Abstract
ProteinCCD (CCD for Crystallographic Construct Design) aims to facilitate a common practice in structural biology, namely the design of several truncation constructs of the protein under investigation, based on experimental data or on sequence analysis tools. ProteinCCD functions as a meta-server, available online at http://xtal.nki.nl/ccd, that collects information from prediction servers concerning secondary structure, disorder, coiled coils, transmembrane segments, domains and domain linkers. It then displays a condensed view of all results against the protein sequence. The user can study the output and choose interactively possible starts and ends for suitable protein constructs. Since the required input to ProteinCCD is the DNA and not the protein sequence, once the starts and ends of constructs are chosen, the software can automatically design the oligonucleotides needed for PCR amplification of all constructs. ProteinCCD outputs a comprehensive view of all constructs and all oligos needed for bookkeeping or for direct copy-paste ordering of the designed oligonucleotides.
INTRODUCTION
The production of soluble proteins in amounts suitable for structural studies has been a common bottleneck in structural biology and structural genomics alike (1,2). For X-ray crystallographic studies an additional goal is to obtain not only soluble protein, but also a specific construct with a high propensity to crystallize. Similarly, for NMR studies the soluble protein domain does not only need to be relatively small, but also highly soluble in relatively high concentration to deliver clear spectra. The advent of cloning techniques that are high-throughput, inexpensive and compatible with robotic implementations (3,4) allows parallel construction of tens of expression constructs for each protein under study; a standard practice in many labs.
Expression constructs can be designed based on experimental information, typically limited proteolysis experiments followed by mass spectrometry based identification of the proteolytic fragments (4). Computational design based on sequence analysis is another method of choice. The use of multiple sequence alignments is wide spread and there are a variety of specific tools, e.g. T-Coffee (5) or MUSCLE (6). Based on similarities and differences among family members, the researcher decides what are the likely domain boundaries that will yield soluble, well-behaved proteins. Comparative modeling (7) can also be used if a structure of a homologous protein is known. Finally, a variety of sequence analysis methods aim to deliver structural information from the sequence alone. Although significant progress has been made in sequence analysis, there is no definitive method of choice. Typically, most researchers use a ‘personal’ collection of web-based tools for sequence analysis, based on prior experience. A clear bottleneck arises, as it is cumbersome to display the results of different tools in a condensed form. At present, this requires submitting many queries to different servers, and subsequent copying-pasting to compare the results of different methods.
After a concise and condensed representation of all analysis results is obtained, the researcher typically decides what are promising domain boundaries for the protein in hand. The next step is to design oligonucleotides to be used for PCR-based amplification of all these fragments. At this stage a trivial but time consuming additional bottleneck is encountered: the protein-based analysis has to be transformed back to the DNA sequence. Although the task is by all means trivial, it is time consuming and error prone, since the direct mapping between protein and DNA sequence is lost in the analysis step.
METHODS
We have developed a web-based tool to address these bottlenecks, which provides a simple and practical solution for daily use in the research laboratory. The tool we developed, ProteinCCD, addresses these bottlenecks by:
Acting as a meta-server that combines several sequence analysis methods that are available as web services and commonly used for expression construct design, and displaying all results in a condensed and concise manner.
Requiring as user input the DNA rather than the protein sequence and thus enabling the 'single-click' design of all oligonucleotides needed for PCR amplification of the user-designed constructs.
We chose methods from four groups of sequence analysis tools.
The first group concerns secondary structure prediction servers, aiming to predict stretches of sequences that are likely to be either helices or strands in the three dimensional structure (8) and should not be disrupted. Different algorithms give slightly different results especially at the secondary structure element boundaries. Therefore we have included a few methods from this group, and use the collection available at the Network Protein Sequence @nalysis (9) (NPS@).
The second group of sequence analysis methods concerns algorithms that aim to predict disordered regions in protein sequences. We chose four methods: IUPred (10) which uses the estimated pairwise energy content; RONN (11) which is based on a Bio-Basis Function Neural Network (BBFNN) to predict intrinsically disordered regions in proteins; DisEMBL (12) which uses empirical definitions for disorder predictions; and GlobPlot (13) which uses predictions for globularity against disorder to better identify disordered regions in proteins.
The third group of methods involves specialized servers for specific features of protein sequence. Currently we check for coiled-coil regions (14), and trans-membrane topology prediction combined with signal peptide prediction, as available from the Phobius webserver (15).
The fourth and final group of methods includes two of the many new algorithms that look for domains. The Simple Modular Architecture Research Tool (16) (SMART) aims to identify and annotate genetically mobile domains and to analyze domain architecture. The Domain Linker Predictor (17) in contrast attempts to flag the regions between likely domains.
A user needs to submit a cDNA sequence to ProteinCCD. This entry is first checked for validity, translated to amino-acid sequence, displayed in the output window and sequentially submitted to the selected servers. As results are returned in real time, they are displayed below the query sequence in a multiple-alignment manner allowing the direct comparison of results between different methods. The user can utilize the condensed comparative output, to scroll along the sequence and choose domain boundaries, with simple mouse-clicks for choosing N- and C-terminal boundaries. This step is entirely up to the user and no automated method is provided.
