Abstract
T cell diversity posttransplant is thought to be severely restricted, based on T cell receptor β chain immunophenotyping or spectratyping. Using β chain sequencing, we studied CD4 T cell diversity in two adult patients undergoing “lymphoablative” conditioning with cyclophosphamide, total body irradiation and antithymocyte globulin and autologous transplantation of hematopoietic cells depleted of T cells by enrichment for CD34 cells. The indication for the transplant was systemic sclerosis or multiple sclerosis. Pretransplant, the estimated number of distinct β chains (the minimum number of CD4 T cell clones in the person) was 600,000 to 700,000, which was similar to a healthy control. It was 200,000 to 500,000 at one month and 400,000 to 1,600,000 at twelve months posttransplant. In conclusion, T cells early after lymphoablative conditioning and autologous CD34 cell transplantation may be more diverse than previously appreciated, possibly because many T cell clones survive the conditioning or are reinfused with the graft. Thus, the therapy may not be completely T-lymphoablative.
Introduction
Diversity of T cells posttransplant has been studied primarily by the flow cytometric detection of T cell receptor β chain families (with variable region-specific monoclonal antibodies) or by spectratyping, ie, electrophoretic detection of various lengths of the β chain complementarity determining region 3 (CDR3) segments of rearranged β chain genes.1-17,18,19-21 With both techniques, the repertoire of posttransplant T cells appears skewed, ie, some T cell clones appear overrepresented while others appear absent. This is particularly prominent in the first several months posttransplant and in patients having received T cell-depleted grafts (eg, in vivo with antithymocyte globulin or ex vivo by CD34 cell enrichment of the graft). Normalization of the flow cytometric or spectratypic findings in adults typically takes at least 2 years.14,22-25 This suggests that in the first several months posttransplant the diversity of T cells is severely restricted, and that it takes at least 2 years to restore the diversity to normal. However, the skewed Vβ representation by flow cytometry or spectratyping may also reflect expansion of certain T cell clones rather than restricted T cell diversity.26 The expanded clones may make the detection of non-expanded clones difficult or impossible, especially if specimens contain only a small number of T cells. Here we used a sequencing-based method and specimens containing a large number of T cells (obtained by apheresis) to estimate the number of distinct β chains (surrogate of T cell diversity).
Subjects and Methods
Three individuals were studied: a 46-year-old female with systemic sclerosis (SSc) undergoing autologous CD34 cell transplantation using conditioning with cyclophosphamide (120 mg/kg), total body irradiation (TBI) (8 Gy) and antithymocyte globulin (ATG) (90 mg/kg),27 a 28-year-old male with multiple sclerosis (MS) undergoing autologous CD34 cell transplantation using the same conditioning,28 and a healthy 44-year-old male (control). The two patients were selected from a cohort of 56 patients undergoing transplantation for SSc or MS based on their willingness to undergo research apheresis at 1 and 12 months posttransplant. As the transplantation was performed for autoimmune diseases, it was designed to be maximally lymphoablative. Thus, not only cyclophosphamide, TBI and ATG were used, but also the autologous graft was enriched immunomagnetically for CD34 cells. Typical composition of the graft using this protocol was 261×106 CD34 cells, 2×106 CD4 T cells and 1×106 CD8 T cells.29
Immune reconstitution of the whole cohort has been described in detail.29 The immunological studies of the two patients were representative of the whole cohort in that their CD4 T cell counts were very low at 1 month and moderately low at 12 months (Table 1, 1st column), and their CD4 as well as CD8 Vβ-Jβ spectratypes showed a lower-than-normal median number of peaks both at 1 and 12 months posttransplant (data not shown). For the studies of posttransplant T cell diversity, 8-liter mononuclear cell apheresis (Cobe Spectra) was performed at 1 and 12 months posttransplant. For the SSc patient, pretransplant T cell diversity was also studied, using cells obtained as the flow-through (CD34-negative) fraction during the immunomagnetic selection of CD34 cells from the autologous graft (apheresis product). The flow-through fraction of the graft was unavailable for the MS patient.
Table 1.
