Abstract
Genome-wide association studies have become a popular strategy to find associations of genes to traits of interest. Despite the high-resolution available today to carry out genotyping studies, the success of its application in real studies has been limited by the testing strategy used. As an alternative to brute force solutions involving the use of very large cohorts, we propose the use of the Gene Set Analysis (GSA), a different analysis strategy based on testing the association of modules of functionally related genes. We show here how the Gene Set-based Analysis of Polymorphisms (GeSBAP), which is a simple implementation of the GSA strategy for the analysis of genome-wide association studies, provides a significant increase in the power testing for this type of studies. GeSBAP is freely available at http://bioinfo.cipf.es/gesbap/
INTRODUCTION
Genome-wide association studies (GWAS) use high-resolution maps of markers (Single Nucleotide Polymorphisms - SNPs - and, in some platforms, Copy Number Variations - CNVs - too) along the genome to look for allele frequency differences between cases (e.g. individuals with a certain disease or trait) and controls. A significant frequency difference is taken as an indication of the presence of functional variants in the DNA sequence related to the disease or trait in question within the genomic region corresponding to the markers (1). Despite the high resolution available to interrogate the genomes (e.g. more than 1.8 million markers, half of them SNPs, in the Affymetrix 6.0 chip), the current testing strategies that considers markers independently results in weak, usually non-significant, associations (2). Thus under this conventional testing paradigm, only consortia analysing very large cohorts were successful in finding clear and reproducible results (3).
However, a new paradigm inspired in systems biology has recently been adopted in other areas, such as transcriptomics (4). Since the concept aimed at is the functional association, it might occur that individual genes are not the best proxies for such concept. Actually, it is widely accepted that conventional biological functions, in the way we understand them, can rarely be attributed only to an individual molecule. Instead, most of the biological functionality of the cell arises from complex interactions between their molecular components that define operational interacting entities or modules of functionally related genes (5). In the case of genetic traits, it is generally believed that multigenicity reflects disruptions in proteins that participate in a protein complex or a in a pathway (6). Accordingly, Gene Set Analysis (GSA) methods aim to test the activity of such modules instead of testing the activity of individual genes (7,8). The extrapolation of this concept to other fields, and in particular to the study of complex traits by GWAS, where combinations of mutations in different genes can globally affect a pathway making it difficult to find detectable associations in individual genes but not in the pathway, has recently been suggested (9–11). Actually, the application of this emergent concept (also known as pathway-based analysis) to the analysis of GWAS is already producing new and original results for different pathologies (12–14). Although stand-alone software is available for this type of analysis (9,11,15), no web servers with better user-interface are available for experimentalists to use.
The Gene Set-based Analysis of Polymorphisms (GeSBAP) web server can input lists of SNP or CNV identifiers arranged according to a parameter accounting for the association (P-value) and returns a collection of biological processes significantly associated to the trait studied. Lists of genes belonging to such processes can easily be obtained for further validation.
RATIONALE FOR EXPANDING GENE-SET ANALYSIS CONCEPTS TO THE STUDY OF POLYMORPHISMS
The rationale behind the GeSBAP approach relies on the use of a testing strategy that target damaged functionalities which can produce the final phenotype. Most of the traits analysed in GWAS experiments are multigenic. Even in the cases that such traits depend on a unique main gene, there are other modifier genes that modulate the phenotype. The main problem with GWAS is that testing markers independently results in weak, usually non-significant, associations of them to the trait (2). Since multigenicity is generally caused by different combinations of mutations whose only common feature is their belonging to a pathway (or, generally speaking, to a functional unit), the GeSBAP approach proposed here, where the entity tested is the pathway, seems the natural way of discovering associations in GWAS.
DESCRIPTION OF THE PROGRAM
GeSBAP is a web-server written in Java with a core that essentially conducts the test written C++. The program is running in a high-end cluster with 10 dedicated Intel XEON Quad-Core CPUs at 2.0 GHz (summing up a total of 40 cores) with a large amount of RAM (total 60 GB).
