Abstract
PLecDom is a program for detection of Plant Lectin Domains in a polypeptide or EST sequence, followed by a classification of the identified domains into known families. The web server is a collection of plant lectin domain families represented by alignments and profile Hidden Markov Models. PLecDom was developed after a rigorous analysis of evolutionary relationships between available sequences of lectin domains with known specificities. Users can test their sequences for potential lectin domains, catalog the identified domains into broad substrate classes, estimate the extent of divergence of new domains with existing homologs, extract domain boundaries and examine flanking sequences for further analysis. The high prediction accuracy of PLecDom combined with the ease with which it handles large scale input, enabled us to apply the program to protein and EST data from 48 plant genome-sequencing projects in various stages of completion. Our results represent a significant enrichment of the currently annotated plant lectins, and highlight potential targets for biochemical characterization. The search algorithm requires input in fasta format and is designed to process simultaneous connection requests from multiple users, such that huge sets of input sequences can be scanned in a matter of seconds. PLecDom is available at http://www.nipgr.res.in/plecdom.html.
INTRODUCTION
Modern glycobiology revolves, to a large extent, around the potential biological information stored in cell surface carbohydrates, whose roles in cell growth, differentiation and surface recognition are increasingly being investigated using lectins, a large family of ubiquitous proteins with the ability to bind and agglutinate these sugars. Lectins display an enormous diversity in their sequence, biological activity and mono-/oligosaccharide specificity in addition to an unsurpassed structural versatility (1,2). In plants, lectins play crucial physiological roles in stress responses, defense, symbiotic communication, and are considered one of the most important biological recognition molecules (1,3,4).
Plant lectins have been variously classified into distinct families based on their structure, ligands and evolutionary relationships. Structurally, some of the major folds reported for plant lectins are beta prism-I, beta-prism-II, beta-trefoil, seven-bladed beta-propeller, knottins, jelly-roll (also called the lectin fold) and the P-domain fold of calnexin/calreticulin (5). These structural groups show varying degrees of overlap with the sequence-based classes, and new folds are constantly being identified. Until a few years back, a consensus of seven distinct lectin families were known in plants, based upon their carbohydrate binding domains (6). These were the amaranthins, cucurbitaceae phloem lectins (now called the Nictaba lectins), lectins with hevein domains, jacalin-related lectins, legume lectins, monocot mannose-binding lectins [now called the GNA-related lectins (7)] and type-II ribosome-inactivating proteins (RIPs) also known as the Ricin-B family. However, this classification has become inadequate with the very recent addition of new families or by regrouping of the existing families (8). Likewise, the identification of functional homologs of various animal lectin families in plants (9), as in the case of the galectins (previously called the S-type lectins, having a strong affinity for β-galactosides) and the calnexin/calreticulin lectin families, has further amplified the complexity in plant lectin classification (10,11). Among the newly identified and/or regrouped families, are the agaricus bisporus agglutinins (or ABA domains), class V chitinase homologs with lectin activity (12), the EEA (13), LysM family (14) and cyanovirins (15), all of which emphasize the need to re-address the question of lectin classification in light of new data.
Due to the huge variation in the sequence, structure and specificity of plant lectins and the complexity associated with their classification, methods that seek to improve detection, annotation or assignation of carbohydrate specificity to these proteins would be of immediate interest to researchers. Currently, the determination of fine specificities of lectins remains largely experimental although attempts to understand the sugar recognition mechanisms within families have demonstrated a huge potential for development of bioinformatics-based predictive/automated tools. Some of the significant computational efforts have involved flexible docking between lectin and sugar molecules, structural mapping and pattern recognition in glycan branches via probabilistic sibling-dependent tree markov models (16–18). Efficient algorithms have been developed for training and improvement of the probabilistic models, but these have been tested on binding affinity data for a limited number of families (19). A comparative structural and specificity analysis led to the identification of three crucial residues for carbohydrate recognition in legume lectins (20), providing insights into the molecular interactions of lectins with simple sugars, involving a network of hydrogen bonds and an aromatic residue in the vicinity of the binding site. Despite these breakthroughs and pioneering work being carried out in lectin biology, there is no dedicated program or tool for identification of these domains. This inspired us to develop PLecDom, an online, predictive and interactive web server that can assist in the identification and analysis of these proteins using sequence information alone. The program has a very simple user friendly interface, and help pages, allowing users to submit their own queries or browse available data.
