Humana Press; 2007. p. 199. [DOI] [PubMed] [Google Scholar]
- 3.Aszodi A, Gradwell M, Taylor W. Global fold determination from a small number of distance restraints. J. Mol. Biol. 1995;251:308–326. doi: 10.1006/jmbi.1995.0436. [DOI] [PubMed] [Google Scholar]
- 4.Vendruscolo M, Kussell E, Domany E. Recovery of protein structure from contact maps. Fold. Des. 1997;2:295–306. doi: 10.1016/S1359-0278(97)00041-2. [DOI] [PubMed] [Google Scholar]
- 5.Skolnick J, Kolinski A, Ortiz A. MONSSTER: a method for folding globular proteins with a small number of distance restraints. J. Mol. Biol. 1997;265:217–241. doi: 10.1006/jmbi.1996.0720. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc. Natl Acad. Sci. USA. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vassura M, Margara L, di Lena P, Medri F, Fariselli P, Casadio R. FT-COMAR: fault tolerant three-dimensional structure reconstruction from protein contact maps. Bioinformatics. 2008;24:1313–1315. doi: 10.1093/bioinformatics/btn115. [DOI] [PubMed] [Google Scholar]
- 8.Goebel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18:309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 9.Olmea O, Valencia A. Improving contact predictions by the combination of correlated mutations and other sources of sequence information. Fold. Des. 1997;2:s25–s32. doi: 10.1016/s1359-0278(97)00060-6. [DOI] [PubMed] [Google Scholar]
- 10.Shindyalov I, Kolchanov N, Sander C. Can three-dimensional contacts in protein structure be predicted by analysis of correlated mutation? Protein Eng. 1994;7:349–358. doi: 10.1093/protein/7.3.349. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton N, Burrage K, Ragan M, Huber T. Protein contact prediction using patterns of correlation. Proteins. 2004;56:679–684. doi: 10.1002/prot.20160. [DOI] [PubMed] [Google Scholar]
- 12.Valencia A, Pazos F. Computational methods for the prediction of protein interactons. Curr. Opin. Struct. Biol. 2002;12:368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 13.Halperin I, Wolfson HJ, Nussinov R. Correlated mutations: Advances and limitations. A study on fusion proteins and on the Cohesin-Dockerin families. Proteins. 2006;63:832–845. doi: 10.1002/prot.20933. [DOI] [PubMed] [Google Scholar]
- 14.Kundrotas PJ, Alexov EG. Predicting residue contacts using pragmatic correlated mutations method: reducing the false positives. BMC Bioinformatics. 2006;7:503. doi: 10.1186/1471-2105-7-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fariselli P, Olmea O, Valencia A, Casadio R. Prediction of contact maps with neural networks and correlated mutations. Prot. Eng. 2001;13:835–843. doi: 10.1093/protein/14.11.835. [DOI] [PubMed] [Google Scholar]
- 16.Lund O, Frimand K, Gorodkin J, Bohr H, Bohr J, Hansen J, Brunak S. Protein distance constraints predicted by neural networks and probability density functions. Prot. Eng. 1997;10:1241–1248. doi: 10.1093/protein/10.11.1241. [DOI] [PubMed] [Google Scholar]
- 17.Fariselli P, Casadio R. Neural network based predictor of residue contacts in proteins. Prot. Eng. 1999;12:15–21. doi: 10.1093/protein/12.1.15. [DOI] [PubMed] [Google Scholar]
- 18.Fariselli P, Olmea O, Valencia A, Casadio R. Progress in predicting inter-residue contacts of proteins with neural networks and correlated mutations. Proteins. 2001;45:157–162. doi: 10.1002/prot.1173. [DOI] [PubMed] [Google Scholar]
- 19.Pollastri G, Baldi P, Fariselli P, Casadio R. Improved prediction of the number of residue contacts in proteins by recurrent neural networks. Bioinformatics. 2001;17:S234–S242. doi: 10.1093/bioinformatics/17.suppl_1.s234. [DOI] [PubMed] [Google Scholar]
