Abstract
OmicBrowse is a genome browser designed as a scalable system for maintaining numerous genome annotation datasets. It is an open source tool capable of regulating multiple user data access to each dataset to allow multiple users to have their own integrative view of both their unpublished and published datasets, so that the maintenance costs related to supplying each collaborator exclusively with their own private data are significantly reduced. OmicBrowse supports DAS1 imports and exports of annotations to Internet site servers worldwide. We also provide a data-download named OmicDownload server that interactively selects datasets and filters the data on the selected datasets. Our OmicBrowse server has been freely available at http://omicspace.riken.jp/ since its launch in 2003. The OmicBrowse source code is downloadable from http://sourceforge.net/projects/omicbrowse/.
INTRODUCTION
Omics research involves a comprehensive analysis of phenomes and various biomolecules including genomes, transcriptomes and proteomes. Experimental data are often analysed with reference to other annotation databases. A typical biological data record contains many kinds of attributes such as annotations, gene names, genomic positions, ontology terms, interactions, phenotypes and expression levels. In this article, we discuss our public OmicBrowse server hosted at http://omicspace.riken.jp/ with public data published by RIKEN and others, and details of the functionalities of OmicBrowse.
Furthermore OmicBrowse is a genome browser that integrates multiple heterogeneous databases into a single omic space (1). For this purpose, OmicBrowse can also be installed on a user's PC to deploy users' private and confidential datasets. An OmicBrowse server installed in a user's PC works as a user's private databases and achieves integration of both private and public datasets provided by different OmicBrowse servers. OmicBrowse allows a manager of a server to regulate accesses for each user to each dataset, and connections among multiple OmicBrowse servers securely integrate confidential datasets in multiple OmicBrowse servers. Mechanisms that manage private datasets on the user's side achieve securely robust data integration that prevents loss and leakage of data, compared with other data integration services that simply allow the user's datasets to be uploaded. In order to realize operation and data interoperability between OmicBrowse and other bioinformatics tools, OmicBrowse provides a standardized interface to exchange annotation data. We also provide a OmicDownload server that allows the user to download OmicBrowse datasets using an interactive graphical user interface.
DATA RESOURCE
In OmicBrowse, a database is a set of annotation data associated with each version of a genome. In order to host our OmicBrowse server, we have generated a list of databases for each genome version. The databases include not only RIKEN's public databases such as FANTOM (2), RARGE (3) and tiling array data (4), but also the public databases published by other institutes such as NCBI (5), Ensembl (6), MGI (7), HGNC (8), TAIR (9), WormBase (10), RAP-DB (11) and RGD (12). The original dataset of each database has been converted into our original format including chromosomal interval. Table 1 shows the numbers of integrated databases computed for each species, genome version and omics data types. As of December 2008, 344 databases are integrated into our OmicBrowse servers for six species: Arabidopsis thaliana, Oryza sativa, Homo sapiens, Mus musculus, Rattus norvegicus and Caenorhabditis elegans. When a new version of a genome sequence is released by an original data site, a new set of databases related to a new genome sequence is generated and integrated into OmicBrowse. Furthermore, the databases are updated every 3 months.
Table 1.
