Abstract
Model quality estimation is an essential component of protein structure prediction, since ultimately the accuracy of a model determines its usefulness for specific applications. Usually, in the course of protein structure prediction a set of alternative models is produced, from which subsequently the most accurate model has to be selected. The QMEAN server provides access to two scoring functions successfully tested at the eighth round of the community-wide blind test experiment CASP. The user can choose between the composite scoring function QMEAN, which derives a quality estimate on the basis of the geometrical analysis of single models, and the clustering-based scoring function QMEANclust which calculates a global and local quality estimate based on a weighted all-against-all comparison of the models from the ensemble provided by the user. The web server performs a ranking of the input models and highlights potentially problematic regions for each model. The QMEAN server is available at http://swissmodel.expasy.org/qmean.
INTRODUCTION
In the course of protein structure prediction usually a set of alternative models is produced from which subsequently the final model has to be selected. For this purpose, scoring functions have been developed which aim at estimating the expected accuracy of models. These methods fall into two categories: The first category of scoring functions relies on the analysis of single models based on evolutionary (1,2) or physiochemical criteria, e.g. by comparing models with statistical properties of known structures (3–10). The second category derives a quality score from the information contained in an ensemble of models for a given sequence using an all-against-all structural comparison of the models. These so called clustering or consensus methods are based on the idea that conformations predicted more frequently are more likely to be correct than structural patterns occurring in only a few models (11–13). Both approaches have their advantages and the choice of the method depends on the situation and the availability of information.
The QMEAN server provides access to both kind of methods, giving the user the opportunity to choose between the composite scoring function QMEAN (6) (which stands for Qualitative Model Energy ANalysis) and the clustering method QMEANclust (32) building on it. As highlighted recently during the CASP blind-test experiment (14,15), correctly ranking different models for the same target protein is not a trivial task and structure prediction groups have considerable problems in ranking their own models. The QMEAN scoring function, which calculates both global and local (per-residue) quality estimates on the basis of single models, can be used to assist model selection and to identify problematic regions for subsequent refinement. The QMEANclust scoring function on the other hand needs a certain number of models and structural diversity within the model ensemble in order to work properly and may be used to estimate the quality and the local conformational diversity of multiple models. Such sets of models are for example obtained from meta-servers (16,17) (i.e. servers collecting and integrating results from multiple modelling servers) or as a result of extensive conformational sampling runs typically performed in fragment-based approaches (18,19). QMEANclust differs from other consensus methods such as Pcons (12) or the consensus method included in the ModFOLD server (20) in that it takes advantage of an initial ranking of the individual models obtained by QMEAN in order to weight the contribution of the models in the clustering process. This allows QMEANclust to circumvent inherent limitations of clustering methods and in some cases to identify good candidate models from the ensemble even if they are not part of most dominant structural cluster. The QMEAN scoring function differs from other publicly available model quality assessment servers operating on single models in its constituting terms: e.g. in comparison to the ModFOLD server (20) and the ProQ method (21), QMEAN additionally includes a more detailed distance-dependent all-atom interaction potential as well as a torsion angle potential over three consecutive residues. The individual terms of QMEAN are available in the output and their analysis may reveal possible explanations for the low score of a model, in contrast to the two above mentioned machine learning approaches which return a single score. QMEAN has been compared with a variety of state-of-the-art scoring functions (6) and both QMEAN and QMEANclust have been tested recently at CASP8 (32), where QMEAN was among the top performing non-consensus scoring functions, and QMEANclust showed good results for both global and local quality estimation (http://predictioncenter.org/casp8/).
THE QMEAN SERVER
Required input
The user has the possibility to either submit a single model (in PDB-format), or multiple models (as zip- or tar.gz-archive) and the full-length sequence of the target protein (which is needed for secondary structure and solvent accessibility prediction). In the case of multiple models, the models are mapped on the target sequence and automatically renumbered if necessary. A flag can be set in order to penalize incomplete models. The penalization of short models aims at obtaining a balance between quality of the models and coverage. By setting the flag the model score is additionally multiplied by the fraction of modelled residues with respect to the input sequence. The user can choose between one of the two scoring functions QMEAN and QMEANclust. By default, the QMEAN composite scoring function is selected since QMEAN is able to estimate the quality of single models and small sets of models whereas QMEANclust requires a certain number and diversity of models to work properly (see below).
