Abstract
Understanding the structure and the dynamics of the complex intercellular network of interactions that contributes to the structure and function of a living cell is one of the main challenges of today's biology. SNOW inputs a collection of protein (or gene) identifiers and, by using the interactome as scaffold, draws the connections among them, calculates several relevant network parameters and, as a novelty among the rest of tools of its class, it estimates their statistical significance. The parameters calculated for each node are: connectivity, betweenness and clustering coefficient. It also calculates the number of components, number of bicomponents and articulation points. An interactive network viewer is also available to explore the resulting network. SNOW is available at http://snow.bioinfo.cipf.es.
INTRODUCTION
It is widely accepted that most of the biological functionality of the cell arises from complex interactions between their molecular components that define operational interacting entities (1). Understanding the structure and the dynamics of the complex intercellular network of interactions that contribute to the structure and function of a living cell is one of the main challenges of today's biology (2) and constitutes the objective of systems biology (3). Alterations in the network of protein interactions are of special relevance in many diseases. For example, it has recently been demonstrated that tumorigenesis takes place in a specific and organized way that encompasses the precise down-regulation of groups of topologically-associated proteins (4). For the last years, there has been an enormous interest in the exploration of the interactome of model organisms such as Saccharomyces cerevisiae (5–7), Drosophila melanogaster (8,9), Caenorhabditis elegans (10) or human (11,12), just to cite a few examples. Thus, for several model organisms, a reasonable and operative description of the interactome is available. Several tools exist for network representation and calculation of network parameters, such as the popular Cytoscape (13) and its plugins (14) and other programs (15), but most of them are stand alone applications [see a recent review in (16)]. Also, several web tools for network analysis and visualization have been reported (see Supplementary Table 1) with functionalities that rank from mere network viewers to more sophisticated programs that perform different network parameter calculations (17–22). Tools of this type are enormously useful for the interpretation of genomic experiments. For example, if a number of genes has been found to be activated in a microarray experiment, their analysis in the context of the interactome can give clues on their possible role as a protein complex, as a signalling pathway, etc. However, the conclusions extracted from the simple visualization of a network of the calculation of some of its parameters are subjective without the proper statistical support. This feature, the proper statistical analysis of networks, still remains to be addressed by a web tool. Here we introduce Studying Networks in the Omics World (SNOW), a unique tool specifically designed to offer visualization and analysis of protein–protein interacting (PPI) networks, including their statistical analysis.
DESCRIPTION OF THE TOOL
Functionality of SNOW
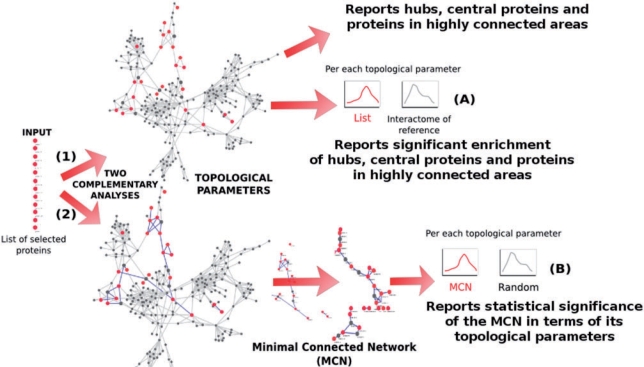
Essentially, SNOW takes a list of proteins (or genes) and maps them onto an interactome of reference. This interactome can be the human interactome (in two versions, see databases subsection) or any other user-defined interactome. Once the list is mapped, SNOW calculates several relevant network parameters for the proteins in the contexts of the interactome and the minimum connected network (MCN) defined by the proteins. The corresponding tests are performed to assess the significance of the parameters calculated (see below). Alternatively two lists of proteins can be compared by testing for significant deviations in their respective parameters’ distributions. Figure 1 shows a schema of the analysis steps implemented in SNOW.
Figure 1.
Schema of the analysis steps implemented in SNOW. Two complementary analyses are carried out: (1) the distribution of parameters (connection degree, betweenness centrality, clustering coefficient and number of components) of the list of proteins are tested against the rest of proteins of the interactome of reference within the context of connections of the complete interactome of reference and (2) The list of proteins is mapped onto the reference interactome and the MCN is found. The distributions of networks parameters in this MCN are tested against their random expectation (see ‘Testing strategy for the network parameters’ section in the Supplementary Data). See Supplementary Figure 2 for a more detailed schema.
