Abstract
‘Protinfo PPC’ (Prediction of Protein Complex) is a web server that predicts atomic level structures of interacting proteins from their amino-acid sequences. It uses the interolog method to search for experimental protein complex structures that are homologous to the input sequences submitted by a user. These structures are then used as starting templates to generate protein complex models, which are returned to the user in Protein Data Bank format via email. The server supports modeling of both homo and hetero multimers and generally produces full atomic level models (including insertion/deletion regions) of protein complexes as long as at least one putative homologous template for the query sequences is found. The modeling pipeline behind Protinfo PPC has been rigorously benchmarked and proven to produce highly accurate protein complex models. The fully automated all atom comparative modeling service for protein complexes provided by Protinfo PPC server offers wide capabilities ranging from prediction of protein complex interactions to identification of possible interaction sites, which will be useful for researchers studying these topics. The Protinfo PPC web server is available at http://protinfo.compbio.washington.edu/ppc/
INTRODUCTION
Every biological process in a living cell is mediated by the interaction between proteins. Understanding the mechanism of these interactions is necessary to unravel the complexity of the biological systems. A large volume of experimental data that provides a partial picture of the cellular protein interaction networks has been generated by high throughput technologies such as yeast two-hybrid systems (1) or tandem affinity purification (2). However these laboratory approaches reveal only the interacting protein pairs and do not provide atomic level detail on how these interactions occur. Alternatively, atomic resolution structures of multimeric protein complexes solved by X-ray diffraction and/or NMR spectroscopy provide a wealth of important molecular insights into the functional mechanisms of protein interactions. However, the generation of this unique biological information is extremely tedious and labor intensive, and still not possible for most protein complexes (3). Complementary computational methods that are capable of extrapolating the existing structure data to predict three-dimensional (3D) structures of other protein complexes are therefore necessary and useful to bridge this data gap.
Computational methods for predicting 3D structures have been extensively explored over the past decade and can generally be classified into two categories, which are template free modeling (de novo prediction) (4) and template based modeling (threading and comparative modeling) (5). In particular, the possibility of using comparative modeling to predict the structures of multimeric protein complexes has been recently investigated along with the lines pioneered by Chothia and Lesk (6–8). These studies show that the sequence and structural similarity principal holds true for most of multimeric protein complexes suggesting that it is possible to extend comparative modeling protocols to predict protein complex structures (7,8). A number of web servers that utilize the experimental protein complex structures to predict interactions between a pair of protein sequences and/or suggest interacting partners of an input protein sequence exist, such as InterPreTS (9), 3D-partner (10) and HOMCOS (11).
Here, we present Protinfo PPC, a server for predicting 3D atomic level structures of protein complexes from their amino-acid sequences. The server generates 3D models based on our multimeric comparative modeling protocol. Briefly, this involves using the interolog method (12) to search for experimental protein complex structures that are homologous to the target sequences. These structures are used as templates to generate the protein complex models, which are energy minimized and returned to the user in Protein Data Bank (PDB) format via email. The Protinfo PPC server differs from other related tools in that it uses a combination of template based and template free approaches to predict protein complex models. In addition, it natively supports comparative modeling of both homo and hetero multimers of up to five sequences and generally produces full atomic level 3D models (including insertion/deletion regions) of protein complexes as long as at least one putative homologous template for the target sequences is found. We have also performed a rigorous assessment to benchmark the structural and interface accuracy of protein complex models produced by our method. Each model is accompanied by structure and interface confidence scores as well as several parameters that are useful in assessing the reliability of each prediction. Finally, a list of interacting residues is provided with each model for an easy identification of residues that are mediating the protein complex interaction.
