Abstract
SplitPocket (http://pocket.uchicago.edu/) is a web server to identify functional surfaces of protein from structure coordinates. Using the Alpha Shape Theory, we previously developed an analytical approach to identify protein functional surfaces by the geometric concept of a split pocket, which is a pocket split by a binding ligand. Our geometric approach extracts site-specific spatial information from coordinates of structures. To reduce the search space, probe radii are designed according to the physicochemical textures of molecules. The method uses the weighted Delaunay triangulation and the discrete flow algorithm to obtain geometric measurements and spatial patterns for each predicted pocket. It can also measure the hydrophobicity on a surface patch. Furthermore, we quantify the evolutionary conservation of surface patches by an index derived from the entropy scores in HSSP (homology-derived secondary structure of proteins). We have used the method to examine ∼1.16 million potential pockets and identified the split pockets in >26 000 structures in the Protein Data Bank. This integrated web server of functional surfaces provides a source of spatial patterns to serve as templates for predicting the functional surfaces of unbound structures involved in binding activities. These spatial patterns should also be useful for protein functional inference, structural evolution and drug design.
INTRODUCTION
Protein functions are usually fulfilled via molecular interactions on the protein surfaces. A functional surface of a protein is a local region where the protein interacts with ligands, substrates or proteins. Identifying the functional surfaces of a protein is therefore an important step toward understanding the binding properties of the protein. A large number of structures, including novel proteins, have been accumulated largely through the efforts of structural genomics projects (1). Most of these structures have been solved without knowing their binding regions and key residues involved in binding activities. Therefore, identifying the functional surfaces of proteins is an urgent task.
Recently, a variety of web servers or databases have been developed for locating regional surfaces involved in protein functions. These methods including 3D-SURFER (2), Q-SiteFinder (3), eF-site (4) and SURFACE (5) can identify regions of the protein that are potentially involved in biological activities. However, most of these methods use a fixed radius probe or heavily rely on heuristic algorithms like grid points or discretization. None of them comprehensively provides analytical geometric measurements such as length, solvent accessible area and molecular volume over specific radius probes. In contrast, our approach uses specific information on geometric, physicochemical and evolutionary characteristics for local surface assignments. Another major difference between our method and the others is that our approach is analytical in modeling the shape of a binding surface instead of modeling the shape of a binding ligand. The difference becomes important when there is a large size difference between the binding surface and the ligand.
In the method we developed recently (6), we work directly on the protein functional surfaces and study their spatial patterns in local regions associated with binding activities. Instead of using heuristic algorithms, we use the purely geometric concept of a split pocket, which is a pocket split by a binding ligand. This new approach uses an exact algorithm and does not require any data training. We compute detailed geometric measurements, using the Alpha Shape Theory (7–9), which is one of the most useful analytical approaches to describe molecular surfaces (10). Our finding indicates that a split pocket has a high propensity to be a surface that mediates molecular functions (6). In addition, we use customized probe radii to model the shapes of dynamic molecules, so that we can quickly eliminate trivial surface patches, thus expediting the process of surface partitioning. Importantly, these spatial patterns of functional surfaces can be applied to predict the binding sites of unbound forms by geometric matching (6). Furthermore, our method can be used to infer protein biological functions and to classify proteins; for an overview (6,11–13). SplitPocket provides spatial patterns related to protein biochemical functions and underlying physicochemical characteristics, which can be used to identify proteins with similar functions. The system is also applicable to other studies such as virtual screening in drug discovery, protein-surface classification and protein-structural evolution.