QMEAN and QMEANclust scoring functions
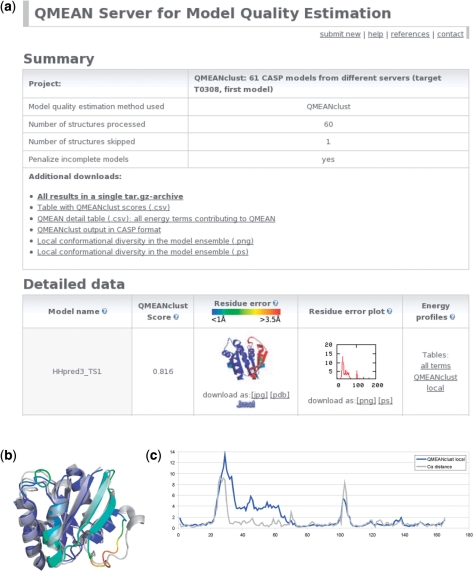
The QMEAN scoring function estimates the global quality of the models on the basis of a linear combination of six structural descriptors, four of them are statistical potentials of mean force: The local geometry is analysed by a torsion angle potential over three consecutive amino acids. Two distance-dependent interaction potentials based on Cβ atoms and all atoms, respectively, are used to assess long-range interactions. A solvation potential describes the burial status of the residues. Two terms reflecting the agreement between predicted and calculated secondary structure (22) and solvent accessibility (23) are included. A table containing for each model its QMEAN score and the values of the six contributing terms is included in the summary section of the results page (Figure 1a). These data allow the user to inspect differences between the models, which help understanding which terms contributed most to the low quality estimate of a certain model. The ranking of the models on the results page is based on the QMEAN or QMEANclust score which reflects the predicted global model reliability ranging from 0 to 1.
Figure 1.
Example results page for the 61 CASP7 server models of target T0308 together with local details for model HHpred3_TS1: (a) screenshot of the example results page with the summary section at the top and the first model from the details section. (b) Superposition of the best predicted model HHpred3_TS1 according to QMEANclust (colour-coded according to predicted local error based on QMEANclust local; from blue to red) and the experimental structure (in grey, PDB identifier 2h57). (c) Comparison of predicted error (in blue based on QMEANclust local) versus measured Cα distance between model and native structure (in grey).
The per-residue error estimates of QMEANlocal are based on a linear combination of eight terms smoothed over a sliding window of nine residues around the given amino acid. Local versions of the six terms used in QMEAN are combined with two additional terms which take into account the fact that the conformation of solvent exposed residues and residues outside regular secondary structure elements are potentially predicted less reliably. For each model, a table containing the QMEANlocal score together with all contributing terms is provided in the last column of the details section on the results page. A closer inspection of the terms per position may help to explain high energy peaks (e.g. as a consequence of unfavourable torsion angles or clashes).
The performance of clustering methods such as QMEANclust typically depends on the composition of the set of models to be assessed. Clustering methods have been shown to outperform physics-based energy functions in situation when the ensemble contains a variety of models from different sources covering a wide quality range such as given at CASP or at meta-servers. On the other hand, if the set of models does not include any good models or the best models are outliers, clustering methods are prone to fail (24). In order to counteract the limitations of pure clustering methods, QMEANclust combines clustering information with knowledge of the quality of the single models as estimated by QMEAN. Unlike for other clustering methods, in which all models contribute equally to the consensus score, QMEANclust incorporates knowledge of the quality of single models to weight each model in the consensus calculation. This combination allows the inherent limitations of pure consensus methods, which are designed to select models from the most highly populated structural cluster, to be circumvented.
The local error estimates by QMEANclust are derived in analogy to the global scores by analysing the local variability among the models based on pairwise superpositions. In a hierarchical approach, QMEAN is used to prioritize the models in the calculation of the QMEANclust score. The QMEANclust score is subsequently used to weight each model in the derivation of the local clustering scores. This means that models predicted to be more reliable contribute more in the calculation of the local score. This approach to local quality estimation has been shown to perform statistically significantly better than pure consensus methods as indicated by preliminary results from the CASP8 assessment (see assessment results by Anna Tramontano on the CASP8 website: http://predictioncenter.org/casp8/doc/presentations/CASP8_QA_Tramontano.pdf).
However, the absolute estimated per-residue error in Ångstrom as predicted by the two scoring functions has to be treated with caution. Statistical potentials in general are well-suited for identifying regions of small to medium deviations from ideal geometries whereas they are unable to discriminate between serious and very serious deviations (e.g. between 5 Å and 15 Å) both being geometrically incorrect. As a consequence, the residue error predicted by QMEANlocal rarely exceeds 5 Å. Nevertheless, the main purpose of the local quality estimation is to help identify potentially incorrect regions. For this purpose, the estimation of the relative local quality as provided by QMEAN is a good starting point.