METHODS
Modeling of protein complexes
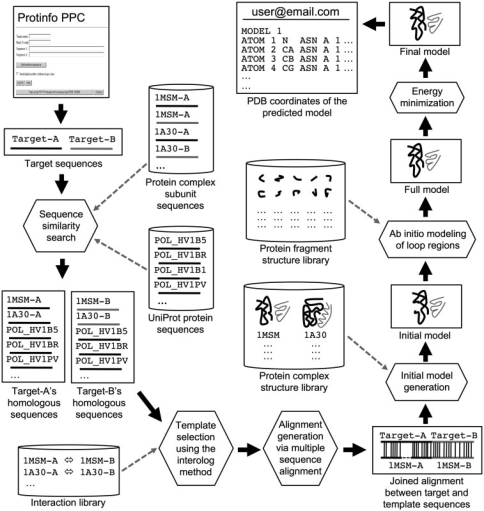
Protein sequences submitted to the Protinfo PPC server are sent to our multimeric comparative modeling pipeline as depicted by the flowchart in Figure 1. The modeling process starts with the comparison of each target sequence to our protein complex subunit sequence database using two similarity search tools, PSI-BLAST (13) and SSEARCH (14) The BLOSUM62 matrix is used and details about each database are provided below. All significant hits (as defined by e-value <0.01) from both tools are combined to produce a non-redundant set of protein complex subunits that are homologous to each target sequence. Possible protein complex templates are then selected by scanning through each target's ‘hits list’ for cases where individual subunit of a protein complex template is homologous to each target sequence. This is analogous to the interolog method (12).
Figure 1.
Flow chart depicting the Protinfo PPC multimeric comparative modeling procedure. The Protinfo PPC webpage serves as a portal that enables users to submit their sequences to our multimeric comparative modeling pipeline. The web page is made up of basic hypertext markup language (HTML) and javascript to maximize browser compatibility. The modeling pipeline is composed of several programs and scripts written in C, Shell scripting and the Perl programming language.
A pairwise sequence alignment between targets and each protein complex template is generated through a multiple sequence alignment. This involves independently using the ClustalW program (15) to generate a multiple sequence alignment between each target sequence and its homologs from the protein complex subunit sequence database. Additional homologous sequences of each target from the Uniprot database (16) are also added to the multiple sequence alignment input file to improve the alignment result. The pairwise alignment between the targets and each protein complex template is then extracted directly from the multiple sequence alignments and are concatenated based on the protein chain order presented in the template PDB file to produce a single ‘joined’ pairwise alignment. The joined alignment allows our server to model all target sequences as a single protein complex and take into account the effect of atoms involved in protein–protein interactions. In particular, it prevents our method from generating protein complex models that contain atom clashes especially those that are in the interaction sites.
The protein complex ‘initial’ models are generated based on the alignment results by using the following procedure. The main chain coordinates of residues in the target that are similar (alignable) to the template are copied over from the template PDB file. Similarly, the side chain coordinates are also copied over if the aligned residues are conserved (identical). For the substituted residues (non identically aligned residues), our χ angle equivalence matrix method is used to predict the side chain's coordinates (17). The remaining residues that are in the insertion and deletion (indel) regions (as specified by dashes in the alignment) are left unmodeled at this stage. Finally, the side chains of the initial models are repacked by using the side chains with a rotamer library (SCWRL) program (18). After all initial models are generated, they are scored by using both simple sequence similarity/identity metric and our residue-specific all atom conditional probability scoring function (RAPDF) (19). The top five models (based on the combined sequence and structure scores) are then selected for the subsequent modeling of indel regions using our de novo methods.
An exhaustive search of possible main chain conformations that fit best to the given indel region (20) or, alternately, a segment matching technique (21) is used to model residues in the indel regions depending on the size of the indel. Specifically, if the indel size is less than 10 residues, our method generates a number of loop conformations for a given region by exhaustively enumerating all possible main chain conformations for that indel using a discrete n-state φ/ψ model and selecting ones that fit best to the given indel region in the initial model, as measured by our all atom (RAPDF) scoring function (20). We limit the usage of this method to small indels due to its computationally intensive nature. For larger indel regions, we applied our segment matching and folding technique to generate possible indel conformations. This method is based on inserting small (three residues) fragments randomly and using a Monte Carlo/simulated annealing procedure to find combinations of these fragments that have the best score (21). After all indel regions have been modeled, the Energy Calculation and Dynamics (ENCAD) method (22) is used to energy optimize or ‘relax’ the full protein complex models. Finally, remarks describing various scores and parameters, such as structure/interface confident, sequence and structure scores, indel regions and interacting residues, are added into the final model PDB files before they are sent to the user via email.