Determination of CD4 T cell diversity in a healthy control and two patients.
| Subject (time-point) [CD4 T cells per μl blood*] | No. of Vβ17+ CD4+ T cells analyzed | Rearrangement | Frequency of Vβ17 gene (%) | Frequency of Jβ (1.4, 1.5 or 2.6) gene (%) | Peak size (%) | No. of distinct sequences within the peak(s) | Diversity (×106) |
|---|---|---|---|---|---|---|---|
| Healthy control | 105 | Vβ17-Jβ1.5 | 1.6 | 13 | 18.5 | 140 | 0.4 |
| 106 | Vβ17-Jβ1.4 | 1.6 | 2.9 | 20.2 | 60 | 0.6 | |
| 105 | Vβ17-Jβ1.4 | 1.6 | 2.9 | 20.2 | 60 | 0.6 | |
| SSc patient (pretransplant) | 106 | Vβ17-Jβ1.4 | 1.0 | 0.6 | 35.1 | 12 | 0.7 |
| 105 | Vβ17-Jβ1.4 | 1.0 | 0.6 | 35.1 | 11 | 0.6 | |
| SSc patient (1 mo posttransplant) [CD4 = 26] | 105 | Vβ17-Jβ1.4 | 0.7 | 6.7 | 100.0 | 12 | 0.3 |
| 105 | Vβ17-Jβ2.6 | 0.7 | 2.0 | 100.0 | 25 | 0.2 | |
| SSc patient (12 mo posttransplant) [CD4 = 347] | 105 | Vβ17-Jβ1.4 | 1.5 | 2.1 | 100.0 | 16 | 0.5 |
| 105 | Vβ17-Jβ2.6 | 1.5 | 1.1 | 100.0 | 70 | 0.4 | |
| MS patient (1 mo posttransplant) [CD4 = 5] | 105 | Vβ17-Jβ1.4 | 0.5 | 3.3 | 100.0 | 74 | 0.4 |
| 105 | Vβ17-Jβ2.6 | 0.5 | 1.7 | 100.0 | 45 | 0.5 | |
| MS patient (12 mo posttransplant) [CD4 = 186] | 105 | Vβ17-Jβ1.4 | 0.5 | 2.8 | 100.0 | 230 | 1.6 |
| 105 | Vβ17-Jβ2.6 | 0.5 | 1.8 | 100.0 | >112** | >1.2 |
5th-95th percentile reference range, 416-1437/μl.
Plateau of the curve showing the number of distinct clones vs the number of clones sequenced had not been reached by the time 180 clones were sequenced.
Clinically, the underlying disease was at least partially stabilized. The SSc patient’s Rodnan skin score (RSS) was 48 pretransplant, 39 at one year, 32 at two years and 42 at four years posttransplant, and lung function (limited pretransplant) was stable for four years posttransplant. The MS patient’s extended disability status scale (EDSS) was 2.0 at one year pretransplant, 7.0-7.5 immediately pretransplant, 8.0 at one, two and three years posttransplant and 7.5 at four years posttransplant. There were no new T2 magnetic resonance imaging lesions on all available determinations (3, 6 and 12 months posttransplant). The study was Institutional Review Board-approved.
For the estimation of the number of distinct β chains, the method of Arstila et al. was used,30 with minor modifications. The focus was on CD4 T cells, as they appear to play an important role in the pathogenesis of SSc and MS and their quantitative deficiency appears to play an important role in transplant recipients’ susceptibility to infections.31-34 The frequency of cells utilizing Vβ17 among CD4 T cells was determined by flow cytometry, using Vβ17 antibody-fluorescein conjugate (Beckman-Coulter/Immunotech) and CD4 antibody-phycoerythrin conjugate (BD Biosciences) (Table 1, 4th column). To maximize sensitivity and specificity of the downstream PCR, Vβ17+CD4+cells were sorted to >95% purity using FACS Vantage (BD Biosciences). After centrifugation, the cells were resuspended into 100 μl solution D, prepared by mixing 7 μl β-mercaptoethanol (14.2 M) with 100 μl Lysis Buffer from Absolutely RNA Microprep kit (Stratagene). RNA was extracted using the same kit, and cDNA was synthesized using Superscript II RNase H- Reverse Transcriptase Kit (Invitrogen) and Oligo(dT)12-18 primers (Invitrogen). Each reverse transcription yielded 60 μl cDNA. The cDNA was used for real-time PCR to determine the frequency of cells utilizing Jβ1.4 (or Jβ1.5 or Jβ2.6) (Table 1, 5th column), for spectratyping to determine the size of the CDR3 peak(s) of interest (Table 1, 6th column), and for gel electrophoresis for cloning and sequencing to determine the number of different sequences within the peak(s) (Table 1, 7th column). For the details of the determinations of the frequency of cells utilizing Jβ1.4 (or Jβ1.5 or Jβ2.6), the size of the CDR3 peak(s) of interest and the number of different sequences within the peak(s), see the next 3 paragraphs. Diversity (total number of distinct Vβ chains among CD4 T cells, ie, the minimum number of CD4 T cell clones – Table 1, last column) was calculated as
As the determination of the diversity gave a similar result when 105 or 106 sorted Vβ17+CD4+ cells were used, for most determinations 105 cells were used (Table 1, 2nd column). Note that per the ImMunoGeneTics nomenclature,35 Vβ17 is TRBV19, Jβ1.4 is TRBJ1-4, Jβ1.5 is TRBJ1-5 and Jβ2.6 is TRBJ2-6.