As soon as the user has selected domain boundaries, ProteinCCD outputs a list of the resulting protein sequences and all the oligonucleotides needed for the PCR amplification of these sequences. Oligonucleotides are chosen based either on the simple rule that N nucleotides are needed for annealing (default N = 20) or by choosing enough nucleotides to reach a user defined annealing temperature Tm (default Tm= 65) based on the formula:
| 1 |
where GCnr is the number of guanine and cytosine nucleotides in the oligonucleotide sequence. User-defined overhangs are appended both on the 5′ and the 3′ oligonucleotide, to facilitate cloning and quick cut-and-paste ordering of the final oligonucleotides. ProteinCCD does not check oligonucleotides for secondary structure or for false-annealing sites.
EXAMPLE
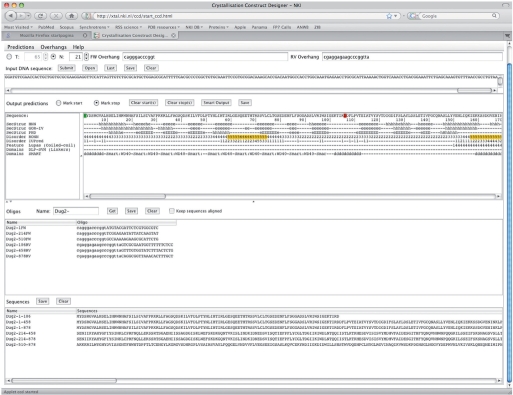
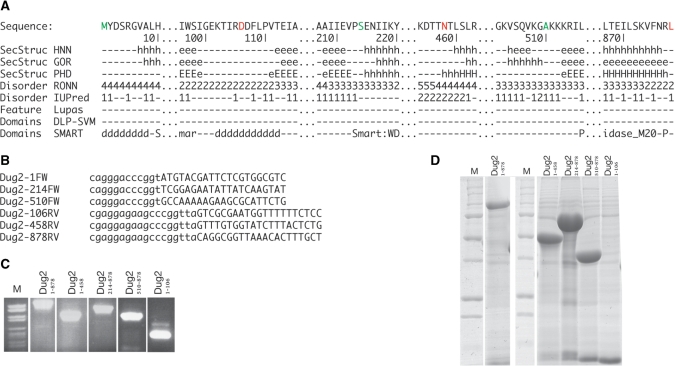
We will now consider a very simple example describing the steps to analyze a new sequence and design five simple truncation constructs. For the example we will use the protein Dug2 from yeast (18). In Figure 1 there is an overview of the ProteinCCD web server after the submitted job was finished. The only input was the DNA sequence of Dug2. After pasting the sequence to the top field and pressing the ‘Submit’ button the servers selected by default returned the analysis results for the protein sequence. The translated protein sequence and the ‘aligned’ predictions appear in the panel below the input. Subsequently, we selected with the mouse three N-termini: The N-terminal residues 1, 214 and 510; and three C-termini: residues 106, 458 and the terminal residue 878. In Figure 2A, we display the predictions in the area of all the six termini. In brief, SMART (16) shows that Dug2 contains four WD40 repeats, spanning the N-terminal half and a peptidase domain spanning the C-terminal half. Here we discuss how we made the exact choices for the N- and C-termini of all truncation constructs.
Nterm -1: The first WD40 repeat starts essentially at the very start of the protein, thus the natural N-terminus was considered as a good selection for an expression construct.
Cterm-106: The first pair of WD40 repeats ends at residue 98. Since a consensus strand prediction runs to residue 99, we chose to include a few more additional residues to be sure that the C-terminal predicted strand interactions are maintained.
Nterm-214: This residue is right at the start of the helix of the third predicted WD40 domain.
Cterm-458: The fourth predicted WD40 domain predicted by SMART ends at residue 396. However, the three secondary structure predictions contradict each other: they indicate a short strand followed by helix, long but weak strand prediction, short but strong strand prediction. On top, this residue is within a region predicted by RONN (11) to be disordered and—contradicting that prediction—just before a region predicted to be a domain linker (17). Finally, we noticed that two of the secondary structure prediction programs predict two helices for the next ∼50 residues. Thus, we decided in this case to use as a C-terminus residue 458 and include these two secondary structure elements.
Nterm-510: The peptidase SMART domain is predicted to start at residue 516. However, a consensus strand is predicted to start residue 513. To be sure we don't interrupt the secondary structure elements, which might be important for domain folding, we chose to start that construct at residue 510.
Cterm-878: The peptidase SMART domain ends up right at the natural C-terminus and choosing exactly that was an easy choice.
Figure 1.
An overview of the ProteinCCD server after all predictions have been collected, user choices have been made, and the oligonucleotides have been suggested, for the example discussed in this paper.
Figure 2.