Input
SNOW inputs a collection of protein (or gene) identifiers in plain text. Most of the standard protein and gene identifiers are accepted given that the tool uses the database of identifiers of Babelomics (23).
Alternatively, a user-defined interactome can be provided to SNOW. It can be uploaded either as a list of protein–protein (or gene–gene) interactions tab-delimited in plain text or in the popular SIF format, used by Cytoscape (see http://www.cytoscape.org/cgi-bin/moin.cgi/Cytoscape_User_Manual/Network_Formats).
Calculation of network parameters and statistics
In particular, SNOW tests four topological parameters: (i) Node connections degree, which was computed as the number of edges (interaction events) for a node; (ii) Betweenness, which depends on the number of shortest pathways passing through a given node; (iii) Clustering coefficient, which measures the connectivity of the neighbourhood and (iv) Number of components of the network (see Additional methods for a detailed description on the calculation of these parameters). SNOW also finds the number of bicomponents and the articulation points and gives detailed descriptions of them. The parameters calculated account for different network properties. For example, signalling networks tend to have high connectivity and low clustering coefficient while metabolic networks have higher clustering coefficients.
The three first parameters (degree, betweenness and clustering coefficient) are calculated for all the nodes in the network. Thus, a distribution of values for any of these parameters can be derived for each particular network, which can give information on the network properties. Consequently, the comparison of two networks by contrasting how different they are in terms of their characteristic parameters is straightforward by means of a Kolmogorov–Smirnov test. The potential biological relevance of a particular network can therefore be obtained by comparing the network parameter distributions to the corresponding empirical distributions derived from ten thousand networks with the same number of nodes and a random protein composition (see ‘Testing strategy for the network parameters’ section in the Supplementary Data). By default the network is compared to the interactome of reference and to a network of random composition. However, the direct comparison of two networks is also possible.
In addition, the same analysis can be performed by adding one (or two or even three) intermediate nodes (proteins originally not included in the list that can actually link two or more proteins of the list), which is useful in proteomics analysis where often not all of the proteins can be identified in an experiment.
Output
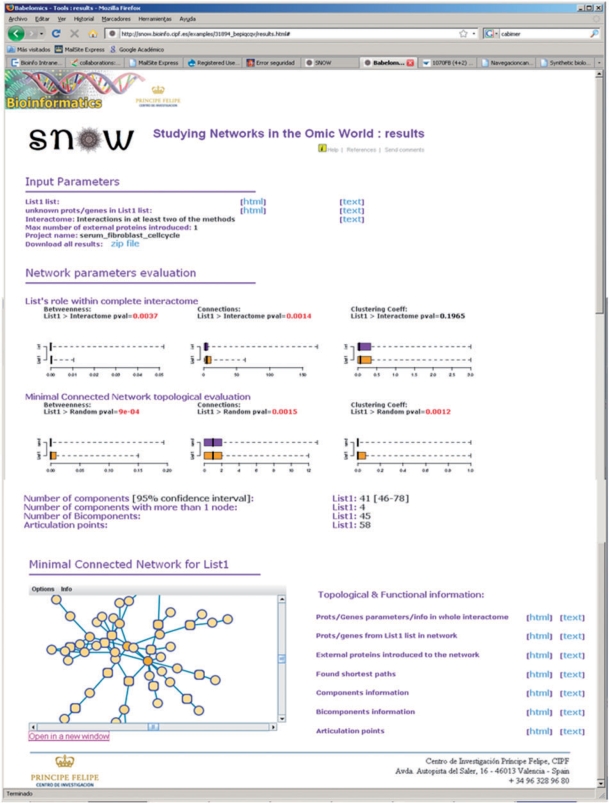
The program outputs the average parameter values, their significance and boxplots representing the comparison of their actual distributions in the network studied to the complete interactome (top boxplots Figure 2) and to a random network of the same size (bottom boxplots, Figure 2). SNOW outputs the number of components, bicomponents and articulation points. The program also provides exhaustive information on these parameters as well as functional information on the proteins in the list. Moreover, information about the shortest paths and articulation points is also available. A low resolution viewer of the network is present in the main results web page (Figure 2). This viewer can be opened in a new window (see below). SNOW includes 12 examples in which different sub-networks along with their corresponding parameters can be visualized (see Tutorial link in the main menu of the SNOW program). A particularly interesting example can be found in the additional case study information. SNOW was used to analyze 168 genes induced by the over-expression of BRCA1, one of the most studied cancer-related genes. The application of conventional functional enrichment analysis (24) failed to detect any functionality significantly over-represented among the genes. Nevertheless, SNOW detected a significant concentration of highly connected (P < 0.0001) and central (P = 0.0017) proteins in the resulting MCN (see Additional Case Study for details).
