Abstract
TOPCONS (http://topcons.net/) is a web server for consensus prediction of membrane protein topology. The underlying algorithm combines an arbitrary number of topology predictions into one consensus prediction and quantifies the reliability of the prediction based on the level of agreement between the underlying methods, both on the protein level and on the level of individual TM regions. Benchmarking the method shows that overall performance levels match the best available topology prediction methods, and for sequences with high reliability scores, performance is increased by ∼10 percentage points. The web interface allows for constraining parts of the sequence to a known inside/outside location, and detailed results are displayed both graphically and in text format.
INTRODUCTION
Genome-wide estimations indicate that alpha-helical transmembrane (TM) proteins comprise roughly 20–30% of the genes in a typical organism (1,2). These proteins are essential for vital biological functions such as cell communication and signaling, active and passive transport of molecules across the membrane, energy-transduction and cell–cell adhesion.
Prediction of membrane protein topology (i.e. the positions and in/out orientation of the membrane-spanning regions) serves to quickly obtain fundamental structural knowledge of TM proteins in silico. For TM proteins, computational methods are particularly important since structural knowledge is difficult to attain experimentally. Therefore, a correctly predicted topology provides an excellent template for further studies in the laboratory and might facilitate and improve functional and structural classification of protein sequences on a genomic level.
A number of different methods have been developed over the last decades that predict topology with high accuracy. Many of these methods are freely available as web servers, both individually (2,3) and combining the results from several methods (4). With prediction algorithms based on different principles, it is not a surprising observation that for a fair amount of proteins, different prediction methods disagree about the final result, causing uncertainty about the correct topology. Earlier studies have stated, for example, that topology predictions are more likely to be correct when individual methods agree in their prediction than when they do not (5), but so far only a few attempts have been made at combining individual topology predictions into one consensus prediction (6–8).
Here we present TOPCONS, a fundamental algorithm that combines an arbitrary number of topology predictions into one consensus prediction and quantifies the reliability of the prediction based on the level of agreement between the underlying methods, both on the protein level and on the level of individual TM regions.
We also present an implementation of TOPCONS as a web-server based on the individual topology prediction methods OCTOPUS (9), PRO-TMHMM and PRODIV-TMHMM (10), SCAMPI-single and SCAMPI-multi (11). During the development a large set of combinations using many different topology predictors as an input to TOPCONS was tested. However, no combination performed significantly better than the one used here and therefore we decided to only use methods developed in house in the current version of the TOPCONS webserver.
METHODS
TOPCONS algorithm
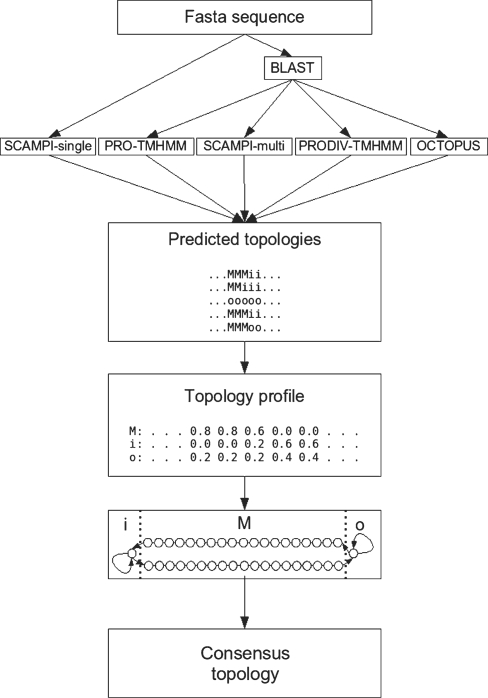
An overview of the different steps of the TOPCONS algorithm is presented in Figure 1. As input TOPCONS uses a set of topology predictions which are combined into a topology profile by letting each residue be represented by three values representing the fraction of methods that predict that residue to be situated in the membrane (M), on the inside of the membrane (i) or on the outside of the membrane (o). This topology profile is used as input to a dynamic programming algorithm similar to a hidden Markov model that has an alphabet consisting of the characters ‘M’, ‘i’ and ‘o’. The final topology corresponds to the highest scoring state path through this model using a Viterbi-like algorithm. In each state, the emission score for the structural category modeled by that state (i, o or M) is equal to 1.0 and for all other structural categories it equals 0.0. All transition probabilities are equal to 1.0. Thus, the final prediction equals the state path with the highest geometric mean score with respect to the topology profile and the grammar of the model.
Figure 1.
TOPCONS workflow: four of the topology predictors make use of multiple sequence information and require a sequence profile as input, created using BLAST (18), whereas the fifth method (SCAMPI-single) only requires the protein sequence. The topology predictions are used to construct a topology profile, which is fed into the TOPCONS hidden Markov model.
Reliability score
A reliability value is calculated for each residue in a sequence by taking the average over a 21 position window of the topology profile value for the consensus prediction of that position (i, o, M). A reliability score on the protein level is calculated by taking the minimum value as calculated above.
Dataset
A dataset of 163 sequences with known topology, compiled by combining the datasets from (10) and (3) and homology reducing at 30% sequence identity using cd-hit (12), was used to evaluate the performance of TOPCONS.
BENCHMARK RESULTS
Using a dataset of 163 sequences with known topology, the performance of TOPCONS was benchmarked against eight other topology prediction algorithms (Table 1). ConPredII (7) is an earlier consensus transmembrane topology prediction approach, and the other methods [OCTOPUS (9), MEMSAT3 (13), SCAMPI (11), HMMTOP (14) and PRO- and PRODIV-TMHMM (10)] are frequently used single topology prediction methods. TOPCONS outperforms the only other consensus prediction method ConPredII, partly because the underlying topology prediction methods are more recent, and achieves accuracy similar to that of the best available individual methods. However, the performance of TOPCONS is not significantly better than the best single server included in the predictions, possibly because the limit of prediction accuracy given the data available today is close to have been reached.