Database construction
Protein complex template library
The biological units from the PDB (23) that are made up of at least two protein chains, each of which contains more than 10 interacting residues, are used to generate our protein complex template library. PDB biological units are macromolecular structures that have been shown to be or are believed to be the functional version of the corresponding monomeric units. We defined interacting residues to be those that have at least one atom of any type that is closer than 5 Å to another atom of another residue from a different polypeptide chain. Since some protein complexes can exhibit different interaction/binding modes, we do not discard templates that share high sequence similarity because some of them represent biologically relevant alternative conformational states or binding modes that can occur as a result of events such as evolution, point mutations, binding of different ligand, flexibility, and/or altered experimental conditions (24). Having these complex structures in our template library allows our modeling pipeline to generate models with diverse conformations and provide information about possible effects of the environment on protein complex of interest. Currently, there are over twenty thousand protein complex templates that the Protinfo PPC server can use for modeling. The protein complex template library is regularly updated.
Protein complex subunit sequence database
The protein complex subunit sequence database is generated by extracting the amino-acid sequences from every chain of every protein complex template in our structure library. Specifically, the sequences are taken from the ATOM records of each template PDB file. We do not use sequences from the SEQRES records because some of the residues may have missing 3D coordinates due to technical problem or lack a fixed tertiary structure (25). A PSI-BLAST database is finally generated from these protein complex subunit sequences using the NCBI-BLAST package (26).
Interaction library
The protein–protein interaction library is generated from the chain information in each template PDB file. For example, an interaction between 1msm-A sequence and 1msm-B sequence is derived from a PDB template 1msm, which is a dimer complex consisting of chain A and B. This information is used when we search for all possible protein complex structure templates for the given target sequences based on the assumption that most homologous proteins are likely to have similar interactions.
Accuracy assessment and expected accuracy measures
The modeling pipeline underlying the Protinfo PPC server has been rigorously benchmarked to assess its ability to produce models that are accurate both in their structures and interface regions. Specifically, the structural accuracy is measured using the all atom root mean square deviation (RMSD) between the predicted model and the corresponding experimental structure. The interface accuracy is defined by the percentage of correct interacting residues (according to the known structure) in the models. The benchmark results in Figure 2 show that the vast majority of the predicted models (a total of 38 463 protein complex models predicted for 10 707 dimer targets) are extremely accurate, both in their structure and interface. The median all atom RMSD and percentage of correct contact residues across 38 463 protein complex models (>3 models per target) are 3.2 Å and 89% respectively (Figure 2A and Supplementary Figure S1). The predicted models at this level of accuracy can potentially provide useful insights into the functional and mechanistic details of the protein complexes. In addition, the extremely accurate interacting residues predicted in these models will provide useful information to guide experiments that focus on the interfaces of protein complexes. We found that the indel regions, which are modeled by a different procedure, only slightly increased the overall RMSD of the models (Supplementary Figure S2). This suggests that the structural accuracy is largely dependent on the identification of correct templates. Interestingly, we also found that higher percentage of hetero dimer targets are more accurately modeled relative to the homo dimer targets (Supplementary Figure S3). This may be because it is harder to pick an incorrect template for hetero dimer targets whereas mistakes in selecting the template for homo dimers can be more costly as it will double the error.
Figure 2.
Structure and interface accuracy assessment of our multimeric comparative modeling method. The results show that the vast majority of the protein complex models predicted by our method are extremely accurate, both in their (A) overall structure and (B) interface residues. (C) A sample predicted model produced by the Protinfo PPC server showing highly accurate overall structure (cartoon representation) and interface accuracy (stick representation colored in red).
In addition to pipeline benchmarking, the predicted models were used to derive expected accuracy measures based on the percentage of sequence identity, normalized all atom RAPDF score, total insertion/deletion length, and target/template length ratio as summarized in Figure 3 and Supplementary Figure S4. The users of the Protinfo PPC server can use these parameters, which will be provided with each prediction, to assess the reliability of the predicted models. For instance, a model with a percentage of sequence identity to the template of 85% has about 60% likelihood of being less than 5 Å all atom RMSD from the correct structure. Similarly, the same model has about 80% likelihood that more than 75% of all interacting residues in the models are correct. For the current version of Protinfo PPC server, data in Figure 3 are used to provide the structure and interface confidence scores.