- 20.Pollastri G, Baldi P. Prediction of contact maps by GIOHMMs and recurrent neural networks using lateral propagation from all four cardinal corners. Bioinformatics. 2002;18(Suppl. 1):S62–S70. doi: 10.1093/bioinformatics/18.suppl_1.s62. [DOI] [PubMed] [Google Scholar]
- 21.MacCallum R. Striped sheets and protein contact prediction. Bioinformatics. 2004;20(Suppl. 1):i224–i231. doi: 10.1093/bioinformatics/bth913. [DOI] [PubMed] [Google Scholar]
- 22.Shao Y, Bystroff C. Predicting inter-residue contacts using templates and pathways. Proteins. 2003;53(Suppl. 6):497–502. doi: 10.1002/prot.10539. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Karypis G. Prediction of contact maps using support vector machines. Proc. IEEE Symp. Bioinformatics BioEng. 2003:26–36. [Google Scholar]
- 24.Punta M, Rost B. PROFcon: novel prediction of long-range contacts. Bioinformatics. 2005;21:2960–2968. doi: 10.1093/bioinformatics/bti454. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J, Saigo H, Baldi P. Large-scale prediction of disulphide bridges using kernel methods, Two-dimensional recursive neural networks, and weighted graph Matching. Proteins: Struct. Funct. Bioinformatics. 2006;62:617–629. doi: 10.1002/prot.20787. [DOI] [PubMed] [Google Scholar]
- 26.Vullo A, Walsh I, Pollastri G. A two-stage approach for improved prediction of residue contact maps. BMC Bioinformatics. 2006;7:180. doi: 10.1186/1471-2105-7-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller CS, Eisenberg D. Using inferred residue contacts to distinguish between correct and incorrect protein models. Bioinformatics. 2008;24:1575–1582. doi: 10.1093/bioinformatics/btn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Tegge AN, , , Cheng J. Evaluating the absolute quality of a single protein model using support vector machines and structural features. Proteins. 2008;75:638–647. doi: 10.1002/prot.22275. [DOI] [PubMed] [Google Scholar]
- 29.Wu S, Zhang Y. A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics. 2008;24:924–931. doi: 10.1093/bioinformatics/btn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng J, Randall A, Sweredoski M, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:w72–w76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bau D, Martin A, Mooney C, Vullo A, Walsh I, Pollastri G. Distill: a suite of web servers for the prediction of one-, two- and three-dimensional structural features of proteins. BMC Bioinformatics. 2006;7:402. doi: 10.1186/1471-2105-7-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng J, Baldi P. Improved residue contact prediction using support vector machines and a large feature set. BMC Bioinformatics. 2007;8:113. doi: 10.1186/1471-2105-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shackelford G, Karplus K. Contact prediction using mutual information and neural nets. Proteins. 2007;69(Suppl. 8):159–164. doi: 10.1002/prot.21791. [DOI] [PubMed] [Google Scholar]
- 34.Kundrotas, Alexov Predicting residue contacts by correlated mutation method. BMC Bioinformatics. 2006;7:503. doi: 10.1186/1471-2105-7-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction - round VII. Proteins. 2007;69(Suppl. 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldi P, Pollastri G. The principled design of large-scale recursive neural network architectures–dag-rnns and the protein structure prediction problem. J. Machine Learning Res. 2003;4:575–602. [Google Scholar]
- 37.Cheng J, Baldi P. Three-stage prediction of protein beta-sheets by neural networks, alignments, and graph algorithms. Proceedings of the 2005 Conference on Intelligent Systems for Molecular Biology (ISMB 2005) Bioinformatics. 2005;21(Suppl. 1):i75–i84. doi: 10.1093/bioinformatics/bti1004. [DOI] [PubMed] [Google Scholar]