Abstract
Computational microRNA (miRNA) target prediction is one of the key means for deciphering the role of miRNAs in development and disease. Here, we present the DIANA-microT web server as the user interface to the DIANA-microT 3.0 miRNA target prediction algorithm. The web server provides extensive information for predicted miRNA:target gene interactions with a user-friendly interface, providing extensive connectivity to online biological resources. Target gene and miRNA functions may be elucidated through automated bibliographic searches and functional information is accessible through Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The web server offers links to nomenclature, sequence and protein databases, and users are facilitated by being able to search for targeted genes using different nomenclatures or functional features, such as the genes possible involvement in biological pathways. The target prediction algorithm supports parameters calculated individually for each miRNA:target gene interaction and provides a signal-to-noise ratio and a precision score that helps in the evaluation of the significance of the predicted results. Using a set of miRNA targets recently identified through the pSILAC method, the performance of several computational target prediction programs was assessed. DIANA-microT 3.0 achieved there with 66% the highest ratio of correctly predicted targets over all predicted targets. The DIANA-microT web server is freely available at www.microrna.gr/microT.
INTRODUCTION
MicroRNAs (miRNAs) are approximately 22-nt long endogenously expressed RNA molecules which regulate gene expression, preferentially by binding to the 3′-untranslated region (3′-UTR) of protein coding genes (1) and have been found to confer a novel layer of genetic regulation in a wide range of biological processes. Since their initial identification in 1993 (2), there have been several efforts for the identification of miRNA targeted genes (miTGs), but biological experiments have uncovered only a small fraction of all miTGs. Due to this, computational target prediction remains one of the key means to analyze the role of miRNAs in biological processes.
In the last 5 years, more than two dozen miRNA target prediction programs have been published (3). Most of these programs are mainly based on sequence alignment of the miRNA seed region (nucleotides 2–7 from the 5′-end of the miRNA) to the 3′-UTR of candidate target genes leading to the identification of putative binding sites. Their specificity is usually increased by exploiting the commonly observed evolutionary conservation of the binding sites or by using additional features such as structural accessibility (4,5), nucleotide composition (6) as well as location of the binding sites within the 3′-UTR (7). Recently, Selbach et al. (12) determined the complement of all the genes targeted by five miRNAs induced independently in HeLa cells using microarrays and pulsed stable isotope labeling with amino acids in cell culture (pSILAC). Based on this dataset, they performed a comparative assessment of several commonly used target prediction programs which showed that only three [DIANA-microT 3.0, PicTar (9) and TargetScanS (13)] achieved precision levels (the fraction of the predicted targets that were actually downregulated) >60%. DIANA-microT 3.0 predicted 294 targets total out of which 194 were correct and thus reached a precision of 66%.
The DIANA-microT 3.0 algorithm is based on parameters that are calculated individually for each miRNA, and for each miRNA recognition element (MRE), depending on binding and conservation levels. The total predicted score of a miRNA:target gene interaction is the weighted sum of conserved and unconserved MREs of a gene. We also provide a signal-to-noise ratio (SNR) and a precision score specific for each interaction that can be used as a helpful confidence estimation of the ‘correctness’ and the false positive rate of each predicted miTG. This information can be easily looked up on the user-friendly DIANA microT web server where prediction results are organized in expandable tabs to group the available information, reduce the presentation complexity and show additional prediction details only on demand. Cases where a predicted interaction is registered as experimentally supported or predicted by other programs are also noted. The server offers an efficient search engine allowing multiple gene nomenclatures or queries based on gene involvement in specific biological pathways. The analysis of predicted interactions is supported by significance evaluation measures, extensive linkage to several online biological resources and automated bibliographic searches in PubMed. The server also supports prediction requests based on user-defined miRNA sequences and is integrated in a platform with two further miRNA functional analysis tools: mirPath, a pathway analysis tool of predicted targets and mirExTra, a miRNA analysis based on differential expressed mRNA profiles.
METHODS AND RESULTS
The DIANA microT web server
The web server may be accessed through a search engine with several options. The upper search box is used for browsing target genes predicted for a single miRNA. In this field, the miRNA name may be provided explicitly or partially. The second search box is used for identifying miRNAs which might be targeting a specific gene. In this case, the gene may be provided either based on Ensembl gene ID, RefSeq gene ID, common name or as part of the Ensembl description. If the search criteria correspond to more than one possible match, a list of alternatives is presented to the user to choose from. The lower search box combines the two search criteria offering the capability to identify if a specified miRNA targets a specified gene. For presenting the results, the web server results page (Figure 1) is divided in two parts. In the upper region, the user may find information concerning the provided search term; whereas, the prediction results are presented in the lower part.
