Abstract
Members of the conserved family of eukaryotic RNA-dependent RNA polymerases (Rdrs) synthesize double-stranded RNA (dsRNA) intermediates in diverse pathways of small RNA (sRNA) biogenesis and RNA-mediated silencing. Rdr-dependent pathways of sRNA production are poorly characterized relative to Rdr-independent pathways, and the Rdr enzymes themselves are poorly characterized relative to their viral RNA-dependent RNA polymerase counterparts. We previously described a physical and functional coupling of the Tetrahymena thermophila Rdr, Rdr1, and a Dicer enzyme, Dcr2, in the production of ∼24-nucleotide (nt) sRNA in vitro. Here we characterize the endogenous complexes that harbor Rdr1, termed RDRCs. Distinct RDRCs assemble to contain Rdr1 and subsets of the total of four tightly Rdr1-associated proteins. Of particular interest are two RDRC subunits, Rdn1 and Rdn2, which possess noncanonical ribonucleotidyl transferase motifs. We show that the two Rdn proteins are uridine-specific polymerases of separate RDRCs. Two additional RDRC subunits, Rdf1 and Rdf2, are present only in RDRCs containing Rdn1. Rdr1 catalytic activity is retained in RDRCs purified from cell extracts lacking any of the nonessential RDRC subunits (Rdn2, Rdf1, Rdf2) or if the RDRC harbors a catalytically inactive Rdn. However, specific disruption of each RDRC imposes distinct loss-of-function consequences at the cellular level and has a differential impact on the accumulation of specific 23–24-nt sRNA sequences in vivo. The biochemical and biological phenotypes of RDRC subunit disruption reveal a previously unanticipated complexity of Rdr-dependent sRNA biogenesis in vivo.
Keywords: Rdr, Dicer, poly(U) polymerase, Tetrahymena thermophila
INTRODUCTION
Endogenous eukaryotic RNA-templated RNA polymerases represent a new enzyme family with an evolutionary origin distinct from that of other eukaryotic or viral polymerases (Iyer et al. 2003; Wassenegger and Krczal 2006). Interest in the Rdrs has grown with increasing recognition of their roles in RNA interference (RNAi) and RNA-mediated silencing. RNAi and related pathways exploit ∼20–30-nucleotide (nt) sRNAs as sequence-specific guides for regulation of gene expression, heterochromatin assembly, and defense against the disruptive impact of viruses and mobile elements (Farazi et al. 2008). Current evidence suggests that dsRNA products of Rdr are processed by the cleavage activity of Dicer(s), and/or by helicase(s), and ultimately assembled into Argonaute-family effector RNPs. Rdr-family polypeptides are encoded in the genomes of a broad range of eukaryotes including amoebae, plants, fungi, and nematodes (Cerutti and Casas-Mollano 2006; Wassenegger and Krczal 2006). Despite the genetically critical roles established for Rdrs in many organisms, much remains to be determined about their biochemical activities and biological regulation.
Mechanisms that govern the in vivo specificity of single-stranded RNA (ssRNA) template selection by an Rdr are largely unknown. One example of a template selection strategy was revealed through studies of endogenous small-interfering RNA (siRNA) biogenesis in Arabidopsis thaliana, in which transcripts targeted by specific microRNAs (miRNAs) were then subject to endonucleolytic cleavage, dsRNA synthesis, and subsequent processing by Dicer (Allen et al. 2005; Yoshikawa et al. 2005). In general, the biological specificity of Rdr function is proposed to require interacting factors. Biochemical purification of the Schizosaccharomyces pombe Rdr, Rdp1, revealed that it assembles as an RDRC with Hrr1, a putative helicase, and Cid12, a protein with predicted noncanonical nucleotidyl transferase motifs, both of which are required for Rdp1 function in heterochromatin silencing (Motamedi et al. 2004). For this S. pombe RDRC, individual subunit roles were not possible to discern due cooperative subunit requirements for RDRC integrity (Motamedi et al. 2004). The Caenorhabditis elegans Rdr-family protein RRF-1 copurified the putative helicase DRH-1, and the Rdr RRF-3 was copurified as one of several proteins associated with DCR-1 (Duchaine et al. 2006; Aoki et al. 2007). In Neurospora crassa, perinuclear localization and biological function of the Rdr SAD-1 depend on SAD-2 (Shiu et al. 2006). These findings reveal coordination of Rdr function by other cellular factors, but the specific biochemical properties and biological roles of Rdr-associated proteins have not been well characterized.