METHODS
Segmenting a protein with customized probes
Using the weighted-Delaunay triangulation, we partition a protein into local regions, such as protein cores and surfaces (14,15). In partitioning a protein into regions, we adopt the discrete flow algorithm (8,15,16) with customized probes (6) (solvent radii) to obtain the pockets on each individual structure. Specifically, the probe of a polar atom such as O, N or S is assigned a radius of 1.29 Å and that of an apolar atom such as C is assigned a radius of 1.96 Å, while the probe of the hydroxyl group is assigned a radius of 1.08 Å. For each pocket, we further obtain the length, solvent-accessible area and volume. For a mouth of a pocket, we are interested in the solvent accessible area and classify the mouth residues into hydrophobic and hydrophilic. In addition, the component residues of each pocket are concatenated into a linear string, called the pocket sequence. Basically, these residues provide a primary source of spatial patterns. In the SplitPocket system, the coordinates of a structure are the only input.
SplitPocket algorithm: detection of split pockets
A geometric concept was introduced to detect the functional surface(s) of a bound structure. A pocket is defined by a set of empty Delaunay triangles with at least one sink (an acute triangle) and can be detected by an exact algorithm. We first predict (i) all pockets in the structure with ligand(s) and (ii) all pockets in the same structure when the ligand(s) is removed (i.e. in the absence of ligand). We then compare these two sets of pockets. If there is a pocket that shows an altered composition, this pocket is split. The hint of atomic content change indicates that this pocket interacts with a ligand. Hence, a split pocket provides an intuitive geometric feature to detect protein binding sites. For a detailed description, see (6).
Computing the conservation index of a pocket
From the entropy measure of sequence variability, we first take the HSSP (homology-derived secondary structure of proteins at http://www.sander.embl-ebi.ac.uk/hssp/) (17) pre-computed conservation weights of all sites in a query structure. We derive surface conservation indices for evaluating the evolutionary conservation of protein surface patches. Specifically, we collect a set of conservation weights in spatial positions of a pocket to compute the conservation index by normalizing the length of the pocket sequence. Further, we obtain the site-specific, surface conservation index for each predicted pocket to assess its evolutionary conservation.
RESULTS
A case study
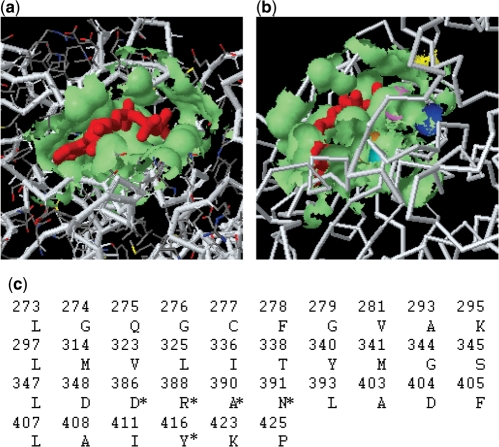
We now use a case study with the SplitPocket system to show the use of our web sever. The example is the human tyrosine–protein kinase (PDB2src): its functional surface is identified by detecting the ligand-binding site and its geometric measurements are also obtained (Figure 1). First, SplitPocket predicts 24 pockets on this kinase. Among them, the binding-ligand ANP (red) splits the pocket (the 23th) into smaller patches. In particular, this functional surface has a solvent-accessible area (681.01 Å2) and a molecular volume (986.38 Å3) that includes the ligand (363.50 Å3) (Figure 1a). Next, we extract the important residues on the wall of this functional surface and concatenate them into a pocket sequence of 36 residues (Figure 1c). Importantly, SplitPocket provides spatial information such as the catalytic residues on the binding surface (Figure 1b). In the present case, the key residues have previously been identified by experimental assays (18).
Figure 1.
Identification of the functional surface in human tyrosine–protein kinase PDB2src. (a) The binding site is split by a binding ligand (red). (b) The key residues responsible for biochemical reactions, such as D386 (blue), R388 (violet), A390 (orange), N391(cyan) and Y416 (yellow) on the functional surface, interact with the binding ligand. (c) Important surface residues are clustered on the protein binding site; the key residues are indicated with an asterisk.