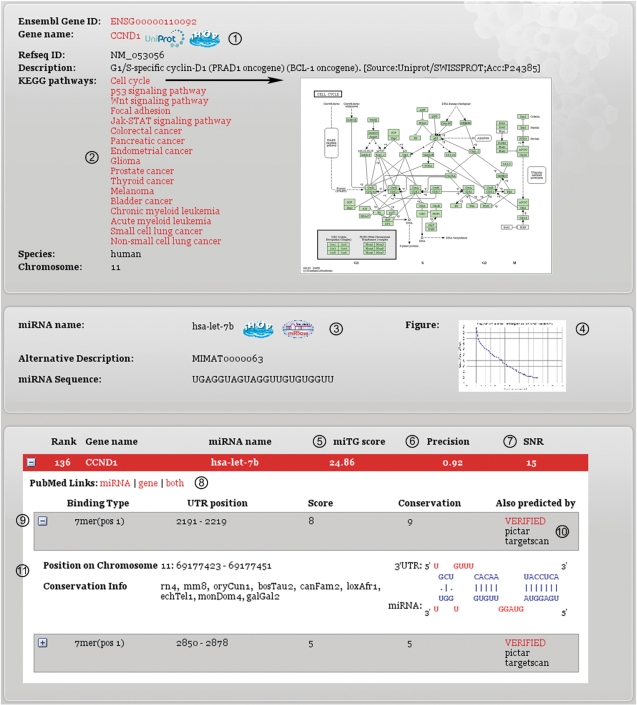
Figure 1.
DIANA-microT web server results page. The key features have been marked in the figure and are explained below. (1) Gene names and corresponding links to UniProt (protein information) and iHOP (functional and bibliographic information). (2) KEGG pathways in which the gene of interest is involved. (3) MiRNA names and corresponding links to miRBase (sequence information) and iHOP. (4) A graph showing the SNR of the miRNA. The SNR is calculated by the DIANA-microT algorithm and is based on a comparative analysis of the real miRNA versus a set of mock miRNAs. (5) The prediction score. Higher miTG scores correspond to higher possibility of correct prediction. (6) SNR score of the interaction. Greater values correspond to better distinction from the mock background. This attribute must be examined in combination with the SNR diagram provided for each miRNA. (7) The precision score of the interaction. This score ranges from 0 to 1, and it estimates the significance of the prediction. (8) Literature links that perform an automated search in PubMed for the gene, the miRNA or for the combination of the two. (9) Binding site info: (a) binding type indicates the number of the binding nucleotides in the 5′-end of the miRNA; (b) UTR position indicates the position of the binding site on the 3′-UTR; (c) score indicates the contribution of each binding site to the overall miTG score; and (d) conservation indicates the number of species in which the binding site is conserved. (10) This field indicates if the interaction may be found in the database of experimentally supported targets (TarBase) or if it has additionally been predicted by another target prediction program (TargetScan or Pictar). (11) Additional binding site info: (a) position on chromosome shows the position of the binding site on the chromosome; (b) conservation info indicates the species in which the binding site is conserved; and (c) graphic representation of the miRNA binding on the 3′-UTR.
Figure 1 presents a typical results page based on a combined search for a miRNA and a gene. To assess the significance of the predicted interactions, the web server offers evaluation measures such as the precision score and the SNR. The information for each MRE score including conservation and binding structure of the MRE:mRNA interaction is also provided. Cases where an interaction is registered in the database of experimentally supported miRNA targets [TarBase, (8)] are highlighted with a link to the database. Moreover, all the interactions which are also predicted by PicTar (9) or TargetScan 4.2 (6) are noted in the web page. For each predicted interaction, the results page offers extensive linkage to multiple online biological resources [UniProt, Ensembl, miRBase, iHOP and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (11)] as well as automated bibliographic searches in PubMed for the miRNA, the target gene or the combination of the two.