The numbers of databases computed for each version of the genome
| Species | Genome version | Omics types |
Other types |
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Transcriptome | Proteome | Phenome | SNP | Marker | Ontology | Homology | Others | |||
| At | tair6 | 2 | 24 | 1 | 1 | 2 | 4 | 34 | |||
| tair7 | 2 | 1 | 1 | 1 | 2 | 6 | 13 | ||||
| tair8 | 2 | 8 | 1 | 1 | 12 | ||||||
| MIPSv200307 | 2 | 10 | 1 | 2 | 15 | ||||||
| Os | IRGSP build3 | 1 | 1 | 2 | |||||||
| IRGSP build4 | 1 | 1 | 2 | ||||||||
| Hs | NCBI35 | 4 | 2 | 5 | 1 | 1 | 1 | 7 | 5 | 26 | |
| NCBI36 | 5 | 6 | 4 | 1 | 2 | 1 | 38 | 2 | 59 | ||
| Mm | NCBIm34 | 2 | 4 | 5 | 2 | 1 | 1 | 8 | 1 | 24 | |
| NCBIm36 | 6 | 7 | 4 | 2 | 1 | 3 | 1 | 34 | 1 | 59 | |
| NCBIm37 | 6 | 2 | 4 | 3 | 15 | ||||||
| Rn | RGSC3.4 | 4 | 2 | 4 | 2 | 3 | 1 | 16 | |||
| Ce | WS116 | 1 | 6 | 1 | 1 | 9 | |||||
| WS150 | 4 | 2 | 4 | 1 | 35 | 2 | 48 | ||||
| WS190 | 4 | 2 | 4 | 10 | |||||||
| Total | 46 | 72 | 43 | 7 | 4 | 16 | 11 | 123 | 22 | 344 | |
As of December 2008, on our public OmicBrowse server the 344 databases of six species including A. thaliana (At), O. sativa (Os), H. sapiens (Hs), M. musculus (Mm), R. norvegicus (Rn) and C. elegans (Ce) are integrated. The set of databases includes not only databases of omics types namely genomes, transcriptomes, proteomes and phenomes, but also non-omic databases such as SNPs, markers, ontologies, homologies including syntenies, and others including literal and metabolic pathways. For detailed information about integrated databases, refer our OmicBrowse server URL http://omicspace.riken.jp/
IMPLEMENTATION
OmicBrowse is a web-oriented tool designed with a client-server model. The server-side program of OmicBrowse is implemented as a Java servlet, which retrieves a query against a set of databases related to a version of the genome stored in MySQL. The client and server securely communicates by exchanging XML documents, including a data request query and its response. On the client side, in order to present multiple databases located in an arbitrary range of chromosomal intervals in a graphical way, we have implemented a dataset viewer as an Adobe Flash component. Thanks to a capability of Flash, we have realized an OmicBrowse client including a dataset viewer with the following advantages. Since Flash can manipulate vector graphics controlled via a script language that does not depend on computational platforms, the OmicBrowse client realizes a high-speed graphical view of data records without image degradation against magnification of the image in the various computational environments on the client side. The OmicBrowse client can be launched in most conventional web browsers and as a stand-alone local program. These functionalities cannot be realized by employing other technologies, such as AJAX that have a high dependency on web browsers and operate at a slower speed.
Integration of personal databases in a user's PC and of public databases among multiple OmicBrowse servers
As a communication function among multiple OmicBrowse servers, OmicBrowse provides a remote search function that can be integrated with a search result from the local server. More specifically, an OmicBrowse server imports a catalogue of datasets published by a remote server and performs a data search against both the local and the imported remote datasets simultaneously, displaying all the results to the client. Because OmicBrowse allows the user to set a remote access policy for each database and data exchange using a secure protocol, the user can set up a private server on a user's PC, including his/her confidential datasets and browse them with public databases by other servers. The user can publish the datasets as a database by just changing their access control policy. For collaborative research among different laboratories, secure data integration and exchange via a peer-to-peer connection or a network of these groups are achieved. This functionality can be set up in the installation phase of OmicBrowse server described in http://omicspace.riken.jp/omicBrowse/OmicBrowseRegister.html.
Presentational function
The Flash component provides a high-performance graphics interface that allows a user to browse the integrated databases as shown in Figure 1. Each database is displayed as a horizontal line, and each data item of forward and reverse strands included in the database is displayed over and under the line, respectively. A set of databases selected by user is displayed as a vertically integrated set of lines. In another view, OmicBrowse is able to show a whole genome allowing the display of other data from a simple collection of all chromosomes. This integrated view realizes the browsing of various omics datasets at once with an exon–intron structure as shown in Figure 1F. Thanks to the advanced graphic functions of Flash, a smooth magnification of chromosomal interval from 20 bp to chromosome wide and a high-resolution printing of the view are realized. When a large number of items in a database are included in the view area of the chromosomal interval, OmicBrowse automatically generates histograms and displays them with numbers of items for each histogram bar.
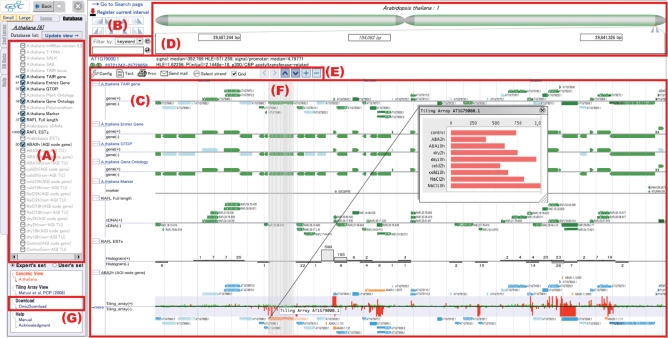
Figure 1.