METHODOLOGY
Data collection
Published reports of characterized sequences assigned to distinct plant lectin families were searched from literature followed by keyword search of the major protein databanks as well as the plant lectin database (21). Homologs of animals origin, wherever available, were also compiled for these families. In all, 845 and 487 sequences, from plant and animal origins, respectively, were compiled for various families. Families for which a minimum of 30 representative sequences were not available were discarded from further analysis. These included the cyanovirins, ABA lectins, LysM, EEA, Amaranthins and class V chitinase homologs with lectin activity. The filtered starting dataset was composed of the eight remaining families, namely, the (i) GNA-related lectin domains, (ii) lectins with Hevein domains, (iii) Jacalin related lectins, (iv) Legume lectins, (v) Ricin–B lectins, (vi) Galectins, (vii) Calreticulin/Calnexins and (viii) Nictaba lectins. This dataset was called the curated sequence (CS) dataset (available as online Supplementary Data). Apart from the CS dataset, protein sequence prediction data was downloaded for 10 completed plant genome projects from NCBI, TIGR (22) and JGI web sites. These include four dicots (Arabidopsis, Poplar, Grape and Soybean), four chlorophytes (Chlamydomonas, Volvox and two species of Ostreococcus), a bryophyte (Physcomitrella) and Rice—a monocot. This data comprised of a total of 357 139 protein sequences and was called the ‘Protein Complete Genome’ (PCG) dataset. In addition, EST sequence data for 38 incomplete plant genomes, was downloaded from the TIGR database (22). These include eight monocots (Allium, Festuca, Hordeum, Wheat, Maize, Rye, Sorghum and Sugarcane), two gymnosperms (Pinus and Spruce), and 28 dicots including Apple, Aquilegia, Beetroot, Brassica, Capsicum, Cocoa, Coffee, Euphorbia, Sunflower, Ice plant, Ipomoea, Lotus, Medicago, Petunia, Phaseolus, Potato, Prunus, Tomato and two species each of Citrus, Gossypium, Nicotiana, Lactuca and Triphysaria. Full species details and scientific names can be found on the web server. This dataset includes 42% clustered ESTs and 57% individual ESTs and a fraction of ETs, adding to a total of 1 873 460 sequences in all. This was called the ‘EST Incomplete Genome’ (EIG) Dataset.
Analysis and program development
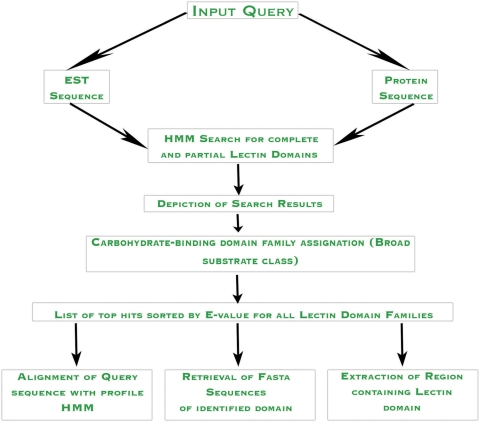
Multiple alignments and phylogenetic reconstruction of the sequences in the CS dataset were carried out using CLUSTALW (23). Profile HMMs were built using HMMER version 2.3.2 (24). The first set of profile HMMs were built using plant sequences and were trained on the animal data to strengthen the predictability and to enable identification of distant homologs. Different build-and-search parameters were tested, allowing only one parameter change at a time. The GNA-related lectins and calreticulin sequences gave optimal results when weighed using the Krogh/Mitchinson maximum entropy algorithm, a slightly more robust form of the Eddy/Mitchinson/Durbin maximum discrimination algorithm, giving an increase in sensitivity. For all other families, the default weighing method, i.e. the Gerstein/Sonhammer/Chothia tree-weighting algorithm gave best results. The BLOSUM62 scoring matrix was used for all families except the jacalins and legume lectins, both of which performed better with a heuristic PAM60 matrix. In the second step, the plant and animal sequences from the CS dataset were combined and profiles were rebuilt using parameters optimized in the first stage. This strategy offered us a larger dataset and greater representation of each family, and was thenceforth treated as the training dataset, having a total of 1198 sequences from eight families described in the previous section. For testing EST sequences, which often contain partial domains rather than full domains, the fragmentary search option was used in addition to the optimized parameters described earlier, to enable short fragments of target domains to be captured. An E-value filter of 0.01 was applied when using these HMMfs profiles. The EIG dataset was subjected to a six-frame translation using EMBOSS version 6.0.1 (25). For protein sequence search, default E-value was used with the optimized profile HMMs. Domain boundaries were identified using the alignments of sequences with profiles so that individual domains could be extracted for further analysis. Database match with query was done using a local version of BLAST program (26). In-house Fortran programs were used to streamline and automate the entire process and shell scripts were added for the testing of input sequences and to allow multiple users to use the program simultaneously. The program thus developed was converted into a web server using html and back-end CGI coding. Figure 1 depicts a schematic overview of the PLecDom query submission protocol.