Table 1.
Prediction accuracy for a benchmark set of 163 membrane proteins with known topology
| Method | Accuracy (%) |
|---|---|
| TOPCONS | 83 |
| OCTOPUS | 82 |
| PRODIV-TMHMM | 78 |
| MEMSAT3 | 76 |
| SCAMPI-multi | 75 |
| ConPredII | 74 |
| PRO-TMHMM | 72 |
| HMMTOP | 70 |
| SCAMPI-single | 68 |
The accuracy is measures as the fraction of correct topologies at the protein level. A correct topology should have the correct number of TM regions at approximately correct locations and the correct location of the N and C-termini.
Studying the reliability scores (Figure 2); it is evident that higher reliability scores (i.e. in principle the number of methods that agree for the sequence position with least agreement across the protein) correspond to higher probability that a prediction is correct. In our benchmark set, 71% of the sequences achieved reliability scores above 70, and among those sequences, the accuracy of TOPCONS is 93%, i.e. 10 percentage points higher than the overall accuracy for the complete dataset.
Figure 2.
Estimated probability of correct topology prediction as a function of reliability score. Of the sequences in the benchmark set, 71% have a reliability score >70, and among those, 93% of the predictions are correct. The probability of a correct topology prediction (y-axis) was estimated as the prediction accuracy among proteins with reliability score ± 10 from the value given by the x-axis.
Compared to an earlier derived reliability score for the individual prediction method TMHMM (15), the reliability score of TOPCONS is similar (although the overall prediction accuracy is higher). A z-value for the reliability scores were calculated using the Wilcoxon rank sum test to get a quantified comparison between the two reliability scores independent of the overall prediction accuracy. The z-value for TMHMM is −6.1 and for TOPCONS −4.8.
THE TOPCONS AND SCAMPI WEBSERVERS
TOPCONS
To make TOPCONS available to a broad audience, a web server implementing the algorithm has been developed and can be freely accessed at http://topcons.net/. Given the amino-acid sequence of a putative membrane protein, the server outputs the predicted topology using the individual methods, as well as the consensus prediction (TOPCONS) (Figure 3). In addition, ZPRED (16) is used to predict the Z-coordinate (i.e. the distance to the membrane center) of each amino acid, and a scale describing the free-energy contributions of translocon-mediated membrane insertion (17) is used to predict a ΔG value for a window of 21 amino acids centered around each position in the sequence. Optionally, parts of the sequence can be constrained to a known Inside/Outside/Membrane-location, by using the Restrainment options, allowing for any combinations of restraints on one or more parts of the sequence. All results are both displayed graphically and are available for download in text format. The BLAST output, which is used as input to the methods, and high-resolution versions of the images are also available for download.
Figure 3.
Example output from the TOPCONS webserver, based on the Bacteriorhodopsin sequence from Halobacterium species (SwissProt-ID: BACR_HALS4). (A) Topologies predicted by the individual methods, predicted Z-coordinates, and predicted ΔG-values across the sequence. (B) The consensus prediction, which is based on the individual methods, and reliability score across the sequence.
SCAMPI
Due to the computational limitations arising from the need to run BLAST (18) (Figure 1), only one sequence per query is allowed using TOPCONS, and the prediction typically takes 10–30 s. For large benchmark sets and full proteome scans, the SCAMPI server, implementing only the single-sequence version of SCAMPI (11), may be used instead, which is able to process around 20 000 protein sequences per minute (http://scampi.cbr.su.se/). Here, prediction results are made available in easily parsable text files.
FUNDING
Swedish Research Council, the Foundation for Strategic Research, EU 6th Framework Program [Biosapiens, Contract LSHG-CT-2004-512092] and 7th Framework program [EDICT Contract No FP7-HEALTH-F4-2007-201924]. Funding for open access charge: EDICT program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 3.Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Amico M, Finelli M, Rossi I, Zauli A, Elofsson A, Viklund H, von Heijne G, Jones D, Krogh A, Fariselli P, et al. PONGO: a web server for multiple predictions of all-alpha transmembrane proteins. Nucleic Acids Res. 2006;34:W169–W172. doi: 10.1093/nar/gkl208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsson J, Persson B, von Heijne G. Consensus predictions of membrane protein topology. FEBS Lett. 2000;486:267–269. doi: 10.1016/s0014-5793(00)02321-8. [DOI] [PubMed] [Google Scholar]
- 6.Parodi LA, Granatir CA, Maggiora GM. A consensus procedure for predicting the location of alpha-helical transmembrane segments in proteins. Comput. Appl. Biosci. 1994;10:527–535. doi: 10.1093/bioinformatics/10.5.527. [DOI] [PubMed] [Google Scholar]
- 7.Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis MJ, Zhang F, Yuan Z, Teasdale RD. MemO: a consensus approach to the annotation of a protein's membrane organization. In Silico Biol. 2006;6:387–399. [PubMed] [Google Scholar]
- 9.Viklund H, Elofsson A. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics. 2008;24:1662–1668. doi: 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- 10.Viklund H, Elofsson A. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 2004;13:1908–1917. doi: 10.1110/ps.04625404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A. Prediction of membrane-protein topology from first principles. Proc. Natl Acad. Sci. USA. 2008;105:7177–7181. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 13.Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 14.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 15.Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- 16.Granseth E, Viklund H, Elofsson A. ZPRED: predicting the distance to the membrane center for residues in alpha-helical membrane proteins. Bioinformatics. 2006;22:e191–e196. doi: 10.1093/bioinformatics/btl206. [DOI] [PubMed] [Google Scholar]
- 17.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]