The frequency of cells utilizing Jβ1.4 (or Jβ1.5 or Jβ2.6) was determined by real-time PCR as follows: Forward primer was AAGGGTACAGCGTCTCTCGG (Vβ17), reverse primer was AGACAGAGAGCTGGGTTCCA (Jβ1.4), CAGCATTTTGGTGATGGGAC (Jβ1.5), ACTTTCGGGGCCGGCAGCAG (Jβ2.6) or GAGGTCGCTGTGTTTGAGCC (C), and probe was FAM- TCCTTTCCTCTCACTGTGACATCGGC-TAMRA (MegaBases, Chicago, IL). Each PCR reaction contained 5 μl cDNA as template, 1U Platinum-Taq polymerase (Invitrogen), 2.5 mM MgCl2, 500 μM dNTPs and 500 nM each primer, 250 nM probe and 200 nM Blue-636 reference (MegaBases). The reactions were run at 95°C/5 min, then 94°C/30 sec, 58°C/30 sec and 72°C/30 sec for 40, using ABI Prism 7700 Sequence Detector (PE Biosystems, Norwalk, CT). Endpoint normalized fluorescence (Rn) was detected. Vβ17-VC was considered as standard. Jβ1.4 percentage was calculated as RnVβ17-Jβ1.4 / RnVβ17-VC × 100 (analogous for Jβ1.5 and Jb2.6) using the ABI7700 software. Vβ17-Jβ1.4 (Vβ17-Jβ1.5, Vβ17-Jb2.6) and Vβ17-VC were assumed to have the same amplification rate. The correctness of this assumption was verified by multiple 10-fold dilutions of the templates.
The size of the CDR3 spectratyping peak(s) of interest was determined as follows: Vβ17-Jβ1.4, Vβ17-Jβ1.5 or Vβ17-Jβ2.6 segments were separately amplified by PCR, using forward primer FAM-AAGGGTACAGCGTCTCTCGG (Vβ17) and the same reverse primers as in real-time PCR. Each PCR reaction contained 5 μl cDNA, 1U Platinum-Taq polymerase (Invitrogen), 2.5 mM MgCl2, 500 μM dNTPs and 500 nM each primer. The reactions were run at 95°C/9 min, then 94°C/30 sec, 58°C/30 sec and 72°C/30 sec for 35 cycles, and finally 72°C/30 min. The dye-labeled PCR products were detected on an ABI 3100 sequence analyzer; peaks were visualized and their height quantified using GeneScan software (Applied Biosystems). If for cloning and sequencing only one peak was excised from gel and analyzed, the peak size value is <100%. If all peaks were excised and analyzed, the value is 100%. The decision whether to excise one or all peaks was based on whether the peak heights showed a Gaussian distribution (one peak excised) or not (all peaks excised), because only in case of Gaussian distribution it is reasonable to assume that the peak height is proportional to the diversity within the peak.
The number of different (distinct) sequences within the peak(s) was determined by cloning and sequencing as follows: Vβ17-Jβ1.4, Vβ17-Jβ1.5 or Vβ17-Jβ2.6 PCR products were separated by 2% agarose gel and 12% polyacrylamide gel, and extracted and purified using QIAEX®II Gel Extraction Kit (Qiagen) after cutting single or multiple band(s) under UV light. Purified PCR products were inserted into vector pCR®2.1-TOPO and transformed into DH5αT1R competent E.coli by using TOPO TA cloning Kit (Invitrogen). After culture on LB plates with 100μg/ml ampicillin, 25μg/ml kanamycin, 0.1 mM IPTG and 60μg/ml X-gal for over 12 hours, white or light blue colonies were picked as positive clones. Plasmid DNA was sequenced using Bigdye® dNTP (AppliedBiosystems), ABI373 sequencer and ABI-PRISM software. Clones were sequenced in batches of 20 until it was obvious that no new distinct sequences would be found (100-500 clones per each subject, time-point and Vβ17-Jβ combination) (Fig. 1).
Fig. 1.
