Abstract
In recent years, a lot of efforts have been made in conformational epitope prediction as antigen proteins usually bind antibodies with an assembly of sequentially discontinuous and structurally compact surface residues. Currently, only a few methods for spatial epitope prediction are available with focus on single residue propensity scales or continual segments clustering. In the method of SEPPA, a concept of ‘unit patch of residue triangle’ was introduced to better describe the local spatial context in protein surface. Besides that, SEPPA incorporated clustering coefficient to describe the spatial compactness of surface residues. Validated by independent testing datasets, SEPPA gave an average AUC value over 0.742 and produced a successful pick-up rate of 96.64%. Comparing with peers, SEPPA shows significant improvement over other popular methods like CEP, DiscoTope and BEpro. In addition, the threshold scores for certain accuracy, sensitivity and specificity are provided online to give the confidence level of the spatial epitope identification. The web server can be accessed at http://lifecenter.sgst.cn/seppa/index.php. Batch query is supported.
INTRODUCTION
With growing need of monoclonal antibodies and vaccines, B cell epitope prediction has become more and more desirable especially for new proteins isolated from pathogens. A lot of efforts have been put for this purpose, but primarily on continuous epitopes. However, crystallographic studies have shown that most of the epitopes in protein antigens are discontinuous (1), while only a few methods have been designed for this condition. For instance, the first server CEP was erected in 2005 by introducing ‘accessibility of residues’ based on the 3D nature of the antigen proteins (2). Subsequently, DiscoTope prediction method was designed by combining propensity scale matrixes with the spatial proximity and surface exposure (1). Recently, BEpro improved DiscoTope method by introducing spatial attribute of half sphere exposure (3). While these tools provided a lot of help in designing molecular experiments, benchmark and reviews have shown that the prediction of spatial epitope remained difficult for protein antigens (4,5). As being pointed out, the possible improvement could lie in new features characterizing 3D structures of epitope and better training data (4).
This article introduces a new computational server, SEPPA, for spatial epitope prediction of protein antigens. In the method of SEPPA, a novel concept of ‘unit patch of residue triangle’ is introduced to better describe local spatial context in protein antigen surface. Typical network parameter of spatial clustering coefficient is also incorporated to reflect the 3D characteristic of epitopes. In addition, comprehensive training data of non-redundant spatial epitopes is curated from PDB database (6) with unique representatives covering the vast diversity of known epitopes. SEPPA was rigorously trained by 82 antigen–antibody protein complexes, which contained 84 unique epitopes. One hundred and nineteen independent spatial epitopes of protein antigens were collected as testing dataset. SEPPA's performance in detecting potential spatial epitopes was evaluated and compared with popular peer methods.
DATASET
Antigen–antibody complexes were extracted from PDB database dated August 2008. Only those with resolution better than 3.0 Å and protein antigen length with more than 25 residues were retained. Redundant epitopes were removed by 60% similarity. Eighty two structures were finally retained as the training data which included 84 unique epitopes.
The testing data were collected from the training dataset of DiscoTope (1), databases of IEDB (7) and Epitome (8). A testing dataset of 119 antigens was set up by deducting SEPPA's training data from above testing data. The PDB IDs included in the training and testing datasets can be found in the Supplementary Data (Table S1).
METHODS
Definition for unit patch of residue triangle
Solvent accessible surface areas (SASA) were calculated (Naccess V2.1.1.) for each residue in antigen proteins. Surface residues were those with more than 1 Å2 SASA, while those with SASA loss in binding of more than 1 Å2 were classified as epitope residues. The unit patch of residue triangle was defined among any three surface residues if the distance for every two of them was within 4 Å atom distance. Based on the training data, unit patches containing more than two epitope residues were termed as epitope unit patches; otherwise were classified as non-epitope unit patches.
Derivation of propensity indices for unit patches
Considering that various residues may have similar functional moieties of R-groups in antigen–antibody interaction, the 20 residues were consolidated into 13 functional subgroups according to the conformational epitope research of Erez et al. (9). Four hundred and fifty five combination patterns of subgroups were observed out of 13 × 13 × 13 for unit patches. Propensity index (trii) of the unit patch pattern i is calculated as the ratio of the number of pattern i among all epitope unit patches (fi*/Σi fi*) compared with that ratio in the non-epitope unit patches (fi/Σi fi):
| 1 |
where fi* is the number of unit patch pattern i in epitope unit patches, while Σi fi* is the number of all epitope patches. Denominator indicates those for non-epitope unit patches.
For a certain surface residue r, the propensity score of it (avgr) is predominantly determined by its local neighboring environment. Thus avgr is calculated as the averaged propensity indices of all possible unit patches around residue r:
| 2 |
where Σtrii is the sum of propensity indices for neighboring unit patches within certain distance of residue r, and N is the number of these neighboring unit patches.
Definition of residue neighbor and clustering coefficient
Clustering coefficient is introduced to describe the compactness of the neighboring residues around one residue. It reflects the probability that the neighbors of residue r are also neighbors with each other (10). For one residue r, all residues within 15 Å of r are defined as residue neighbors of r. kr is the total number of residues neighbors for r.