An example data view of OmicBrowse client. In this example, the set of data including seven databases is the genomic interval of length 100 000 bp located at chromosome 1 from 29 687 244 bp to 29 841 326 bp of the genome sequence tair 6 in Arabidopsis. First, (A) is a list of databases mapped to the genome sequence tair 6 of Arabidopsis. It allows users to select databases that they would like to view in the annotation viewer (C). By specifying a keyword or an ontology term in (B), only the annotation data having the keyword or the ontology term is displayed in (C), respectively. The annotation viewer (C) provides an integrated view of annotation data stored in the selected databases in (A) and located in the interval specified in the interval selector (D). The interval selector (D) allows a user to select an interval on the currently displayed chromosome. The length of interval specified in (D) is from 20 bp to whole chromosome. The control bar (E) provides icons respecting functions that control the annotation viewer (C), such as icons for launching the sequence viewer in text format, printing the current view, sending a URL indicating the current view via an e-mail to other users. The sequence viewer allows the user to download the data currently displayed in (C). The control bar also includes panel scrolling buttons indicated by arrowheads <,>, ∨ and ∧, and panel magnification buttons indicated by + and −. When the mouse cursor is located over the data item having an exon–intron structure, vertical lines that indicate the structure are displayed as (F) in order to compare other structured data located in the same genomic interval. On the annotation data display in the annotation viewer (C), a histogram is automatically generated when too many data are located in the interval to display, such as the RAFL ESTs database displayed in the second last line of this example. By clicking a histogram bar, the interval is set from the start address to the end address of that histogram bar. OmicBrowse supports numerical values mapped to the chromosomal axis. The last line of this example shows a dataset of tiling array in Arabidopsis and a result of the ARabidopsis Tiling Array-based Detection of Exons (ARTADE) (4) program which statistically predicts an exon–intron structure utilizing the dataset of the tiling array. The tiling array data are displayed as a bar chart in green and red that indicate whether the data are greater or lower than a threshold. The reference gene and predicted genes with the exon–intron structure are shown just outside of the bar chart. By selecting a gene, the stress intensity data of the tiling array of the gene is displayed in a pop-up window. A link to launch OmicDownload is displayed in the user's menu (G).
As shown in Figure 1, the user can select databases to be displayed, and specify a filter to select the data records included in the selected databases. OmicBrowse provides a keyword filter that selects data records with a keyword, and an ontology filter that selects data records associated with an ontology term.
OmicBrowse provides a URL that links to a view and sends it to the user's mail program. It allows the view displayed in the OmicBrowse client to be shared with other researchers by sending them the URL and bookmarking it as the user's personal record.
We provide another client named OmicDownload server, available at http://omicspace.riken.jp/gps/download, which implements an interactive graphical interface for data downloads shown in Figure 2. Since OmicDownload implements interactive procedures based on the data integration model of OmicBrowse, it allows a user to download annotation data selected by a keyword search and a chromosomal interval check as well as other data stored in various omic databases located in the chromosomal interval.
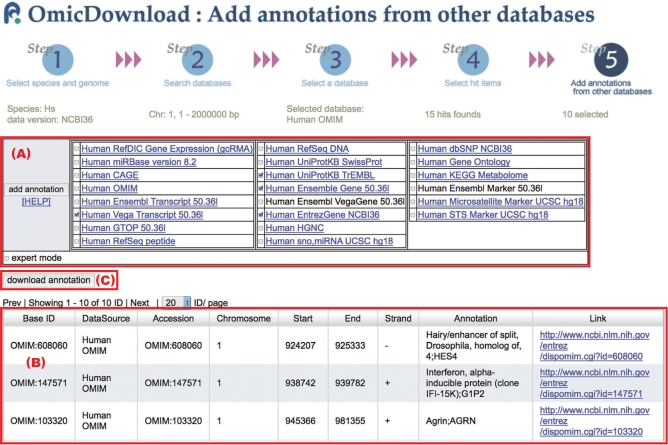
Figure 2.