The prediction of the absolute residue error with QMEANclust highly depends on the quality of the set of models in the ensemble (diversity, fraction and distribution of near native models in the set). For model ensembles containing useful structural density information, the predicted local error can be quite accurate as shown in the example, Figure 1b and c.
For each submitted model the predicted local (per-residue) error is displayed as line plot or as colour-coded PDB file with the local error in the B-factor column. The molecular graphics viewer Jmol (http://www.jmol.org/) can be directly used on the website to interactively inspect the problematic regions in the colour-coded structure. In the case of QMEANclust, the summary section additionally contains a visualization of the local conformational diversity within the ensemble of models. The plots (provided in two different formats) show the median QMEANclust score per position as a measure of diversity.
Calculation time of the method is typically on the order of a few minutes for small sets and is mainly limited by the time needed for predicting secondary structure and solvent accessibility from the sequence. For larger test sets, the clustering process in the QMEANclust mode is the time and memory determining step. The user can optionally specify an e-mail address; in this case a notification Email with a link to the results page as well as a download link to a tar.gz-archive containing all results is sent after completion.
EXAMPLE
The start page of the QMEAN server provides a link to an example results page which allows the user to inspect a typical output of the server. A snapshot of the example results page is given in Figure 1a. The example test set contains 61 models of the CASP7 experiment submitted by automated servers for the target sequence T0308, a 165 residue long target provided by the Structural Genomics Consortium (SGC). The first model of each server has been used here as indicated by the suffix TS1. The models of all previous CASP rounds are publicly available at http://predictioncenter.org/download_area/. In the example, the QMEANclust scoring function has been chosen to rank the models. From the 61 models contained in the set, 60 have been processed. One structure has been excluded by the server: it is a reduced model consisting only of Cα atoms which can not be handled by QMEAN since the torsion angle potential and the all-atom potential at least need the backbone atoms to be present.
Table 1 shows a comparison of the rankings based on the two scoring functions for the example model set. The best ten models are shown sorted according to their QMEANclust score in comparison to GDT_TS score (25), a well-established measure for the similarity between a model and the corresponding experimental structure. Some of the top models in the test set have a quite similar score, but the quality of the remaining models rapidly decreases with the majority of models having a GDT_TS between 70 and 80 (data not shown). The original performance data can be found on the website of the Prediction Center (http://www.predictioncenter.org/casp7/). The model HHpred3_TS1 ranked first by QMEANclust was the fifth best in the corresponding CASP7 ranking which corresponds to a marginal GDT_TS loss of 0.8 units compared to the best available model. The model on the second rank according to QMEANclust (PROTINFO_TS1) was the best model in the set. The QMEAN scoring function is able to recognize the best model in the given test set, but does not recognize all the top models. The two models SAM_T02 and UNI_EID_expm which are ranked poorly by QMEAN are both models without side chains and the latter additionally has several unfavourable torsion angles. Both structural features are not captured by the GDT_TS score which is based on Cα atoms only.
Table 1.
Comparison of model rankings based on QMEANclust and QMEAN for the 61 CASP7 server models of target T0308: from the CASP7 ranking, the top 10 models according to their similarity to the experimental structure (measured in GDT_TS units) are shown together with their rank obtained on the basis of the QMEANclust and the QMEAN scoring function
| Group | GDT_TS | QMEANclust rank | QMEAN rank |
|---|---|---|---|
| HHpred3 | 90.0 | 1 | 20 |
| PROTINFO | 90.8 | 2 | 1 |
| FOLDpro | 90.5 | 3 | 6 |
| BayesHH | 90.5 | 4 | 18 |
| HHpred2 | 90.0 | 5 | 7 |
| PROTINFO-AB | 88.6 | 6 | 5 |
| karypis.srv.2 | 89.2 | 7 | 22 |
| GeneSilicoMetaServer | 88.5 | 9 | 3 |
| SAM-T02* | 88.6 | 10 | 43 |
| UNI-EID_expm* | 87.9 | 11 | 50 |
| Median of 61 models | 75.1 |
Models are sorted by their QMEANclust score. The two models marked by asterisks are backbone-only models.
Figure 1b and c show a comparison of the predicted local error and the calculated local deviation of the model from the experimental structure (Cα distance) for the model ranked first by QMEANclust (HHpred3_TS1). The PDB identifier of the experimental structure, which was published after CASP7, is 2h57. Figure 1b shows a superposition of the model coloured according to the predicted residue error (from blue to red) and the experimental structure in grey. Regions in the model labelled by colours from the red part of the spectrum indeed correspond to residues deviating from the native conformation. The error plot given in Figure 1c shows that both regions with errors above 5 Å with respect to the native structure (grey line) were identified by QMEANclust. A scatter plot showing predicted versus calculated per-residue error for this example can be found in Supplementary Data (correlation coefficient r = 0.67).