(A) Regions of interest from the ProteinCCD output where starts (green letters) and stops (red letters) are marked by the user. Discontinuities in the sequence have been marked with (...) (B) The oligonucleotide sequences for PCR amplifications of the regions of interest as suggested by ProteinCCD. (C) An ethidium bromide stained agarose gel showing the PCR products for the five selected truncation constructs of interest obtained with the designed oligonucleotides under standard experimental conditions (D) A polyacrylamide SDS gel stained with Coomassie blue showing the expressed and IMAC purified truncation constructs designed and cloned above. The (*) denotes the protein of interest in each lane.
After these ‘starts’ and ‘stops’ are selected by the corresponding buttons in the web server application, the ‘Submit’ button is pressed. This automatically calculates the needed oligonucleotides to design all possible constructs between these termini. In this particular case we only wanted constructs Dug21–878, Dug21–458, Dug2214–878, Dug2510–878, Dug21–106. The oligonucleotides that were designed to amplify these constructs and clone them to the NKI-LIC-His-3C vector, are presented in Figure 2B. In Figure 2C we show the amplified DNA under standard PCR conditions, and in Figure 2D the expression experiments, that resulted in all constructs being soluble. In conclusion, in this case study, educated ‘guesses’ for construct design were made very easy by the comprehensive view offered by ProteinCCD, and the experiments designed with the aid of the web server resulted in soluble proteins suitable for structural and functional studies.
CONCLUSION AND PERSPECTIVE
ProteinCCD provides a user-friendly tool to facilitate protein construct design and oligonucleotide ordering. By consolidating tools that are common in structural biology in a single platform ProteinCCD enables comparative analysis of the sequence, and by keeping track of both the protein and DNA sequence it allows the straightforward design of oligonucleotides for PCR amplification of the protein constructs.
The choice of protein constructs for structural studies is not a straightforward task. Although some notable automation attempts exist, there is no widely accepted method. Thus, at this stage we chose to only implement a tool that collects the results of a variety of popular web servers. This enables informed decisions by the user, but at present not further automation is provided, and we are unable to quantify or benchmark the server, since it is clearly dependent on user choices. It provides an enabling technology and not an automated tool.
Among the plans for future work however, is to allow users to submit their constructs choices to a connected database. This could allow for the future development of supervised learning methods to imitate user choices. This would still not allow the development of algorithms that actually predict which constructs end up being soluble (or even crystallizable), since that information is not readily available and is very difficult to collect outside the context of well-managed high-throughput projects. It can be argued however, that a learning system that mimics common user choices for a variety of targets could indeed be a useful direction for future research.
FUNDING
ProteinCCD has been both inspired and tested as part of our participation in the EU FP6 programs 3D-Repertoire (LSHG-CT-2005-512028) and SPINE2-Complexes (LSH-2004-1.1.2-1). Funding for open access charge: Netherlands Cancer Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank all member of the Perrakis and Sixma labs at the NKI for their input, and in particular Dene Littler, Rick Hibbert, and Evangelos Christodoulou for their ideas, beta-testing the software and their comments for this manuscript.
REFERENCES
- 1.Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, et al. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lesley SA. High-throughput proteomics: protein expression and purification in the postgenomic world. Protein Expr. Purif. 2001;22:159–164. doi: 10.1006/prep.2001.1465. [DOI] [PubMed] [Google Scholar]
- 3.Alzari PM, Berglund H, Berrow NS, Blagova E, Busso D, Cambillau C, Campanacci V, Christodoulou E, Eiler S, Fogg MJ, et al. Implementation of semi-automated cloning and prokaryotic expression screening: the impact of SPINE. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1103–1113. doi: 10.1107/S0907444906029775. [DOI] [PubMed] [Google Scholar]
- 4.Banci L, Bertini I, Cusack S, de Jong RN, Heinemann U, Jones EY, Kozielski F, Maskos K, Messerschmidt A, Owens R, et al. First steps towards effective methods in exploiting high-throughput technologies for the determination of human protein structures of high biomedical value. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1208–1217. doi: 10.1107/S0907444906029350. [DOI] [PubMed] [Google Scholar]
- 5.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 6.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginalski K. Comparative modeling for protein structure prediction. Curr. Opin. Struct. Biol. 2006;16:172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Dunbrack RL., Jr. Sequence comparison and protein structure prediction. Curr. Opin. Struct. Biol. 2006;16:374–384. doi: 10.1016/j.sbi.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 10.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 11.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 12.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupas A. Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 1997;7:388–393. doi: 10.1016/s0959-440x(97)80056-5. [DOI] [PubMed] [Google Scholar]
- 15.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction – the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazaki S, Kuroda Y, Yokoyama S. Characterization and prediction of linker sequences of multi-domain proteins by a neural network. J. Struct. Funct. Genomics. 2002;2:37–51. doi: 10.1023/a:1014418700858. [DOI] [PubMed] [Google Scholar]
- 18.Holmstrom K, Brandt T, Kallesoe T. The sequence of a 32,420 bp segment located on the right arm of chromosome II from Saccharomyces cerevisiae. Yeast. 1994;10 (Suppl. A):S47–S62. doi: 10.1002/yea.320100007. [DOI] [PubMed] [Google Scholar]