DIANA-microT 3.0 algorithm description
A typical miRNA is approximately 22-nt long, but the nucleotides close to the 5′-end of the miRNA are crucial for recognizing a target sequence and binding to it. Usually, a strong binding [at least seven consecutive Watson–Crick (WC) base pairing nucleotides] between the first 9 nt from the 5′-end of the miRNA sequence (here called as the miRNA driver sequence) and the target gene is required for sufficient repression of protein production. However, there is experimental (10) evidence that a weaker binding, involving only six consecutively paired nucleotides or including G:U wobble pairs, can also repress protein production if there is additional binding between the miRNA 3′-end and the target gene.
The DIANA-microT 3.0 algorithm considers as MREs, those UTR sites that have 7-, 8- or 9-nt long consecutive WC base pairing with the miRNA, starting from position 1 or 2 from the 5′end of the miRNA. For sites with additional base pairing involving the 3′-end of the miRNA, a single G:U wobble pair or binding of only six consecutive nucleotides to the driver sequence are also allowed. Using as features the MRE binding type and the MRE conservation profile, all identified MREs are scored through comparative analysis versus a set of MREs identified based on mock miRNA sequences. The overall miTG score is calculated as the weighted sum of the scores of all identified MREs on the 3′-UTR. The algorithm uses up to 27 species to assess the MRE conservation profile taking into account both conserved and nonconserved MREs for the estimation of the final miTG score.
For the evaluation of each miRNAs predicted interactions, the program compares them to those predicted for a set of mock miRNAs. Mock miRNAs are independently created for each real miRNA and are designed to have approximately the same number of predicted targets as the real miRNA. This allows for the calculation of miRNA-specific SNR at different miTG score cut-offs as well as for the estimation of a precision score that provides an indication of the false positive rate of a particular miTG interaction.
Target prediction support for novel miRNA sequences
The DIANA-microT server also supports prediction requests for user-defined miRNA sequences. The results of the de novo predictions are stored in a database from which they can later be retrieved and presented to the user who is provided with a unique key via email notification. Support for target prediction based on user-defined sequences remains a computationally intensive task even though the DIANA-microT 3.0 prediction algorithm is mainly based on dynamic programming routines. For this reason, all miRNA target prediction requests are supported by a 256 core cluster consisting of 32 nodes which succeeds close to linear speedup and is hosted at the National Technical University of Athens (NTUA).
Integration of further analysis tools mirPath and mirExTra
In a typical case, the miRNA involved in a biological process is known and there is a need to predict its targets. However, the reverse search may also be relevant in some cases where, for instance, high-throughput data from cDNA arrays indicating changes in the expression of protein coding genes is available. In this case, the putative targets are known whereas the miRNA targeting them is unknown. To this end, an additional pre-processing tool for target prediction (mirExTra) is also available that is able to uncover miRNAs that may be involved in the changes of the transcriptome by processing a list of differentially expressed protein coding genes and a list of genes whose expression is unchanged. The program identifies hexamers that correspond to the driver region of a miRNA starting at position 1 and 2, which are significantly overrepresented in the input list of the overexpressed genes relative to those whose expression levels are constant under the same conditions. The web server is also combined with a post-processing analysis tool of predicted targets (mirPath) regarding their role in biological pathways. To this end, KEGG pathways that are enriched in a group of miTGs are identified and the results are visualized by highlighting the miTGs in the pathway.
CONCLUSION
The miRNA target prediction experiment by Selbach et al. (12) revealed the problem of the large fraction of under predicted or falsely predicted target genes. With lower score thresholds sensitivity can be increased, while trading off specificity and variable score thresholds can help to find best combination of these two measures. It is therefore crucial to give the user the possibility to modify this threshold and simultaneously present all relevant information facilitating the interpretation, the evaluation or even the experimental verification of predicted interactions. We found that most miRNA target prediction programs are insufficient in this respect, even when providing a graphical user interface for their results. Additionally, the search and identification of interactions of interest is complicated by the existence of different gene nomenclatures and may discourage researchers from trying to further elucidate the effects of miRNAs in biological processes. New miRNAs are identified nearly every month and this rate is increasing through the use of the new deep sequencing technologies. MiRNAs may also undergo editing and change the majority of their targets (14). Our approach, the DIANA-microT web server, has been designed with these challenges in mind and provides a user-friendly interface also for unannotated miRNAs by a precise target prediction algorithm.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 4.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 5.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 6.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2008;37(Database issue):D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Kawahara B, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Dictation of silencing targets by adenosine-to-inosine editing of microRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]