A screen grab of the data download wizard accessing our OmicDownload server. OmicDownload provides an interactive interface for the users to navigate through a data download procedure consisting the following five steps: (Step 1) selection of species and genome version, (Step 2) search of databases with the user's keyword and chromosomal interval related to the species and the genome version selected in Step 1, (Step 3) selection of a database from the set of databases selected in Step 2, (Step 4) selection of items that satisfy user's keyword and chromosomal interval specified in Step 2 in the database selected in Step 3 and (Step 5) selection of other databases that satisfy Step 1 for each item selected in Step 4, in order to download other items located in the chromosomal interval of the selected item. This figure is a screen grab of Step 5 reached by specifying the following parameters: (Step 1) H. sapiens (Hs) and NCBI36 were selected for species and genome version, respectively, (Step 2) chromosome 1, from 1 Mbp to 2 Mbp was specified as the chromosomal interval, (Step 3) Human OMIM was selected as the database, and 15 records of Human OMIM were found, (Step 4) 10 records were selected by the user from the 15 records obtained in Step 3. In Step 5, four databases related to species Hs and genome version NCBI36 are shown in the database selection table (A). In the annotation table (B), the annotation of the focused Human OMIM record OMIM:608060 is displayed on the top, and other records stored in the selected databases in (A) and located in the chromosomal interval of OMIM:608060 (Chromosome 1, from 924 207 bp to 925 333 bp) are followed. For the other nine records selected in Step 4, related annotation records are displayed as well as for OMIM:608060. Finally, by clicking the download annotation button (C), all annotation data listed in (B) for the 10 records selected in Step 4 are downloaded in a tab separated values (TSV) format.
For other data presentation functions, refer to the OmicBrowse User Manual published at http://omicspace.riken.jp/tutorial/HowToUseOmicBrowse_Eng.pdf.
INTERFACES AMONG EXTERNAL SERVERS
An OmicBrowse server is deployed not only as a stand-alone server and a communication server among multiple OmicBrowse servers, but also a communication server among external servers.
Supporting standardized interface
OmicBrowse also supports an interface that realizes interoperability among existing systems. We have implemented Distributed Annotation System (DAS) 1.53E (13), which is a protocol for exchanging annotations on genomic or protein sequences, and enables the user to integrate data from multiple data sources. In the implementation, we have mapped a DAS data source to a web-based OmicBrowse database, so that every database can be accessed via DAS at http://omicspace.riken.jp/gps/das/dsn. As a result, OmicBrowse allows a user access through a DAS compliant program or via an arbitrary DAS client viewer such as Ensembl (6) and GBrowse (14) as well as OmicBrowse viewer and OmicDownload.
Solving an in silico positional cloning problem with PosMed
Since OmicBrowse enables us to select candidate genes from numerous genes existing within the wide genetic loci by filtering only those annotated with the user's keyword, it can be applied to positional cloning problem solving. However, OmicBrowse only detects annotations with the user's keyword in the filtering function. In order to solve this restriction, we have also implemented a web-based database system that possesses inference-type full-text search functions named Positional Medline (15,16), or PosMed for short.
At first, PosMed performs a full-text search of the document database such as MEDLINE abstracts using the user's arbitrary keywords. Second, PosMed finds significant gene names or symbols that exist within the returned documents and generates a ranked list of these genes by applying statistical tests. This mechanism allows genes with a keyword within their annotations as well as with an indirect relevance to the keyword to be found. The user can call PosMed from OmicBrowse with a chromosomal interval specified in OmicBrowse, and the user can search genes located in the chromosomal interval by specifying a keyword. For each resultant gene, PosMed can call OmicBrowse to view data records of databases integrated in OmicBrowse located around the position of the gene.
Our PosMed server is available at http://omicspace.riken.jp/PosMed/.
APPLICATIONS
Our OmicBrowse public server in RIKEN has been used to analyse tiling array data in Arabidopsis with the ARTADE program for genome-wide suppression of aberrant mRNA-like non-coding RNAs (17), identification of the candidate genes regulated by RNA-directed DNA methylation (18) and transcriptome analysis in stress conditions (19). The OmicBrowse server has also been used to specify the genes responsible for ENU-mutant mice produced in a large-scale mutagenesis program in RIKEN (20) with PosMed, and over 65 responsible genes have been found so far.
COMPARISON WITH THE PREVIOUS VERSION OF OmicBrowse AND OTHER SIMILAR SYSTEMS
The advances over the previous version of OmicBorwse (21) are the following:
Introduction of datasets for O. sativa and R. norvegicus.