Figure 1.
Flowchart depicting the query submission protocol in PLecDom.
Program testing and prediction accuracy
The PLecDom server has been tested on several browsers and platforms, including Safari, Firefox, Konqueror and IE on Macintosh, Linux as well as Windows workstations thereby making it cross-platform compatible. The validation dataset was separated at the time of data collection and comprised of 128 sequences having representatives from all families. The results of PLecDom on this dataset were compared with those from major annotation databases like Pfam (27), PANTHER (28) and SMART (29). In order to check the precision accuracy of the program, and more importantly, its negative prediction ability, a negative dataset comprising 1146 sequences was added to the 128 positives. The performance of the optimized profiles on this dataset was tested using the statistical concepts of sensitivity, specificity and precision. Sensitivity measures the proportion of actual positives, which are correctly identified as such, and was calculated for each family as the ratio of true positives to combined true positives and false negatives. Specificity is a measure of the proportion of negatives that are correctly identified, and was calculated for each domain family as the ratio of true negatives to combined true negatives and false positives. Precision, or positive predictive value, refers to the fraction of returned positives that are true positives, and is often considered more important than accuracy, which estimates the overall proportion of true positives in the population. Precision was calculated as the ratio of true positives to the combined true and false positives.
RESULTS
PLecDom represents a collection of profile Hidden Markov Models (HMMs) based on a rigorous analysis of eight distinct sugar-binding domain families of plant lectins, namely, (i) GNA-related lectin domains, (ii) lectins with Hevein domains, (iii) Jacalin-related lectins, (iv) Legume lectins, (v) Ricin–B lectins, (vi) Galectins, (vii) Calreticulin/Calnexins and (viii) Nictaba lectins. PLecDom is an online, automated, interactive and predictive search tool, the first of its kind dedicated to lectin domains.
Input and output
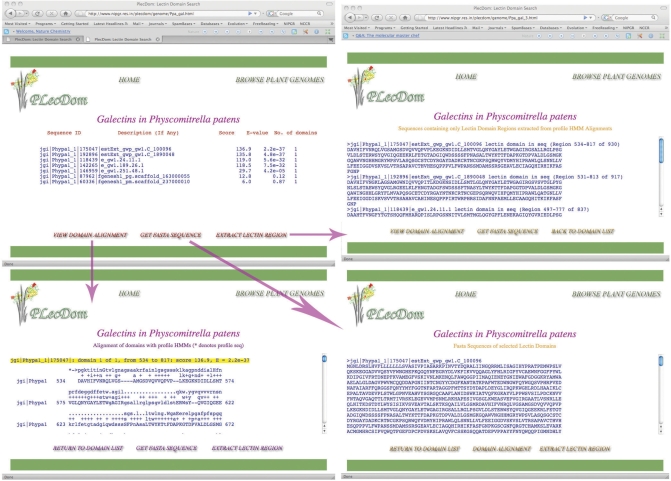
The PLecDom search algorithm requires input in Fasta format. It can accept protein as well as EST data and automatically recognizes the type of input without user intervention. EST data is translated into six frames, and each frame is tested for presence of lectin domains using the optimized fragmentary search profiles. If multiple frames of the same sequence are found to have lectin domains or their parts, they can be viewed in the results window of the relevant lectin family. PLecDom has been designed to process simultaneous connection requests from multiple users, and optimized such that huge sets of input sequences can be scanned in a matter of seconds. A detailed tutorial for query submission has been provided in addition to an example set of test sequences for explaining the submission and search procedure. Figure 2 shows a snapshot of the outcome of a successful search. The identified lectin domains are catalogued by the program into distinct sugar-binding families and users can view the number of domains identified in each family. This feature can be useful to narrow down the spectrum of probable substrates and assist in the prediction of fine specificities of newly identified lectin domains. Users can also check if their input sequences already exist in our database. This aspect of PLecDom can be very useful since researchers with partial sequences may find longer pieces, which could help them in cloning full-length sequences. To analyze the identified domains in detail, users can select the lectin family of interest, as depicted in Figure 3. In case of EST sequences, multiple-frame lectin domains, if detected, can be singled out for analysis on this page, as it shows reading frame numbers within the domain list. Furthermore, alignments of the identified domains with family-specific profile HMMs allow users to estimate the divergence of their data from existing homologs. Domain boundaries captured from this alignment allow users to specifically extract the selected lectin regions. In case of EST data, the domain boundaries enable users to analyze flanking regions on the genome for additional information. For example, putative lectin regions identified from EST sequences can be aligned to genomic DNA to carry out intron analysis and for the presence of signal sequences, providing clues to probable biological function and sub-cellular localization. All sequences including source data, extracted lectin regions, and alignments are available for download by a variety of grouping methods.