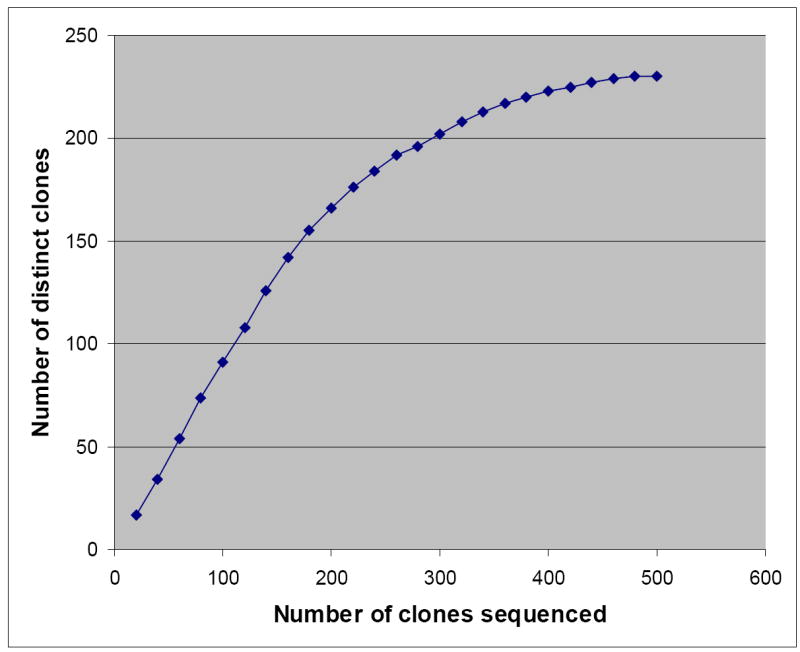
Example of sequencing clones in batches of 20 until it was obvious that no new distinct sequences would be found.
Results
In the healthy control, there were 400,000-600,000 distinct Vβ chains (Table 1), suggesting that the minimum number of CD4 T cell clones in a normal person is approximately 500,000. This result is close to that obtained by Arstila et al. (800,000-1,200,000).30 Our estimate may be lower than Arstila’s estimate because Arstila used total T cells whereas we used CD4 T cells.
At 1 month posttransplant, despite severe CD4 T lymphopenia and very abnormal CD4 T cell spectratypes, the calculated number of distinct Vβ chains was only 2 to 3-times lower than in the healthy control or the same patient pretransplant in the SSc patient, and similar to the healthy control in the MS patient (Table 1). This unexpected finding suggests that multiple T cell clones survive the conditioning or are reinfused with the graft (using this treatment protocol, ~2,000,000 CD4 T cells are typically reinfused with the graft29). Consistent with that, in the SSc patient 2 of 2 dominant (each sequenced >50-times) Vβ17–Jβ1.4 sequences in the pretransplant sample were also detected in the 1 month posttransplant sample. There were also 10 nondominant (each sequenced <10-times) Vβ17–Jβ1.4 sequences in the pretransplant sample which were not detected in the 1 month posttransplant sample. It is unclear whether these clones were located at one month posttransplant in extravascular compartments, were undetectable in blood due to technical reasons or were truly absent in the patient. The near-normal number of distinct Vβ chains at 1 month posttransplant is likely not due to the generation of CD4 T cells de novo (from hematopoietic stem cells) in the first month posttransplant, as T cell receptor excision circles (TRECs) in adult autologous transplant recipients typically become detectable only at ≥3 months posttransplant.24,29 Consistent with that, in the two patients studied here, TRECs were undetectable in CD4 T cells at 1 month posttransplant (see ref.29 for method).
By 12 months posttransplant, the diversity of β chains in both patients became normal or near-normal (Table 1). This can be at least in part attributed to T cell generation de novo.24,25,29 Consistent with that, in the SSc patient, of the 16 distinct Vβ17–Jβ1.4 sequences at 1 year posttransplant, 15 were new sequences compared to 1 month posttransplant. In the MS patient, of the 230 distinct Vβ17–Jβ1.4 sequences at 1 year posttransplant, 196 were new compared to 1 month posttransplant. Also consistent with the de novo generation is the fact that both patients showed a marked increase of phenotypically naïve (CD45RAhigh) CD4 T cell counts from one month to one year posttransplant – from 1 to 72/μl in the SSc patient and from 1 to 109/μl in the MS patient (see ref.29 for method). (Naïve T cells are more diverse than memory/effector T cells.30)
CD8 T cell diversity was not the focus of this study. Nevertheless, we wished to determine whether, analogous to CD4 T cells, pretransplant CD8 clones can persist after the “lymphoablative” conditioning with autologous CD34 cell transplantation. This was studied using cytomegalovirus (CMV) and Epstein-Barr virus (EBV)-specific CD8 clones. As shown in Fig. 2, in the MS patient, the same EBV-specific CD8 clone that was present pretransplant was also present at 1 and 12 months posttransplant. Similarly, in the SSc patient, the same EBV-specific and CMV-specific CD8 clones that were present pretransplant were also present posttransplant. TRECs in CD8 T cells at 1 month posttransplant were undetectable. Thus, it is likely that all 3 CD8 clones studied survived the “lymphoablative” therapy.