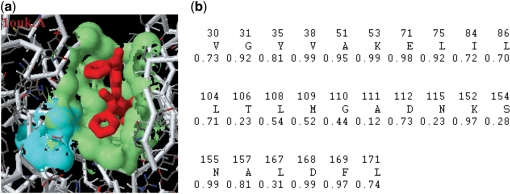
From the evolutionary perspective, it is informative to obtain the degree of conservation for a protein surface. In particular, the members of a protein family often carry out similar biochemical functions under various physicochemical constraints and selection pressures. Moreover, local structures such as functional surfaces tend to be evolutionarily more conserved than other regions of the protein (13). Therefore, we use the evolutionary conservation indices of the protein functional surfaces to characterize their spatial patterns. We take the human mitogen-activated protein kinase (PDB1ouk.A) to illustrate the conservation of a functional surface. The surface conservation index for the functional surface (the 22th pocket: 26 amino acids) is 0.703, which is the highest index among all of the putative pockets with >8 residues (Figure 2a). In comparison, the conservation index is 0.626 for the 23th pocket (the largest cavity: 33 amino acids), 0.299 for the 21th pocket (18 amino acids) and 0.377 for the 20th pocket (9 amino acids). In addition, the catalytic residues such as K152 (0.97) and N155 (0.99) are highly conserved but S154 (0.28) is not (Figure 2b). Our finding indicates that the conservation index is a useful evolutionary feature to distinguish a protein functional surface (binding site) from other local regions.
Figure 2.
Surface conservation index of the split pocket in the human mitogen-activated protein kinase (PDB1ouk.A). (a) The split pocket (the 22th pocket) has the highest conservation index (0.703). Catalytic residues {K152, S154, N155} colored in cyan are located on the protein binding surface. (b) The spatial pattern of (a) consists of 26 residues with conservation weights for assessing the degree of evolutionary conservation.
Collection of functional surfaces
A test of our method on a benchmark from Nissink et al. (19) showed that our method achieved a success rate of 96% (6). This encouraged us to compute the pockets of all X-ray structures (47 350 entries) in PDB and we have been able to find 47 950 entries (chains) from 26 213 structures each of which has at least one split pocket due to ligand binding. All the geometric measurements, surface conservation indices and spatial patterns are included in the SplitPocket system. These split pockets are useful templates for finding other similar spatial patterns of protein surfaces, particularly in homologous unbound structures (6).
WEB SERVER
SplitPocket has a friendly interface for users to obtain spatial information. The web sever is freely accessible at http://pocket.uchicago.edu/.
Input: PDB code
The SplitPocket system requires the coordinates of the protein structure under study in Protein Data Bank (PDB, http://www.rcsb.org/). However, a user needs to input only a PDB code, e.g. 2src. In addition, the system allows users to upload their specific coordinate files in the PDB format to perform a computational task.