CONCLUSIONS
Identifying the most accurate model among a set of alternatives is a crucial step in protein structure prediction. Here we present the QMEAN server which makes two methods for model quality estimation publicly available: QMEAN and QMEANclust. The QMEAN server addresses both users of protein structure models as well as method developers. The user of the web server has the possibility to either assess a single model or multiple models. Each model is assigned a quality score which is used to rank the structures. Additionally, a per-residue error estimate is provided and visualized in several ways allowing the user to inspect the protein models in more detail.
QMEAN can also be used for assessing individual models within SWISS-MODEL Workspace (26,27) together with other tools such as ProQres (28), ANOLEA (29) and WhatCheck (30). This allows the comparison of the quality estimation of the different approaches which potentially improves the reliability of the prediction. Models from the SWISS-MODEL Repository (31) can be directly sent to the quality estimation section of the Workspace.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Swiss Institute of Bioinformatics.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Chen H, Kihara D. Estimating quality of template-based protein models by alignment stability. Proteins. 2008;71:1255–1274. doi: 10.1002/prot.21819. [DOI] [PubMed] [Google Scholar]
- 2.Tress ML, Jones D, Valencia A. Predicting reliable regions in protein alignments from sequence profiles. J. Mol. Biol. 2003;330:705–718. doi: 10.1016/s0022-2836(03)00622-3. [DOI] [PubMed] [Google Scholar]
- 3.Sippl MJ. Boltzmann's principle, knowledge-based mean fields and protein folding. An approach to the computational determination of protein structures. J. Comput. Aided Mol. Des. 1993;7:473–501. doi: 10.1007/BF02337562. [DOI] [PubMed] [Google Scholar]
- 4.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 5.Melo F, Feytmans E. Novel knowledge-based mean force potential at atomic level. J. Mol. Biol. 1997;267:207–222. doi: 10.1006/jmbi.1996.0868. [DOI] [PubMed] [Google Scholar]
- 6.Benkert P, Tosatto SCE, Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Prot. Struct. Funct. Bioinformatics. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 7.Tosatto S. The victor/FRST function for model quality estimation. J. Comput. Biol.: J. Comput. Mol. Cell Biol. 2005;12:1316–1327. doi: 10.1089/cmb.2005.12.1316. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samudrala R, Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol. 1998;275:895–916. doi: 10.1006/jmbi.1997.1479. [DOI] [PubMed] [Google Scholar]
- 10.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 11.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 12.Lundstrom J, Rychlewski L, Bujnicki J, Elofsson A. Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Sci. 2001;10:2354–2362. doi: 10.1110/ps.08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuffin LJ. Benchmarking consensus model quality assessment for protein fold recognition. BMC Bioinformatics. 2007;8:345. doi: 10.1186/1471-2105-8-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction – Round VII. Prot. Struct., Funct. Bioinformatics. 2007;69:3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cozzetto D, Kryshtafovych A, Ceriani M, Tramontano A. Assessment of predictions in the model quality assessment category. Proteins. 2007;69(Suppl. 8):175–183. doi: 10.1002/prot.21669. [DOI] [PubMed] [Google Scholar]
- 16.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallner B, Fang H, Elofsson A. Automatic consensus-based fold recognition using Pcons, ProQ, and Pmodeller. Proteins. 2003;53(Suppl. 6):534–541. doi: 10.1002/prot.10536. [DOI] [PubMed] [Google Scholar]
- 18.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuffin LJ. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008;24:586–587. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- 21.Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 23.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallner B, Elofsson A. Prediction of global and local model quality in CASP7 using Pcons and ProQ. Proteins. 2007;69(Suppl. 8):184–193. doi: 10.1002/prot.21774. [DOI] [PubMed] [Google Scholar]
- 25.Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 27.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 28.Wallner B, Elofsson A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 2006;15:900–913. doi: 10.1110/ps.051799606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melo F, Devos D, Depiereux E, Feytmans E. ANOLEA: a www server to assess protein structures. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:187–190. [PubMed] [Google Scholar]
- 30.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 31.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benkert P, Tosatto S, Schwede T. Global and local model quality estimation at CASP8 using the scoring functions QMEAN and QMEANclust. Proteins. 2009;(Suppl) doi: 10.1002/prot.22532. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.