Fig. 2.
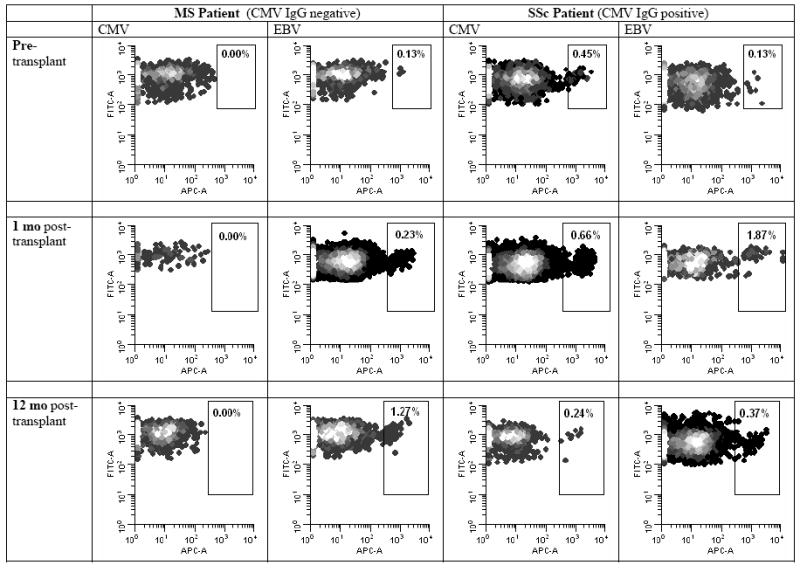
Persistence of cytomegalovirus (CMV) and Epstein-Barr virus (EBV)-specific CD8 T cell clones, using allophycocyanin (APC)-labeled HLA-viral peptide tetramer analysis. Method: Both patients were positive for HLA-A*0201. Thus, we used NLVPMVATV (CMV peptide) conjugated to HLA-A*0201-APC tetramer and GLCTLVAML (EBV peptide) conjugated to HLA*0201-APC tetramer (both purchased from Beckman Coulter) to stain CMV and EBV-specific clones in blood mononuclear cell specimens by flow cytometry. The cells were also stained by CD3-phycoerythrin antibody and CD8-FITC antibody. The density dotplots, showing tetramer-APC fluorescence on the x-axis and CD8-FITC fluorescence on the y-axis, were gated on CD3+CD8+ lymphocytes. The CMV tetramer staining of the cells from the MS patient, who was CMV-seronegative, served as a negative control for setting the border between tetramer+ and tetramer- cells. The percentages of the tetramer+ cells (of total CD8 T cells) are shown, indicating that even though the percentages varied, the same EBV-specific CD8 clones in both patients and the same CMV-specific CD8 clone in the CMV-seropositive patient were present both pretransplant and posttransplant.
Discussion
The unexpectedly large number of CD4 T cell clones early posttransplant and the persistence of herpesvirus-specific CD8 T cell clones from pretransplant to posttransplant may explain why infections in autologous transplant recipients following neutrophil engraftment are relatively rare.29 It also questions whether clinical stabilization or improvement of patients with autoimmune diseases after autologous transplantation is due to the conditioning-induced elimination of most T cell clones (including those causing the disease). On the one hand, conditioning intensity appears to be inversely related to the likelihood of autoimmune disease progression,36 suggesting that debulking of pathogenic T cell clones may be beneficial. On the other hand, Moore et al. showed that clinical improvement of patients with rheumatoid arthritis posttransplant was not inversely related to the number of T cells infused with the autologous graft.37 Alternative explanations for clinical stabilization or improvement of autoimmune diseases after autografting may include functional alteration of the pathogenic T cell clone(s) by the conditioning, faster reconstitution of regulatory than effector T cells38,39 or B-lymphoablation40 (B cells are undetectable on day 7 of this kind of transplant29).