Figure 3.
Expected accuracy based on the percentage of sequence identity between targets and templates. 38 463 predicted protein complex models were used to derive expected (A) structure and (B) interface accuracy. The structure confidence score is calculated by mapping each model's identity score to the likelihood that the model is less than 10 Å all-atom RMSD to the native structure. Likewise, the interface confident score is calculated by mapping the identity score to the likelihood that more than 50% of the interacting residues are correct.
USING THE PROTINFO PPC SERVER
The Protinfo PPC server web page is created using the Hyper Text Mark up Language (HTML) with minimal embedded javascript to ensure maximum compatibility with all web browsers. The server supports modeling of both homo and hetero multimer protein complexes of up to five sequences. In addition, the server allows users to upload a custom protein complex template for modeling of their target sequences. This provides flexibility in the case where a user's template of interest does not exist in our library. Sections below explain the required input formats as well as different ‘remarks’ in a prediction result.
Input format
The Proinfo PPC server requires the user to enter the target amino-acid sequences into separate input boxes. The submitted sequences can be in FASTA format or the amino-acid sequences without the sequence identifiers. A unique identifier will automatically be assigned to each amino-acid sequence, such as ‘TargetA’, ‘TargetB’, according to the sequence order submitted. A name and an email address must be provided with the submission for job identification and result delivery. Since modeling a large protein complex that consists of several protein chains usually takes a considerable amount of time, a maximum of five sequences are allowed. Users who are interested to use our protocol to model a protein complex that is made up of more than five chains are encouraged to contact us directly so separate resources can be allocated appropriately. Users can instruct the Protinfo PPC server to mark interacting residues in the output PDB files by selecting ‘Mark interacting residues using temperature factor column’ check box. The value in the temperature factor column will be 99.99 if the residue is interacting with other residues (based on our criteria described above) and 0.00 otherwise. The server also provides users with the option to receive the coordinates of the corresponding initial models that were used to create the final models. Finally, the server accepts a custom template that conforms to all PDB standards and contains the same number of chains to the number of sequences submitted. The uploaded PDB file will be preprocessed by the server before it is used to model the target sequences. In the case where the server failed to use the uploaded file, a message explaining the error will be sent to the user's email.
Output format
The Protinfo PPC server emails the resulting protein complex models in a standard PDB CASP format. For each submitted job, a maximum of five complex models will be returned (each in a separate email). The amount of time needed to model each submitted job generally depends on the total length of the target sequences where larger targets require more time (Supplementary Figure S5). The PDB text can easily be saved into a PDB file using any text editor program and the predicted structure can be visualized using a visualization tool such as PyMOL (http://www.pymol.org). The ‘REMARK’ lines describing various model confidence scores and parameters, such as the percentage of sequence identity between template and target, the normalized all atom RAPDF score, the template-target map, the total length of insertion/deletion and insertion/deletion regions, are provided with each complex model (Figure 4A–E). As described above, some of these parameters can be mapped back to our expected accuracy analyses to derive the confidence level of the predicted models (Figure 3 and Supplementary Figure S4). In addition to model parameters, a summary of interacting residues is also provided in the REMARK section of each predicted model as a tab delimited list where the first column shows the residue on one protein chain and the remaining columns are other residues on other protein chains that it is interacting with (Figure 4F). This unique information can be used to suggest residues that are mediating the protein complex interaction.
Figure 4.
A sample PDB output from the Protinfo PPC server. The predicted protein complex models are returned to the user in PDB format via email. (A) Structure and interface confidence scores are provided with each prediction. Each model is also accompanied by several parameters, such as (B) sequence identity scores, (C) total target/template length ratio, (D) total length of insertion/deletion regions and (E) List of all insertion/deletion regions, which are useful in assessing the reliability of the predictions. (F) A summary of interacting residues is also provided as a tab delimited list where the first column represents a residue on one protein chain and the remaining columns shows other residues on other protein chain that it is interacting with. This information can be used to suggest residues that are mediating protein complex interactions.
While previous studies have shown that protein complexes with similar sequence tend to have similar structure or binding modes, completely different interaction topologies or large conformational changes have also been reported between complexes that share high sequence similarity. We have done an all against all sequence and structure comparison to identify these biologically relevant alternate conformation templates (data not shown). A remark will be provided within the output PDB text to inform the user when the predicted model is created from such templates (Supplementary Figure S6).