Figure 2.
Snapshots of query submission results in PLecDom.
Figure 3.
Genome browsing snapshots for a specific lectin family. Screendumps showing details of Galectins identified by PlecDom in the moss genome.
Performance
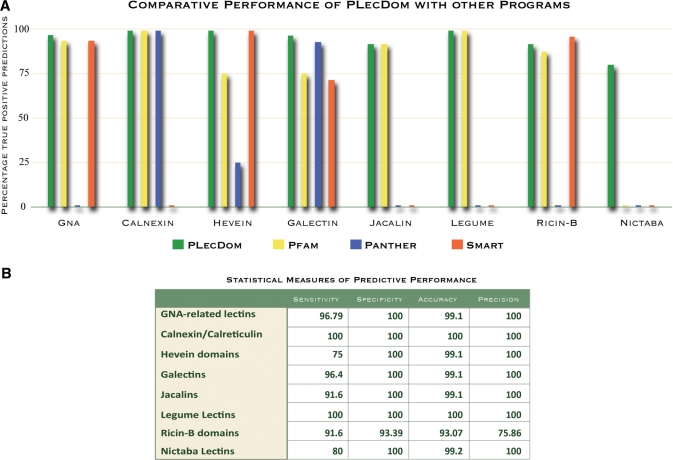
Figure 4A shows the relative performance of PLecDom as compared with three major public databases, namely, Pfam, PANTHER and SMART. As can be seen, PLecDom performs equally well or even better than any of these programs, and most remarkably so for the Nictaba lectins. We would like to emphasize that PLecDom is currently the only program available for detection and characterization of these lectins, since no existing database or annotation program is able to identify them. In case of the Ricin-B family and Hevein domain lectins, PLecDom performs better than PANTHER predictions but marginally less than Pfam.
Figure 4.
(A) Assessment of PLecDom predictions. The chart shows performance of PLecDom as compared to other databases for the validation dataset. Databases having null predictions are shown as base line stubs. PLecDom (in green) shows the best performance in predicting plant lectin domains. Note the Nictaba family predictions where PLecDom is the only program that can detect these domains. (B) Statistical performance measures of PLecDom. All values are reported as percentages.
The second test of PLecDom performance was statistical and Figure 4B shows the various measures of prediction accuracy in each of the eight lectin families analyzed. It is interesting to note that all families reveal very high specificities and prediction accuracy, the lowest values being 75% sensitivity in case of Hevein domains and 75.86% precision in case of Ricin-B family. The implications of these observations are discussed in the last section.
Browsing PLecDom
Encouraged by the high prediction accuracy of PLecDom (see Figure 4), and its ease with handling high-density data, we applied the program to the available protein predictions and EST sequence data from 48 sequencing projects. This exercise resulted in the identification of more than 7000 lectin domains and their assignation to various known plant lectin families. It may be noted that, approximately 5000 of the identified sequences are novel, i.e. previously un-annotated or, in a few cases, annotated as ‘putative’ or ‘conserved hypotheticals’. This data thus represents a major enrichment of plant lectin domain homologs known till date. The identified sequences have been made available at the web server and can be browsed by species name, followed by the domain family of interest. In each case, the output is similar to that described in Figure 3 so as to maintain consistency with query submission outputs. For incomplete genomes, we have additionally made the EST sequences of detected lectins available for download to users. A genome browsing tutorial has also been provided on the web page to explain the search pages in detail.