Technical limitations should be kept in mind when interpreting the results presented in Table 1. First, the number of distinct Vβ chains is lower than the number of T cell clones, as each β chain can combine with various α chains. Thus despite the number of Vβ chains at 1 month posttransplant is near-normal, the number of T cell clones at 1 month may theoretically be significantly lower compared to the same patient pretransplant or a healthy control. Nevertheless, the lack of a marked difference in the number of distinct Vβ chains between 1 month posttransplant and pretransplant/healthy status suggests that the difference in the number of T cell clones is not extreme. Second, the mathematical calculation of the diversity of Vβ specificities presented here assumes that the diversity within the Vβ17-Jβ1.4 segment is similar to the diversity within other Vβ-Jβ segments. Theoretically, this may not be true. However, the fact that the calculated diversity using Vβ17-Jβ1.5 or Vβ17-Jβ2.6 was similar to that using Vβ17-Jβ1.4 (Table 1) suggests that in one individual at one time point the diversity within various Vβ-Jβ segments is similar. Third, given the skewed repertoire of posttransplant T cells, during the sequencing of the cloned CDR3 segments it is easy to miss underrepresented sequences, unless many clones are sequenced. We attempted to overcome this problem by sequencing clones until it was obvious that no new distinct sequences would be found (100-500 clones per each subject, time-point and Vβ17-Jβ combination). Packer and Muraro recently determined in autologous transplant recipient that if only 75-100 clones are sequenced, the true number of distinct sequences is underestimated with an error of 5-7%.41 Thus, as we analyzed 100-500 sequences, it is likely that we have underestimated the true number of distinct sequences with an error of less than 7%.
In conclusion, the diversity of CD4 T cells one month after cyclophosphamide+TBI+ATG conditioning and autologous CD34 cell transplantation for autoimmune diseases may be near-normal. The treatment may be only partially T-depleting rather than completely T-ablative. As the treatment is not selective for certain T cell clones, from most T cell clones at least one or few cells appear to be spared.
Acknowledgments
We are grateful to the staff of the Seattle Cancer Care Alliance Apheresis Unit and the patients for their willingness to undergo research aphereses. We also thank Dr. Paolo A. Muraro for valuable input, and the Seattle Cancer Care Alliance Clinical HLA Laboratory for HLA typing.
The work was supported by Grant AI46108 from the National Institutes of Health (USA). J.S. is a recipient of Canada Research Chair and Alberta Heritage Foundation Clinical Scholar Awards.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Villers D, Milpied N, Gaschet J, et al. Alteration of the T cell repertoire after bone marrow transplantation. Bone Marrow Transplant. 1994;13:19–26. [PubMed] [Google Scholar]
- 2.Gorski J, Yassai M, Keever C, Flomenberg N. Analysis of reconstitution T cell receptor repertoires in bone marrow transplant recipients. Arch Immunol Therap Exp. 1995;43:93–97. [PubMed] [Google Scholar]
- 3.Gorochov G, Debre P, Leblond V, Sadat-Sowti B, Sigaux F, Autran B. Oligoclonal expansion of CD8+CD57+ T cells with restricted T cell receptor beta chain variability after bone marrow transplantation. Blood. 1994;83:587–595. [PubMed] [Google Scholar]
- 4.Gaschet J, Denis C, Milpied N, et al. Alterations of T cell repertoire after bone marrow transplantation: charactrization of over-represented subsets. Bone Marrow Transplant. 1995;16:427–435. [PubMed] [Google Scholar]
- 5.Masuko K, Kato S, Hagihara M, et al. Stable clonal expansion of T cells induced by bone marrow transplantation. Blood. 1996;87:789–799. [PubMed] [Google Scholar]
- 6.Smith FS, Rencher SD, Heslop HE, Hurwitz JL. T cell receptor repertoire of CD4+ and CD8+ T cell subsets in the allogeneic bone marrow transplant recipients. Cancer Immunol Immunother. 1995;41:104–110. doi: 10.1007/BF01527406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roux E, Helg C, Chapuis B, Jeannet M, Roosnek E. T cell repertoire complexity after allogeneic bone marrow transplantation. Human Immunol. 1996;48:135–138. doi: 10.1016/0198-8859(96)00085-7. [DOI] [PubMed] [Google Scholar]