The default mode of use is the traditional ‘anonymous user’. In this case, the results are maintained only for 1 day and then deleted. The program also allows registered users. An account can be created, which can be later used after the corresponding login. The results of the sessions are preserved in the account and can be recovered in future sessions after login.
Input
The program inputs a tab-delimited list of polymorphisms (SNP and/or CNV) along with the corresponding P-value of the association. It is also possible to enter more processed data, such a list of genes with the corresponding P-value of association. Any of the most common gene identifiers can be used given that GeSBAP uses internally the INFRARED engine for gene ID conversion used in the Babelomics package (16). Finally, data can also be input in the format of the popular PLINK program (17), which performs the association test and obtains the corresponding list of P-values.
At present only human, mouse and rat SNPs can be used in the program. The user can choose to test one or several functional categories among Gene Ontology (GO) (18), KEGG (19) and Biocarta (http://cgap.nci.nih.gov/Pathways/BioCarta_Pathways) pathways. Several filters can be applied to use subsets of any of these categories, which essentially involve filtering for maximum and minimum number of genes in the terms tested, filtering by keywords and, in the case of GO, filtering by levels in the GO hierarchy.
Testing strategy
Given a list of genes ranked by any criteria, GSA is used to find enrichments of gene sets significantly associated to high (or low) values of the rank. Here, the ranking criterion for the genes is derived from the associations of the SNPs to the trait studied. In particular, the program uses the –log(P-value) of the association test corresponding to the case–control comparison. The program selects the polymorphisms (SNPs or CNVs) that map into genes or in their neighbourhoods (±5 kb). Among all the polymorphisms corresponding to each gene, the one with the highest association to the trait studied is taken as a proxy of the gene. Then, all the genes represented in the GWAS (usually the complete genome) are mapped to the corresponding functional categories previously selected by the user and ranked accordingly to their proxy polymorphisms. Finally, a GSA test (10) is used to check for functional categories showing significant association to the trait studied. The significant functional terms along with the corresponding P-values [adjusted for multiple testing (20)] are listed in the output.
Output and an example
The output of the programs provides a general overview on the functional categories found as significantly associated to the trait studied. Figure 1 shows the representation of such summary. A table including the significant functional categories along with their corresponding P-values and the genes included in each one is provided. In addition, when GO terms are being analysed, a graphical representation of the significant terms within the GO hierarchy is also provided. The GO viewer implemented in the Babelomics package (16) is used for this purpose.
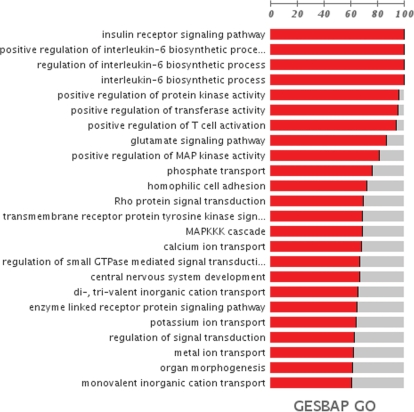
Figure 1.
Plot representing a summary of the GO terms significantly associated to the breast cancer in the case–control analysed (see text). The length of the red bar represents the relative proportion of genes of the GO term in the partition at which the enrichment was significant (16,21).