Theoretically, the number of all possible links among kr residue neighbors is kr(kr–1)/2. However, as only those links within certain distance can be called residue neighbors of each other, the observed actual number of residue neighbor pairs among kr is recorded as er. The clustering coefficient (ccr) is given below:
| 3 |
Algorithm of SEPPA
For each antigen protein from input, SEPPA will:
Step 1: Determine all the surface residues in the protein antigen;
For each surface residue r:
Step 2: Search all possible unit patches within 15 Å atom distance of residue r, map the pre-calculated propensity indices (trii) to above unit patches, and calculate the propensity index avgr using Equation (2);
Step 3: Calculate the clustering coefficient (ccr) for residue r using Equation (3);
Step 4: Summarize avgr and ccr as the antigenicity score for residue r;
Step 5: Give the antigenicity score for each residue, and highlight those residues with scores higher than a threshold. Visualize the subsets of predicted epitope area graphically.
Performance of SEPPA
The Area Under the Curve (AUC) value and successful pick-up rate have been introduced to assess the performance of SEPPA. SEPPA achieved the average AUC value of 0.742 on the 119 independent testing dataset (Table S2). With default threshold 1.80, a sensitivity of 0.580 and a specificity of 0.707 were got on this testing dataset. The performance of SEPPA was also compared to popular tools of CEP, DiscoTope and BEpro under the 119 testing dataset (Table S3). By default thresholds, CEP, DiscoTope and BEpro produced average estimated AUC values (defined as (sensitivity+specificity)/2) of 0.521, 0.601 and 0.563, respectively. In the case of SEPPA, an average estimated AUC value of 0.644 was achieved under its default threshold of 1.80, which was significantly better than those of other tools. It should be reminded that this testing dataset is only new to SEPPA. Hence, this comparison may be biased in favor of other tools.
On the other hand, it would be worth to compare among the tools the ability of picking up true epitope residues from random surface residues. For SEPPA, DiscoTope and BEpro, unpaired t-tests were done to the prediction scores between the true epitope residues and other non-epitope surface residues. If the mean prediction score for all the true epitope residues in a protein antigen is significantly higher than that of non-epitope surface residues (P < 0.05; unpaired t-test, one-tailed), this prediction will be considered as a successful pick-up. The pick-up rates were compared between the three tools on the 119 testing dataset. SEPPA gave the best performance of 96.64%, while DiscoTope achieved 89.08% and BEpro gave 90.76% (Table S4). It is reminded that CEP did not attend this round of evaluation because it produces qualitative patch prediction results instead of antigenicity scores.
IMPLEMENTATION
Input
SEPPA requires a 3D protein structure in PDB format as input. Users can submit the query with a released PDB ID or upload a structural file in PDB format. The format of input file is provided as an example. It is recommended to specify the chain(s) ID if not all peptides are antigen proteins to be queried in the structure file. Otherwise, each chain will be assumed as an antigen protein and calculated for antigenicity scores.
Output
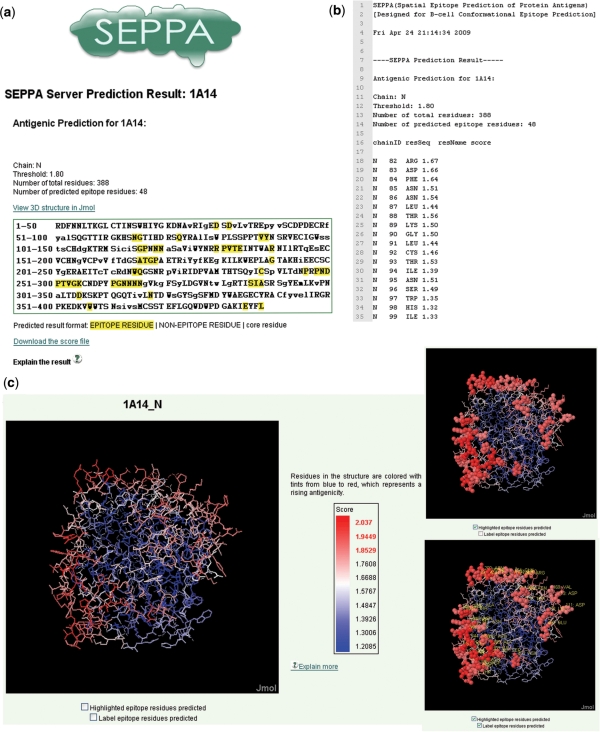
The results of prediction are displayed in html format. As seen in Figure 1, the predicted epitope of the influenza virus (PDB code 1A14:N) are shown as an example. The sequence of submitted protein antigen is displayed in single letter code in result window. The core residues are shown in lowercase and surface residues in uppercase. The residues predicted as epitope are highlighted with yellow color background (Figure 1a). The scores of prediction are recorded in another file, which lists the antigenicity scores for individual residue as shown in Figure 1b and this file is downloadable. A link to visualize the prediction result is also provided in the result page. The visualization of result is displayed with Jmol (an open-source Java viewer for chemical structures in 3D), as shown in Figure 1c. Tints from blue to red represent a rising propensity for a residue to be in the epitope.