Figure 2.
Output of SNOW—the web page contains a summary of the input data and the network parameters estimated along with the statistical significance for the connectivity, betweenness and clustering coefficient. Top boxplots represent the comparison of the network to the complete interactome and bottom boxplots account for the comparison of the network to a random network of the same size (see text). Component, bicomponents and articulation points are also provided. In addition, detailed information of genes in the list, genes included in the analysis, shortest pathways is also accessible from the web page. Finally an interactive network viewer can be launched from the page.
Visualization
An interactive viewer for the connections defining the network studied can also be opened from the results page. All the components of the network are displayed in different layouts. The network orientation can be interactively changed. Gene names can be displayed and connecting proteins (intermediate nodes) can be included or excluded from the graph. Supplementary Figure 1 shows two possible layouts of the same network obtained from cell-cycle dependent genes regulated following exposure to serum in a variety of human fibroblast cell lines (data available from the third example included in the Tutorial and examples link of the main page of the SNOW program, see section above). The topmost view shows the menu with the options and the functional (gene ontology) and topological information on any protein which is displayed when placing the mouse over it.
Databases
Human PPI datasets were downloaded from five main public databases: HPRD (25), IntAct (26), BIND (27), DIP (28) and MINT (29). We used this collection of PPI data to generate two scaffold interactomes: a non-filtered scaffold interactome, which includes all the available PPIs; and a filtered, more confident scaffold interactome. The six top categories of experimental methods described in the Molecular Interaction (MI) Ontology (30) plus the categories in vivo and in vitro from HPRD can be used as confidence indicators. Thus, only PPIs verified by at least two of these categories were considered in the filtered scaffold interactome. Protein identifiers can be directly mapped to Ensembl transcript identifiers that can easily be linked to many other gene or protein identifiers. Since in many genomic experiments only data on genes (but not on particular transcripts) is available, we build up another two high and low confidence interactomes by mapping transcripts onto genes. Conceptually, each gene can potentially interact with all the interaction partners of all of its transcripts. Transcript- and gene-based interactomes are similar but not identical.
Implementation details
The SNOW web interface was implemented using Perl and JavaScript languages. The queries to the server are implemented as web services written in Perl and Pyhton. Boost Graph libraries (http://www.boost.org/libs/graph/doc/index.html) were used to perform graph parameters calculation. SNOW accesses these libraries through BGL-Python bindings (http://www.osl.iu.edu/~dgregor/bgl-python/). SNOW uses R programming language to perform the statistics and mysql to store interactome network parameters. The visualization applet was implemented in Java using TouchGraph libraries (http://touchgraph.sourceforge.net).
Other tools
There are several popular stand-alone tools for the visualization and analysis of networks, such as the popular Cytoscape (13,14) and other analogous tools (15,16). Also, in the last years, several web-based applications for this purpose have been reported in the literature (17–19,31). Some tools provide annotations (32), offer functional enrichment analysis (32,33) or are more focused to pathways (19,34,35) or provides some facilities for network comparisons (20). Several databases offer their own network viewers as is the case of STRING (36) or IntAct (26). Other tools allow for the calculation of some network parameters (20–22,37), although in some cases at the expense of losing interactivity in the visualization of the network (22). Supplementary Table 1 contains a list of web tools for network management with some of their most significant features.