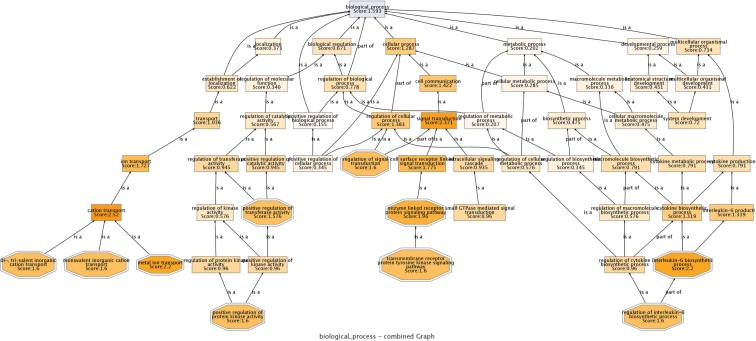
As an example of the application of the program, we show the analysis of a breast cancer case–control from the CGEMs initiative (22), in which a total of 528 173 SNPs were genotyped in 1145 post-menopausal women with invasive breast cancer and 1142 controls. The original study identified four SNPs in intron 2 of FGFR2 (which encodes a receptor tyrosine kinase) that were highly associated with breast cancer (22). The GeSBAP analysis of the resulting list of ∼18 000 genes ranked by –log(P-value) revealed a considerable number of biological processes associated with risk of sporadic postmenopausal breast cancer (23) where the conventional tests only detected a unique gene (22). Figure 1 represents the GO biological processes detected as significantly associated to the cancer, among them ‘transmembrane receptor protein tyrosine kinase signaling pathway’ (GO:0007169, False Discovery Rate (FDR)-adjusted P-value = 1.73 × 10−03) and ‘regulation of signal transduction’ (GO:0009966, FDR-adjusted P-value = 4.45 × 10−03) in which FGFR2 is included. Figure 2 represents a summary of these GO terms mapped within the GO hierarchy. Supplementary Figure 1 displays the complete relationships among all the GO terms found. Notably, some of these processes have been log-standing linked to human neoplasia although not at the germline genetic level, in any case as comprehensively as proposed here. The role of RAS signal transduction in tumorigenesis is well-established (24), but this study will expand its involvement into cancer susceptibility by suggesting that several genes annotated with the corresponding GO term play a key role in risk of breast cancer. This term is a children of Cell Communication and a parent of Rho signaling, which then provides a global view of the cellular alteration in breast cancer susceptibility and a more detailed link to which components of the RAS signaling may be altered, respectively. Notably, previous work have linked the Rho guanine nucleotide exchange factor AKAP13 (significant here as part of the ‘transmembrane receptor protein tyrosine kinase signaling pathway’, GO:0007169, FDR-adjusted P-value = 1.73 × 10−03) with familial breast cancer (25) and two of the most recent world-wide replicated findings include FGFR2 (significant here as previously mentioned as part of the GO terms: ‘transmembrane receptor protein tyrosine kinase signaling pathway’, GO:0007169, FDR-adjusted P-value = 1.73 × 10−03; and ‘regulation of signal transduction’, GO:0009966, FDR-adjusted P-value = 4.45 × 10−03) and MAP3K1 (significant here as part of the GO terms: ‘transmembrane receptor protein tyrosine kinase signaling pathway’, GO:0007169, FDR-adjusted P-value = 1.73 × 10−03; and ‘Rho protein signal transduction’, GO:0007266, FDR-adjusted P-value = 2.26 × 10−02).
Figure 2.
Plot of the relationships in the GO hierarchy of a summary of the main GO terms found as significantly associated to the breast cancer in the case–control experiment analysed (see text). Octagons represent GO terms found as significant. Rectangles represent other GO terms in the hierarchy depicting the functional relationships among the significant terms. Supplementary Figure 1 displays the complete relationships among all the terms found.
DISCUSSION
We have shown how the concept of GSA can easily be extrapolated to the field of polymorphism analysis. An example shows how the application of this test to a breast cancer case–control reveals a considerable number of biological processes associated with risk of sporadic postmenopausal breast cancer (23), where the conventional tests only detected a unique gene association (22).