DISCUSSION
PLecDom is a web server dedicated to plant lectin domains. The program enables identification and analysis based on a computational exploration of the sequence space of currently available and characterized plant lectin families. PLecDom has been designed to accept EST input in addition to protein sequences to assist researchers with preliminary annotation of newly emerging data. Our objective is to build a robust tool for detection of all reported families of plant lectins. However, a few families, mainly the very recently recognized ones, had to be discarded at the data collection stage on account of lack of sufficient number of representative characterized sequences. The families excluded from the current version of PLecDom are the Amaranthins, ABA, EEA, cyanovirins, LysM families, and class V chitinase homologs with lectin activity. In future, as more sequences from these and other families become available, we will update the server to broaden its base and scope.
One of the major achievements of PLecDom is its remarkable predictive success. Unlike pairwise or multiple sequence comparisons, which can often result in capturing false homologs, the optimized profile HMMs in PLecDom give distinct outputs, and are very sensitive, thereby enhancing the likelihood of predicting genuine homologs. A comparison of PLecDom predictions with those obtained from general annotation databases currently available, showed that our program performs very well, and is better than several existing programs, most strikingly so in case of the Nictaba lectin family. We believe our program is a more reliable tool for plant lectin annotation. All families except Ricin-B showed 100% specificity and very high precision values (see Figure 4B). The Ricin-B family profiles returned seven false positive identifications, thereby lowering its precision, although the specificity and accuracy of the family remains significantly high. A comparatively low sensitivity was observed in case of PLecDom predictions for the Hevein domains (75%), but the Hevein domain family profiles showed very high specificity and precision (100%), revealing that although a distant homolog of this family may sometimes fail to be recognized, a positive identification would nevertheless be conclusive. Overall the positive predictive value of all families in PLecDom is very high, thereby making it highly suitable for annotation and assignation of domain family to test sequences.
The application of PLecDom to protein and EST sequence data from 48 sequencing projects resulted in a considerable enrichment of the currently annotated plant lectins. This wealth of data can now be used to carry out a comprehensive functional analysis and highlights potential targets for biochemical characterization in many species. Several interesting insights have been gained from the PLecDom outputs. For example, the data show that, contrary to previous assumptions, legume lectins do occur in several non-leguminous species, and the monocot mannose-binding lectin family has many homologs in dicots as well. Preliminary studies reveal that these atypical homologs have different intronic and exonic features, and this may potentially lead to the recognition of new functions or families (G.Yadav unpublished data). Further, the almost complete ‘lectinome’ that has been extracted in this study, for 10 fully sequenced plant genomes, reveals a mixed set of lectin family combinations in each species rather than taxon-specific lectin families probably signifying a unique ‘lectin signature’ of individual genomes.
Taken together, these observations provide fascinating new insights into the diversity of plant lectins and a huge potential to investigate their evolutionary ramifications. To summarize, we believe that PLecDom would be of immediate interest to glycobiologists and researchers involved in the identification, annotation and characterization of plant lectins, as well in the study of plant stress responses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Innovative Young Biotechnologist grant to GY by the Department of Biotechnology, Government of India [BT/BI/12/040/2005] and [BT/PR/5360/Agr/16/483/2004]. Funding for open access charge: USD 2670.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The JGI sequence data for lower plants used in this work were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/in collaboration with the user community. Laboratory resources provided under BTIS grant of DBT, India, are gratefully acknowledged. Authors thank Dr Suresh Babu, CEMDE, University of Delhi for constructive criticism of the manuscript. Mr Sachin Pundhir's assistance with server monitoring is acknowledged.