LIMITATIONS AND FUTURE WORK
Currently, the ability for the Protinfo PPC server to produce a prediction is limited to whether a suitable protein complex template is available in our protein complex library or from the user. However the coverage will only increase when more protein complex structures are solved and deposited to the PDB. Enhancements to our modeling pipeline planned for the near future include the support for the use of different parts of a larger protein complex to model smaller similar complexes. In addition, we are also exploring the possibility of using structures of large multi-domain, single chain proteins to model protein complexes that are made up of the interaction between similar domains, which is analogous to the gene fusion method for sequence based protein–protein interaction prediction. Finally, we are working on a better scoring function that takes into account all parameters of each prediction and their corresponding likelihood to provide a combined confidence score that can be used to assess the reliability of the predicted model.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Searle Scholar Award; a National Science Foundation CAREER award; National Science Foundation [DBI-0217241 to R.S.]; and National Institution of Heath [GM068152-01 to R.S.]. NIH-NIDCR 5R01DE012212 [to R.E.B. and W.K.]. NIH-NCRR 5R24RR021863 [to R.E.B.]. Funding for open access charge: a National Science Foundation CAREER award and NIH-NIDCR 5R01DE012212.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank members of the Samudrala group for helpful discussions and comments.
REFERENCES
- 1.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 3.Service R. Structural biology. Structural genomics, round 2. Science. 2005;307:1554–1558. doi: 10.1126/science.307.5715.1554. [DOI] [PubMed] [Google Scholar]
- 4.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69 (Suppl. 8):57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 5.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69 (Suppl. 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 6.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J. Mol. Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Launay G, Simonson T. Homology modelling of protein-protein complexes: a simple method and its possibilities and limitations. BMC bioinformatics. 2008;9:427. doi: 10.1186/1471-2105-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aloy P, Russell RB. InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics. 2003;19:161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 10.Chen YC, Lo YS, Hsu WC, Yang JM. 3D-partner: a web server to infer interacting partners and binding models. Nucleic Acids Res. 2007;35:W561–W567. doi: 10.1093/nar/gkm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuhara N, Kawabata T. HOMCOS: a server to predict interacting protein pairs and interacting sites by homology modeling of complex structures. Nucleic Acids Res. 2008;36:W185–W189. doi: 10.1093/nar/gkn218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Gen. Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 16.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samudrala R, Huang ES, Koehl P, Levitt M. Constructing side chains on near-native main chains for ab initio protein structure prediction. Prot. Eng. 2000;13:453–457. doi: 10.1093/protein/13.7.453. [DOI] [PubMed] [Google Scholar]
- 18.Bower MJ, Cohen FE, Dunbrack R.L., Jr Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J. Mol. Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 19.Samudrala R, Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol. 1998;275:895–916. doi: 10.1006/jmbi.1997.1479. [DOI] [PubMed] [Google Scholar]
- 20.Samudrala R, Moult J. A graph-theoretic algorithm for comparative modeling of protein structure. J. Mol. Biol. 1998;279:287–302. doi: 10.1006/jmbi.1998.1689. [DOI] [PubMed] [Google Scholar]
- 21.Samudrala R, Levitt M. A comprehensive analysis of 40 blind protein structure predictions. BMC Struct. Biol. 2002;2:3. doi: 10.1186/1472-6807-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levitt M, Hirshberg M, Sharon R, Daggett V. Potential energy function and parameters for simulations of the molecular dynamics of proteins and nucleic acids in solution. Comput. Phy. Commun. 1995;91:215. [Google Scholar]
- 23.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlessman JL, Woo D, Joshua-Tor L, Howard JB, Rees DC. Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum. J. Mol. Biol. 1998;280:669–685. doi: 10.1006/jmbi.1998.1898. [DOI] [PubMed] [Google Scholar]
- 25.Brandt BW, Heringa J, Leunissen JA. SEQATOMS: a web tool for identifying missing regions in PDB in sequence context. Nucleic Acids Res. 2008;36:W255–W259. doi: 10.1093/nar/gkn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.