- 8.Roux E, Helg C, Dumont-Girard F, Chapuis B, Jeannet M, Roosnek E. Analysis of T cell repopulation after bone marrow transplantation: Significant differences between recipients of T cell depleted and unmanipulated grafts. Blood. 1996;87:3984–3992. [PubMed] [Google Scholar]
- 9.Kubo K, Yamanaka K, Kiyoi H, et al. Different T cell receptor repertoires between lesions and peripheral blood in acute GVHD after allogeneic bone marrow transplantation. Blood. 1996;87:3019–3026. [PubMed] [Google Scholar]
- 10.Liu X, Chesnokova V, Forman SJ, Diamond DJ. Molecular analysis of T cell receptor repertoire in bone marrow transplant recipients: evidence of oligoclonal T cell expansion in GVHD lesions. Blood. 1996;87:3032–3044. [PubMed] [Google Scholar]
- 11.Vavassori M, Maccario R, Moretta A, et al. Restricted TCR repertoire and lont-term persistence of donor-derived antigen-experienced CD4 T cells in allogeneic bone marrow transplantation recipients1. J Immunol. 1996;157:5739–5747. [PubMed] [Google Scholar]
- 12.Akatsuka Y, Cerveny C, Hansen JA. T cell receptor clonal diversity following allogeneic marrow grafting. Human Immunol. 1996;48:125–134. doi: 10.1016/0198-8859(96)00082-1. [DOI] [PubMed] [Google Scholar]
- 13.Bomberger C, Singh-Jairam M, Rodey G, et al. Lymphoid reconstitution after autologous PBSC transplantation with FACS-sorted CD34+ hematopoietic progenitors. Blood. 1998;91:2588–2600. [PubMed] [Google Scholar]
- 14.Wu CJ, Chillemi A, Alyea EP, et al. Reconstitution of T-cell receptor repertoire diversity following T-cell depleted allogeneic bone marrow transplantation is related to hematopoietic chimerism. Blood. 2000;95:352–359. [PubMed] [Google Scholar]
- 15.Godthelp BC, van Tol MJ, Vossen JM, van Den Elsen PJ. T-Cell immune reconstitution in pediatric leukemia patients after allogeneic bone marrow transplantation with T-cell-depleted or unmanipulated grafts: evaluation of overall and antigen-specific T-cell repertoires. Blood. 1999;94:4358–4369. [PubMed] [Google Scholar]
- 16.Dumont-Girard F, Roux E, van Lier RA, et al. Reconstitution of the T-cell compartment after bone marrow transplantation: restoration of the repertoire by thymic emigrants. Blood. 1998;92:4464–4471. In Process Citation. [PubMed] [Google Scholar]
- 17.Verfuerth S, Peggs K, Vyas P, Barnett L, O’Reilly RJ, Mackinnon S. Longitudinal monitoring of immune reconstitution by CDR3 size spectratyping after T-cell-depleted allogeneic bone marrow transplant and the effect of donor lymphocyte infusions on T-cell repertoire. Blood. 2000;95:3990–3995. [PubMed] [Google Scholar]
- 18.Bellucci R, Alyea EP, Weller E, et al. Immunologic effects of prophylactic donor lymphocyte infusion after allogeneic marrow transplantation for multiple myeloma. Blood. 2002;99:4610–4617. doi: 10.1182/blood.v99.12.4610. [DOI] [PubMed] [Google Scholar]
- 19.Claret EJ, Alyea EP, Orsini E, et al. Characterization of T cell repertoire in patients with graft-versus-leukemia after donor lymphocyte infusion. J Clin Invest. 1997;100:855–866. doi: 10.1172/JCI119601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eyrich M, Croner T, Leiler C, et al. Distinct contributions of CD4(+) and CD8(+) naive and memory T-cell subsets to overall T-cell-receptor repertoire complexity following transplantation of T-cell-depleted CD34-selected hematopoietic progenitor cells from unrelated donors. Blood. 2002;100:1915–1918. doi: 10.1182/blood-2001-11-0005. [DOI] [PubMed] [Google Scholar]
- 21.Friedman TM, Azhipa O, Zilberberg J, et al. Reconstitution of T cell subset repertoire diversity following multiple antigen-mismatched bone marrow transplantation. Biol Blood Marrow Transplant. 2006;12:1092–1095. doi: 10.1016/j.bbmt.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Roux E, Dumont-Girard F, Starobinski M, et al. Recovery of immune reactivity after T-cell-depleted bone marrow transplantation depends on thymic activity. Blood. 2000;96:2299–2303. [PubMed] [Google Scholar]
- 23.Talvensaari K, Clave E, Douay C, et al. A broad T-cell repertoire diversity and an efficient thymic function indicate a favorable long-term immune reconstitution after cord blood stem cell transplantation. Blood. 2002;99:1458–1464. doi: 10.1182/blood.v99.4.1458. [DOI] [PubMed] [Google Scholar]