REFERENCES
- 1.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:R53–R62. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 2.Peumans WJ, Van Damme EJM. Lectins as plant defense proteins. Plant Physiol. 1995;109:347–352. doi: 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudiger H, Gabius HJ. Plant lectins: occurrence, biochemistry, functions and applications. Glycoconj. J. 2001;18:589–613. doi: 10.1023/a:1020687518999. [DOI] [PubMed] [Google Scholar]
- 4.Chrispeels MJ, Raikhel NV. Lectins, lectin genes, and their role in plant defense. Plant Cell. 1991;3:1–9. doi: 10.1105/tpc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha S, Gupta G, Vijayan M, Surolia A. Subunit assembly of plant lectins. Curr. Opin. Struct. Biol. 2007;17:498–505. doi: 10.1016/j.sbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Van Damme EJM, Rougé P, Peumans WJ. Plant lectins. In: Kamerling JP, Boons GJ, Lee YC, Suzuki A, Taniguchi N, Voragen AJG, editors. Comprehensive Glycoscience – From Chemistry to Systems Biology. Vol. 3. Oxford, UK: Elsevier; 2007. pp. 563–599. [Google Scholar]
- 7.Hester G, Kaku H, Goldstein IJ, Wright CS. Structure of mannose-specific snowdrop (Galanthus nivalis) lectin is representative of a new plant lectin family. Nat. Struct. Biol. 1995;2:472–479. doi: 10.1038/nsb0695-472. [DOI] [PubMed] [Google Scholar]
- 8.Van Damme EJM, Lannoo N, Peumans WJ. Plant Lectins. Adv. Botanical Res. 2008;48:107–209. [Google Scholar]
- 9.Dodd RB, Drickamer K. Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology. 2001;11:R71–R79. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- 10.Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. Introduction to galectins. Glycoconj. J. 2004;19:433–440. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- 11.Van Damme EJM, Barre A, Rougé P, Peumans WJ. Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci. 2004;9:484–489. doi: 10.1016/j.tplants.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Van Damme EJM, Culerrier R, Barre A, Alvarez R, Rougé P, Peumans WJ. A novel family of lectins evolutionarily related to class V chitinases: an example of neofunctionalization in legumes. Plant Physiol. 2007;144:662–672. doi: 10.1104/pp.106.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouquaert E, Peumans WJ, Smith DF, Proost P, Savvides SN, Van Damme EJM. The “old” Euonymus europaeus agglutinin represents a novel family of ubiquitous plant proteins. Plant Physio.l. 2008;147:1316–1324. doi: 10.1104/pp.108.116764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onaga S, Taira T. A new type of plant chitinase containing LysM domains from a fern (Pteris ryukyuensis): roles of LysM domains in chitin binding and antifungal activity. Glycobiology. 2008;18:414–423. doi: 10.1093/glycob/cwn018. [DOI] [PubMed] [Google Scholar]
- 15.Koharudin LM, Viscomi AR, Jee JG, Ottonello S, Gronenborn AM. The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity. Structure. 2008;16:570–584. doi: 10.1016/j.str.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Neumann D, Lehr CM, Lenhof HP, Kohlbacher O. Computational modeling of the sugar-lectin interaction. Adv. Drug Deliv. Rev. 2004;56:437–457. doi: 10.1016/j.addr.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Kerzmann A, Fuhrmann J, Kohlbacher O, Neumann D. BALLDock/SLICK: a new method for protein-carbohydrate docking. J. Chem. Inf. Model. 2008;48:1616–1625. doi: 10.1021/ci800103u. [DOI] [PubMed] [Google Scholar]
- 18.Fujimoto YK, Terbush RN, Patsalo V, Green DF. Computational models explain the oligosaccharide specificity of cyanovirin-N. Protein Sci. 2008;17:2008–2014. doi: 10.1110/ps.034637.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoki KF, Ueda N, Yamaguchi A, Kanehisa M, Akutsu T, Mamitsuka H. Application of a new probabilistic model for recognizing complex patterns in glycans. Bioinformatics. 2004;20(Suppl 1):i6–i14. doi: 10.1093/bioinformatics/bth916. [DOI] [PubMed] [Google Scholar]
- 20.Sharon N, Lis H. How proteins bind carbohydrates: lessons from legume lectins. J. Agric. Food Chem. 2002;50:6586–6591. doi: 10.1021/jf020190s. [DOI] [PubMed] [Google Scholar]
- 21.Chandra NR, Kumar N, Jeyakani J, Singh DD, Gowda SB, Prathima MN. Lectindb: a plant lectin database. Glycobiology. 2006;16:938–946. doi: 10.1093/glycob/cwl012. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J. The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res. 2005;33:D71–D74. doi: 10.1093/nar/gki064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 24.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 25.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.