Output: dynamic visualization
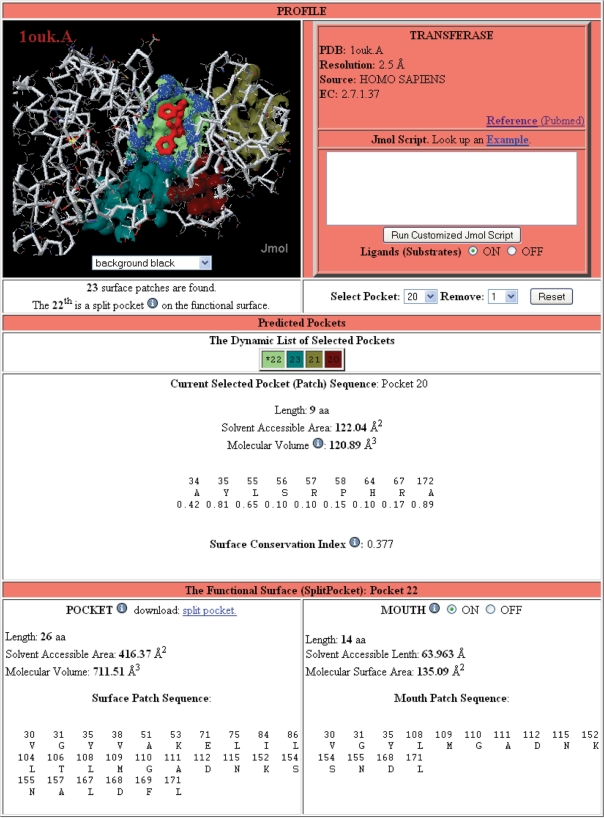
SplitPocket dynamically provides users with geometric measurements of protein pre-computed functional surfaces, including residue composition (spatial patterns), solvent-accessible area and molecular volume of their pockets and mouths. Figure 3 shows a standard output page from the SplitPocket server. Information of surface characteristics is provided at three levels of windows: Profile, Predicted Pockets and The Functional Surface. Note that a predicted pocket is a putative binding site and may not necessarily be a functional surface. In the Profile window, users can find basic structural information and a cross-reference link to the PubMed database. In addition, Splitpocket offers an interactive feature by Jmolscript command lines to manipulate users’ structures in a desired fashion. Users also can optionally use built-in functions (like buttons, a checkbox or menu) to interact with our system as long as the query structure is automatically loaded into a web browser. The corresponding changes will be dynamically reflected in both the Profile and the Predicted Pockets windows. In addition to geometric features, SplitPocket provides site-specific surface conservation indices to explore evolutionary characteristics. SplitPocket thoroughly assesses geometric, physicochemical and evolutionary characteristics of all putative pockets, and finally presents the protein-binding sites in the Functional Surface window.
Figure 3.
Sample functional surface page. In the example of the mitogen-activated protein kinase (PDB1ouk.A) from Homo Sapiens, SplitPocket predicts 23 pockets on the surface with customized probes. Along with the mouth (blue region), the 22th pocket (indicated with an asterisk) is the functional surface (light green) split by a ligand (084, red). Users can interact with the SplitPocket system and dynamically access the geometric and evolutionary measurements of spatial patterns on the pockets and their mouths.
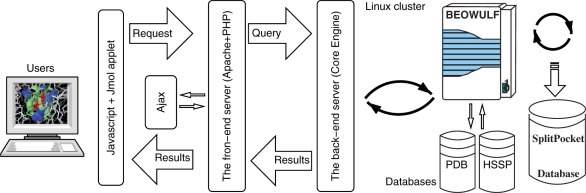
SplitPocket system architecture
SplitPocket incorporates several core techniques to allow users to interact with our system dynamically. Figure 4 shows the implementation of the SplitPocket system architecture. We separate the system into the front-end web server and the back-end core engine. The front-end web server is implemented in Hypertext Preprocessor (PHP) to spontaneously generate Javascript for end-users’ browsers. It allows users to interact with our system for presenting features of functional surfaces by Jmol applets (20). The back-end core engine consists of a 170-processor Beowulf Linux cluster and the databases of functional surfaces, PDB and HSSP. After structures are loaded, users’ requests are passed through the Asynchronous JavaScript + XML (Ajax) scheme to interactively accessing geometric and evolutionary information from the back-end server. Once the back-end server receives users’ requests, its core engine fulfills the designed tasks in a computational pipeline and returns results back to the front-end server after intensive calculations. The system is designed to efficiently utilize the Beowulf's computation power and maximize the capacity of nodes’ rapidest memory.
Figure 4.
SplitPocket system architecture.
SUMMARY
SplitPocket is a web server for identifying functional surfaces with customized probes. We study protein-functional surfaces to gain biological insights into protein structures and functions because protein spatial patterns and physicochemical textures are directly associated with geometrical and biochemical features. Major advantages of the SplitPocket system are: first, it is fully automatic and its only input is the PDB codes (e.g. 2src and 1ouk.A). Indeed, we analytically compute the binding surface solely on the basis of topological notions. Second, we show that functional surfaces have several characteristics such as spatial patterns and evolutionary conservation. Third, the spatial patterns serve as useful templates for predicting binding sites of unbound structures. Since the method is carried out automatically in a computational pipeline, we are able to construct an expandable database of functional surfaces for further shape analysis and binding-site prediction. These distinct features are included in the SplitPocket server.