- 24.Douek DC, Vescio RA, Betts MR, et al. Assessment of thymic output in adults after haematopoietic stem-cell transplantation and prediction of T-cell reconstitution. Lancet. 2000;355:1875–1881. doi: 10.1016/S0140-6736(00)02293-5. see comments. [DOI] [PubMed] [Google Scholar]
- 25.Muraro PA, Douek DC, Packer A, et al. Thymic output generates a new and diverse TCR repertoire after autologous stem cell transplantation in multiple sclerosis patients. J Exp Med. 2005;201:805–816. doi: 10.1084/jem.20041679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Protheroe AS, Pickard C, Johnson PW, et al. Persistence of clonal T-cell expansions following high-dose chemotherapy and autologous peripheral blood progenitor cell rescue. Br J Haematol. 2000;111:766–773. [PubMed] [Google Scholar]
- 27.McSweeney PA, Nash RA, Sullivan KM, et al. High-dose immunosuppressive therapy for severe systemic sclerosis: initial outcomes. Blood. 2002;100:1602–1610. [PMC free article] [PubMed] [Google Scholar]
- 28.Nash RA, Bowen JD, McSweeney PA, et al. High-dose immunosuppressive therapy and autologous peripheral blood stem cell transplantation for severe multiple sclerosis. Blood. 2003;102:2364–2372. doi: 10.1182/blood-2002-12-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storek J, Zhao Z, Lin E, et al. Recovery from and consequences of severe iatrogenic lymphopenia (induced to treat autoimmune diseases) Clin Immunol. 2004;113:285–298. doi: 10.1016/j.clim.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 31.Del Galdo F, Artlett CM. T cells and B cells in the pathogenesis of systemic sclerosis: recent insights and therapeutic opportunities. Curr Rheumatol Rep. 2006;8:123–130. doi: 10.1007/s11926-006-0052-0. [DOI] [PubMed] [Google Scholar]
- 32.Frohman EM, Racke MK, Raine CS. Multiple sclerosis--the plaque and its pathogenesis. N Engl J Med. 2006;354:942–955. doi: 10.1056/NEJMra052130. [DOI] [PubMed] [Google Scholar]
- 33.Kim DH, Sohn SK, Won DI, Lee NY, Suh JS, Lee KB. Rapid helper T-cell recovery above 200 × 10 6/l at 3 months correlates to successful transplant outcomes after allogeneic stem cell transplantation. Bone Marrow Transplant. 2006;37:1119–1128. doi: 10.1038/sj.bmt.1705381. [DOI] [PubMed] [Google Scholar]
- 34.Storek J, Gooley T, Witherspoon RP, Sullivan KM, Storb R. Infectious morbidity in long-term survivors of allogeneic marrow transplantation is associated with low CD4 T cell counts. Am J Hematol. 1997;54:131–138. doi: 10.1002/(sici)1096-8652(199702)54:2<131::aid-ajh6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 35.Lefranc M-P, Lefranc G. The T Cell Receptor FactsBook. San Diego: Academic Press; 2001. [Google Scholar]
- 36.Gratwohl A, Passweg J, Bocelli-Tyndall C, et al. Autologous hematopoietic stem cell transplantation for autoimmune diseases. Bone Marrow Transplant. 2005;35:869–879. doi: 10.1038/sj.bmt.1704892. [DOI] [PubMed] [Google Scholar]
- 37.Moore J, Brooks P, Milliken S, et al. A pilot randomized trial comparing CD34-selected versus unmanipulated hemopoietic stem cell transplantation for severe, refractory rheumatoid arthritis. Arthritis Rheum. 2002;46:2301–2309. doi: 10.1002/art.10495. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Chua KS, Guimond M, et al. Lymphopenia and interleukin-2 therapy alter homeostasis of CD4+CD25+ regulatory T cells. Nat Med. 2005;11:1238–1243. doi: 10.1038/nm1312. [DOI] [PubMed] [Google Scholar]
- 39.de Kleer I, Vastert B, Klein M, et al. Autologous stem cell transplantation for autoimmunity induces immunologic self-tolerance by reprogramming autoreactive T cells and restoring the CD4+CD25+ immune regulatory network. Blood. 2006;107:1696–1702. doi: 10.1182/blood-2005-07-2800. [DOI] [PubMed] [Google Scholar]
- 40.St Clair EW, Tedder TF. New prospects for autoimmune disease therapy: B cells on deathwatch. Arthritis Rheum. 2006;54:1–9. doi: 10.1002/art.21525. [DOI] [PubMed] [Google Scholar]
- 41.Packer AN, Muraro PA. Optimized clonotypic analysis of T-cell receptor repertoire in immune reconstitution. Exp Hematol. 2007;35:516–521. doi: 10.1016/j.exphem.2006.11.007. [DOI] [PubMed] [Google Scholar]