At present, the main limitations for targeting functionality is our limited catalogue of functions, represented mainly by functional annotations contained in repositories such as GO (18), KEGG (19), Biocarta pathways, etc. This fact not only limits the number of testable effects to the content of these repositories, but also conceptually restricts to genes associations that can be found using the available information. It is expectable that projects like ENCODE (26) or the 1000 genomes (http://www.1000genomes.org/) will help to define extra-genic regions with functional significance. Nevertheless, this is not a limitation of the proposed methodology, but of the information available. In any case, the philosophy of testing groups of markers corresponding to functional units instead of markers alone has proven to be superior to the conventional testing schema and will increase its scope as new functional definitions become available in the future.
Despite there are other general purpose programs and web servers for carrying out different flavours of GSA, revised in (7,8,27) (see also http://bioinfo.cipf.es/docus/tools-citations/functional_profiling/), there is no similar application available to GeSBAP, orientated to the analysis of GWAS, that calculates association P-values and maps SNPs or CNVs to genes previously to carry out the GSA.
The program for the GeSBAP has been running for >6 months. The first paper using this approach has recently been published (23). To our knowledge, there are no other web applications offering this type of analysis.
GeSBAP is freely available at http://bioinfo.cipf.es/gesbap/www/index.jsp.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The CIBER de Enfermedades Raras is an initiative of the ISCIII; The RTICC is an initiative of the ISCIII; The National Institute of Bioinformatics (www.inab.org) is a platform of Genoma España.
FUNDING
Spanish Ministry of Science and Innovation (grant BIO2008-04212 to J.D.). Funding for open access charge: BIO2008-04212.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kruglyak L. The road to genome-wide association studies. Nat. Rev. Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dopazo J. Functional interpretation of microarray experiments. Omics. 2006;10:398–410. doi: 10.1089/omi.2006.10.398. [DOI] [PubMed] [Google Scholar]
- 5.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 6.Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat. Rev. Genet. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 7.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dopazo J. Formulating and testing hypotheses in functional genomics. Artif. Intell. Med. 2009;45:97–107. doi: 10.1016/j.artmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Minguez P, Montaner D, Dopazo J. From genes to functional classes in the study of biological systems. BMC Bioinformatics. 2007;8:114. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Zhang L, Zhao Y, Xu L, Shang Y, Wang Q, Li W, Wang H, Li X. Prioritizing risk pathways: a novel association approach to searching for disease pathways fusing SNPs and pathways. Bioinformatics. 2009;25:237–242. doi: 10.1093/bioinformatics/btn613. [DOI] [PubMed] [Google Scholar]
- 12.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Askland K, Read C, Moore J. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum. Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 14.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24:2784–2785. doi: 10.1093/bioinformatics/btn516. [DOI] [PubMed] [Google Scholar]
- 16.Al-Shahrour F, Carbonell J, Minguez P, Goetz S, Conesa A, Tarraga J, Medina I, Alloza E, Montaner D, Dopazo J. Babelomics: advanced functional profiling of transcriptomics, proteomics and genomics experiments. Nucleic Acids Res. 2008;36:W341–W346. doi: 10.1093/nar/gkn318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Yekutieli D. The control of false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 21.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. Discovering molecular functions significantly related to phenotypes by combining gene expression data and biological information. Bioinformatics. 2005;21:2988–2993. doi: 10.1093/bioinformatics/bti457. [DOI] [PubMed] [Google Scholar]
- 22.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonifaci N, Berenguer A, Diez J, Reina O, Medina I, Dopazo J, Moreno V, Pujana MA. Biological processes, properties and molecular wiring diagrams of candidate low-penetrance breast cancer susceptibility genes. BMC Med. Genomics. 2008;1:62. doi: 10.1186/1755-8794-1-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 25.Wirtenberger M, Tchatchou S, Hemminki K, Klaes R, Schmutzler RK, Bermejo JL, Chen B, Wappenschmidt B, Meindl A, Bartram CR, et al. Association of genetic variants in the Rho guanine nucleotide exchange factor AKAP13 with familial breast cancer. Carcinogenesis. 2006;27:593–598. doi: 10.1093/carcin/bgi245. [DOI] [PubMed] [Google Scholar]
- 26.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.