FUNDING
National Institutes of Health (grant GM30998); Academia Sinica, Taiwan. Funding for open access charge: Academia Sinica, Taiwan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Robert M. Hanson at St. Olaf College for the implementation of Jmol isosurface. The authors also thank the two reviewers for valuable comments, Dr Jie Liang at the University of Illinois at Chicago for fruitful discussions and the cutting-edge development of CASTp.
REFERENCES
- 1.Binkowski TA, Joachimiak A, Liang J. Protein surface analysis for function annotation in high-throughput structural genomics pipeline. Protein Sci. 2005;14:2972–2981. doi: 10.1110/ps.051759005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sael L, Li B, La D, Fang Y, Ramani K, Rustamov R, Kihara D. Fast protein tertiary structure retrieval based on global surface shape similarity. Proteins. 2008;72:1259–1273. doi: 10.1002/prot.22030. [DOI] [PubMed] [Google Scholar]
- 3.Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferre F, Ausiello G, Zanzoni A, Helmer-Citterich M. SURFACE: a database of protein surface regions for functional annotation. Nucleic Acids Res. 2004;32:D240–D244. doi: 10.1093/nar/gkh054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng YY, Li WH. Identification of protein functional surfaces by the concept of a split pocket. Proteins. 2009 doi: 10.1002/prot.22402. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 7.Edelsbrunner H, Facello M, Fu P, Liang J. Measuring proteins and voids in proteins. Proc. Ann. Hawaii Int. Conf. Syst. Sci. 1995;5:256–264. [Google Scholar]
- 8.Edelsbrunner H, Facello M, Liang J. On the definition and the construction of pockets in macromolecules. Discrete Appl. Math. 1998;88:83–102. [PubMed] [Google Scholar]
- 9.Edelsbrunner H, Mucke E. Three-dimensional alpha shapes. ACM Trans. Graphics. 1994;13:43–72. [Google Scholar]
- 10.Albou LP, Schwarz B, Poch O, Wurtz JM, Moras D. Defining and characterizing protein surface using alpha shapes. Proteins. 2008 doi: 10.1002/prot.22301. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Binkowski TA, Adamian L, Liang J. Inferring functional relationships of proteins from local sequence and spatial surface patterns. J. Mol. Biol. 2003;332:505–526. doi: 10.1016/s0022-2836(03)00882-9. [DOI] [PubMed] [Google Scholar]
- 12.Tseng YY, Dundas J, Liang J. Predicting protein function and binding profile via matching of local evolutionary and geometric surface patterns. J. Mol. Biol. 2009;387:451–464. doi: 10.1016/j.jmb.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng YY, Liang J. Estimation of amino acid residue substitution rates at local spatial regions and application in protein function inference: a Bayesian Monte Carlo approach. Mol. Biol. Evol. 2006;23:421–436. doi: 10.1093/molbev/msj048. [DOI] [PubMed] [Google Scholar]
- 14.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: I. molecular area and volume through alpha shape. Proteins. 1998;33:1–17. [PubMed] [Google Scholar]
- 15.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sander C, Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 18.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 19.Nissink JW, Murray C, Hartshorn M, Verdonk ML, Cole JC, Taylor R. A new test set for validating predictions of protein-ligand interaction. Proteins. 2002;49:457–471. doi: 10.1002/prot.10232. [DOI] [PubMed] [Google Scholar]
- 20.Jmol: an open-source Java viewer for chemical structures in 3D. [(last accessed date February 20, 2009)]. http://www.jmol.org/