DISCUSSION
SNOW is unique among its genre in the sense that it not only allows visualizing data in an interactive dynamic format while calculating relevant network parameters but it also finds their statistical significance. It can also be used to compare two networks in terms of their corresponding distributions of parameters. To our knowledge there are no other web-based tools with such features.
SNOW constitutes a step forward in our efforts to provide the scientific community with web tools, such as the Babelomics suite (38), for the functional profiling of genomic experiments. SNOW broadens the conventional functional profiling based on gene modules (e.g. gene ontology or KEGG pathway gene sets) to gene sets based on PPIs. SNOW has been operative since last summer and it has been used by projects associated to the Spanish National Bioinformatics Institute (http://www.inab.org), the Spanish Network of Cancer (RTICC; http://www.rticcc.org) and the Network of Centres for Research in Rare Diseases (CIBERER, http://www.ciberer.es). SNOW is running in a high-end cluster with 10 dedicated Intel XEON Quad-Core CPUs at 2.0 GHz (summing up a total of 40 cores) with a large amount of RAM (total 60 GB). The program is available at http://snow.bioinfo.cipf.es.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science and Innovation (BIO2008-04212 to JD). The CIBER de Enfermedades Raras is an initiative of the ISCIII. The RTICC is an initiative of the ISCIII. The National Institute of Bioinformatics (www.inab.org) is a platform of Genoma España. Funding for open access charge: Spanish Ministry of Science and Innovation grant BIO2008-04212.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 2.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 3.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez P, Huerta-Cepas J, Montaner D, Al-Shahrour F, Valls J, Gomez L, Capella G, Dopazo J, Pujana MA. Evidence for systems-level molecular mechanisms of tumorigenesis. BMC Genomics. 2007;8:185. doi: 10.1186/1471-2164-8-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 6.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 13.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 15.Junker BH, Koschutzki D, Schreiber F. Exploration of biological network centralities with CentiBiN. BMC Bioinformatics. 2006;7:219. doi: 10.1186/1471-2105-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suderman M, Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23:2651–2659. doi: 10.1093/bioinformatics/btm401. [DOI] [PubMed] [Google Scholar]
- 17.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2002;3 PREPRINT0012. [PubMed] [Google Scholar]
- 19.Dogrusoz U, Erson EZ, Giral E, Demir E, Babur O, Cetintas A, Colak R. PATIKAweb: a Web interface for analyzing biological pathways through advanced querying and visualization. Bioinformatics. 2006;22:374–375. doi: 10.1093/bioinformatics/bti776. [DOI] [PubMed] [Google Scholar]
- 20.Brohee S, Faust K, Lima-Mendez G, Sand O, Janky R, Vanderstocken G, Deville Y, van Helden J. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 2008;36:W444–W451. doi: 10.1093/nar/gkn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S. Integrated network analysis platform for protein–protein interactions. Nat. Methods. 2009;6:75–77. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- 22.Yip KY, Yu H, Kim PM, Schultz M, Gerstein M. The tYNA platform for comparative interactomics: a web tool for managing, comparing and mining multiple networks. Bioinformatics. 2006;22:2968–2970. doi: 10.1093/bioinformatics/btl488. [DOI] [PubMed] [Google Scholar]
- 23.Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 25.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 31.Iragne F, Nikolski M, Mathieu B, Auber D, Sherman D. ProViz: protein interaction visualization and exploration. Bioinformatics. 2005;21:272–274. doi: 10.1093/bioinformatics/bth494. [DOI] [PubMed] [Google Scholar]
- 32.Reimand J, Tooming L, Peterson H, Adler P, Vilo J. GraphWeb: mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res. 2008;36:W452–W459. doi: 10.1093/nar/gkn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Z, Mellor J, Wu J, Yamada T, Holloway D, Delisi C. VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acids Res. 2005;33:W352–W357. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z, Ng DM, Yamada T, Chen C, Kawashima S, Mellor J, Linghu B, Kanehisa M, Stuart JM, DeLisi C. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 2007;35:W625–W632. doi: 10.1093/nar/gkm295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Snitkin ES, DeLisi C. VisANT: an integrative framework for networks in systems biology. Brief Bioinform. 2008;9:317–325. doi: 10.1093/bib/bbn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prieto C, De Las Rivas J. APID: Agile protein interaction data analyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.