Deployment of DAS server service.
Development of OmicDownload.
Support for the latest version of Flash (version 10).
The advantages of the current version of OmicBrowse compared with other systems such as UCSC Genome Browser Database (22), Ensembl, Anno-J (http://www.annoj.org/) and GBrowse are as follows:
OmicBrowse implements a securely robust data management mechanism by setting an access control policy for each dataset among multiple users for each dataset.
OmicBrowse allows data to be viewed over a wider range of genomic intervals than other systems.
Keyword filtering is implemented in OmicBrowse, but the other systems listed above do not support this.
One of the limitations of OmicBrowse including OmicDownload is in its data download functionality. In Ensemble, BioMart (23) is available and supports data searching by specifying detailed conditions specific to each dataset. However OmicDownload supports only a common condition applicable to various omic datasets.
CONCLUSIONS
We have discussed a genome browser named OmicBrowse, which provides an integrated view of omics datasets based on genomic coordinate axes. OmicBrowse employs a high-performance graphics interface implemented in Adobe Flash that assists effective genome-wide analysis with various data records stored in a multiple databases.
OmicBrowse also provides polished functionalities to support practical biological research activities such as secure integration of in-house private datasets and public datasets, providing a DAS access point as a standardized data access interface, and providing an interactive data download interface named OmicDownload.
FUNDING
Special Coordination Funds of the Japanese Ministry of Education, Culture, Sports, Science and Technology. Funding for open access charge: RIKEN (The Institute of Physical and Chemical Research).
Conflict of interest statement. None declared.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Toyoda T, Wada A. Omic space: coordinate-based integration and analysis of genomic phenomic interactions. Bioinformatics. 2004;20:1759–1765. doi: 10.1093/bioinformatics/bth165. [DOI] [PubMed] [Google Scholar]
- 2.The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 3.Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, Ito T, Konagaya A, Toyoda T, Shinozaki K. RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Res. 2005;33:D647–D650. doi: 10.1093/nar/gki014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyoda T, Shinozaki K. Tiling array-driven elucidation of transcriptional structures based on maximum-likelihood and Markov models. Plant J. 2005;43:611–621. doi: 10.1111/j.1365-313X.2005.02470.x. [DOI] [PubMed] [Google Scholar]
- 5.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povey S, Lovering R, Bruford E, Wright M, Lush M, Wain H. The HUGO Gene Nomenclature Committee (HGNC) Hum. Genet. 2001;109:678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 9.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris TW, Chen N, Cunningham F, Tello-Ruiz M, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Bradnam K, Chan J, et al. WormBase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res. 2004;32:D411–D417. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, Fujii Y, Antonio BA, Nagamura Y, Imanishi T, et al. The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa spp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. The Rat Genome Database, update 2007 – easing the path from disease to data and back again. Nucleic Acids Res. 2006;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The Distributed Annotation System. BMC Bioinformatics. 2001;2(7) doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi N, Toyoda T. Statistical search on the Semantic Web. Bioinformatics. 2008;24:1002–1010. doi: 10.1093/bioinformatics/btn054. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida Y, Makita Y, Heida N, Asano S, Matsushima A, Ishii M, Mochizuki Y, Masuya H, Wakana S, Kobayashi N, et al. PosMed (Positional Medline): prioritizing genes with an artificial neural network comprising medical documents to accelerate positional cloning. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp384. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurihara Y, Matsui A, Kawashima M, Kaminuma E, Ishida J, Morosawa T, Mochizuki Y, Kobayashi N, Toyoda T, Shinozaki K, et al. Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem. Biophys. Res. Commun. 2008;376:553–557. doi: 10.1016/j.bbrc.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 19.Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 20.Masuya H, Yoshikawa S, Heida N, Toyoda T, Wada S, Shiroishi T. Phenosite: a web database integrating the mouse phenotyping platform and the experimental procedures in mice. J. Bioinform. Comput. Biol. 2007;5:1173–1191. doi: 10.1142/s0219720007003168. [DOI] [PubMed] [Google Scholar]
- 21.Toyoda T, Mochizuki Y, Player K, Heida N, Kobayashi N, Sakaki Y. OmicBrowse: a browser of multidimensional omics annotations. Bioinformatics. 2007;23:524–526. doi: 10.1093/bioinformatics/btl523. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart – biological queries made easy. BMC Genomics. 2009;10(22) doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]