Figure 1.
A snapshot of predicted spatial epitope and graphical display of influenza virus (PDB code 1A14:N). (a) Result page for epitope prediction of influenza virus. In result box, the query sequence is displayed in single letter code. Core residues are shown in lowercase, and surface residues are shown in uppercase. Predicted epitope residues are highlighted with yellow color. (b) Antigenicity scores predicted for each residue in influenza virus. (c) Visualization of the predicted spatial epitope. Tints from blue to red represent a rising propensity for a residue to be in the epitope. (c) is generated with Jmol, and predicted epitope residues can be highlighted with solid sphere mode and labeled with their information.
DISCUSSION
Generally there are two strategies to define epitope residues from the structures of immune complexes. One adopts the change of SASA between unbound and bound state of antigen structures to define epitope residues, while the other picks up a distance cutoff between antigen and antibody atoms in complexes, e.g. 4 Å in DiscoTope (1). SEPPA took the first strategy; the distance cutoff one was also simulated to inspect the robustness. Results indicated that these two strategies had given similar results to the performance of SEPPA (data is not shown).
Currently, a parameter of 4 Å was chosen in the definition of unit patch of residue-triangle. Considering that the majority of atom contacts occur at <5 Å (11), different distances of 4, 5 and 6 Å were put under test in our work and 4 Å was found to give the best results. Furthermore, the cutoff for neighborhood was set to be 15 Å because of the following calculation. Cutoff ranges from 5 to 20 Å were scanned according to their impact on prediction. The results showed that the most stable performance was achieved at 15 Å under training data. Thus, this value was selected as the neighborhood cutoff.
ROC curve is often applied to evaluate the performance of predictive methods. In our case, however, CEP is categorized as a discrete classifier, which does not produce the prediction scores. Thus a ROC curve could not be generated for CEP but the AUC value could be estimated as (sensitivity + specificity)/2 following the method of Julia et al. (4), which is also applicable for other three tools under their respective default thresholds. Since the ROC curve and AUC value are highly correlated, we assume that the estimated AUC value could be an alternative vector to compare the performance of those tools in our work.
From this work, the concepts of ‘unit patch of residue triangle’ and ‘clustering coefficient’ seem to make major contribution to the effective prediction of spatial epitopes. As being frequently suggested, epitope residues function as a whole on antigen surface during antibody binding. Correspondingly, the concept of ‘unit patch of residue triangle’ just describes the minimum moiety of surface patches which can better reflect the local spatial context on antigen exterior. Thus SEPPA makes prediction based on both the residual context and the spatial compactness of neighboring residues. Since the propensity index is derived from statistics of training data, these data are very important as well to be representative and unique. With more and more structural data accumulated, we believe that SEPPA would be increasingly improved by incorporating further refined features of spatial epitopes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology China (2004CB720103, 2006AA02312, 2007AA02Z300), National Natural Science Foundation of China (30500107), and Shanghai Municipal Funding (2000236018, 2000236016, 07DZ22901). Funding for open access charge: Ministry of Science and Technology China (2004CB720103).
Conflict of interest statement. None declared.
Supplementary Material
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Haste Andersen P, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulkarni-Kale U, Bhosle S, Kolaskar AS. CEP: a conformational epitope prediction server. Nucleic Acids Res. 2005;33:W168–W171. doi: 10.1093/nar/gki460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweredoski MJ, Baldi P. PEPITO: improved discontinuous B-cell epitope prediction using multiple distance + thresholds and half sphere exposure. Bioinformatics. 2008;24:1459–1460. doi: 10.1093/bioinformatics/btn199. [DOI] [PubMed] [Google Scholar]
- 4.Ponomarenko JV, Bourne PE. Antibody–protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct. Biol. 2007;7:64. doi: 10.1186/1472-6807-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reitmaier R. Review of immunoinformatic approaches to in-silico B-cell epitope prediction. Nature Precedings. 2007 Posted 5 Jul 2007, 10.1038/npre.2007.353.1. [Google Scholar]
- 6.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlessinger A, Ofran Y, Yachdav G, Rost B. Epitome: database of structure-inferred antigenic epitopes. Nucleic Acids Res. 2006;34:D777–D780. doi: 10.1093/nar/gkj053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bublil EM, Freund NT, Mayrose I, Penn O, Roitburd-Berman A, Rubinstein ND, Pupko T, Gershoni JM. Stepwise prediction of conformational discontinuous B-cell epitopes using the Mapitope algorithm. Proteins. 2007;68:294–304. doi: 10.1002/prot.21387. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Kawashima S, Kanehisa M. New amino acid indices based on residue network topology. Genome Informatics. 2007;18:152–161. [PubMed] [Google Scholar]
- 11.McConkey BJ, Sobolev V, Edelman M. Discrimination of native protein structures using atom-atom contact scoring. Proc. Natl Acad. Sci. USA. 2003;100:3215–3220. doi: 10.1073/pnas.0535768100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.