The expressed macronuclear genome of the ciliated protozoan Tetrahymena thermophila encodes a single Rdr, Rdr1, which is genetically essential (Lee and Collins 2007). A putative role for Rdr1 in sRNA biogenesis was inferred from the strand-asymmetric nature of an abundant class of constitutively accumulated 23–24-nt sRNAs in vivo, which derive from the antisense strand of predicted open reading frames lacking EST support (Lee and Collins 2006). The extreme bias in strand origin foreshadowed the discovery of animal germline Piwi-associated RNAs (piRNAs), which share this property (Seto et al. 2007). T. thermophila possesses predicted genes that encode up to 12 Argonaute family members, all of which cluster within the Piwi clade of Argonautes (Cerutti and Casas-Mollano 2006; Seto et al. 2007). The T. thermophila Piwi protein Twi1 and one of three Dicer-like proteins, Dcl1, are induced only in a sexual cycle of conjugation; they are required for biogenesis and stability of conjugation-specific 27–30-nt sRNAs including the functionally defined scan RNAs that act as sequence-specific guides for heterochromatin formation and macronuclear genome maturation (Mochizuki and Gorovsky 2004; Chalker 2008). The multiple size classes of T. thermophila sRNAs, along with the large number of putative genes encoding sRNA biogenesis and effector machinery, suggest that T. thermophila and other ciliates may possess a complexity of RNAi-related pathways comparable to that of multicellular eukaryotes.
We previously showed that T. thermophila Rdr1 assembles with a set of associated proteins into RDRC(s) that interact with the essential Dicer, Dcr2 (Lee and Collins 2007). Affinity purification of endogenously expressed, epitope-tagged Rdr1 under gentle wash conditions copurified Dcr2 and three to four other proteins; with more stringent washing, Dcr2 was released, leaving only Rdr1 and the tightly associated RDRC subunits. Biochemical assays of Rdr1 purified in RDRC context, with or without associated Dcr2, revealed a functional as well as physical coupling of T. thermophila RDRC and Dcr2 in the production of ∼24-nt sRNA: Dcr2 cleavage of only the RDRC product, not other dsRNA substrates, yielded a size of sRNA produced in vivo (Lee and Collins 2006, 2007). With or without associated Dcr2, RDRCs harboring wild-type Rdr1 but not the active-site variant Rdr1–D1004A catalyzed long dsRNA synthesis on a broad spectrum of ssRNA templates (Lee and Collins 2007). Almost the full length of the template was copied to produce dsRNA, as judged by treatment of 32P-NTP incorporation products with the single-strand specific Nuclease S1. In addition to Rdr1-dependent synthesis of dsRNA, RDRC assays also generated radiolabeled ssRNA products that were independent of catalysis by the Rdr1 active site.
Here we examine the contribution of T. thermophila Rdr1-associated proteins to dsRNA synthesis in vitro and to sRNA biogenesis in vivo. We show that two RDRC subunits, Rdn1 and Rdn2, are paralogs that possess the primary sequence motifs of noncanonical ribonucleotidyl transferases, linking together RDRCs of ciliates and other organisms. We also demonstrate, for the first time, the biochemical activity of these conserved RDRC subunits: both Rdn1 and Rdn2 catalyze nontemplated uridine addition to RNA substrates in vitro, expanding the family of known poly(U) polymerases. Although Rdn1 and Rdn2 have a similar specificity of biochemical activity in vitro, the roles of the two proteins differ dramatically in vivo. We demonstrate that the Rdn proteins have opposite developmental mRNA expression profiles, distinct gene knockout or knockdown phenotypes, and mutually exclusive assembly with Rdr1 and the other RDRC proteins Rdf1 and Rdf2. These findings demonstrate separable roles for Rdr1 in the content of functionally specialized RDRCs, which are required to support distinct pathways of 23–24-nt sRNA biogenesis in vivo.
RESULTS
Molecular characterization of four Rdr1-associated RDRC proteins
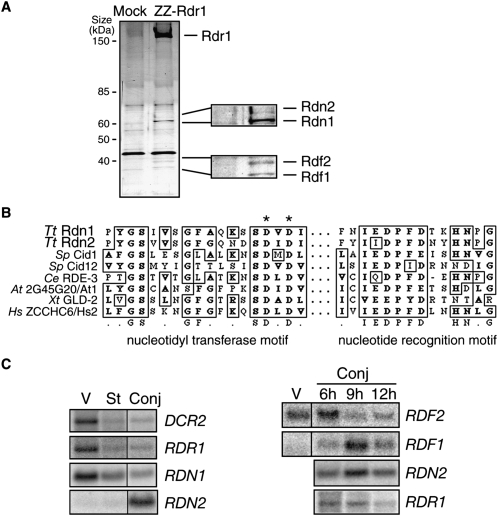
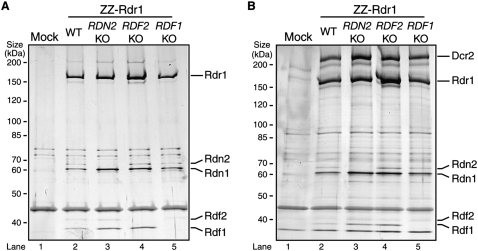
We previously characterized the biochemical activity of T. thermophila Rdr1 complexes isolated by affinity purification of ZZ-Rdr1 (Rdr1 with a N-terminal tandem Protein A domain tag). N-terminally tagged protein expressed from a transgene could functionally substitute for endogenous untagged Rdr1, allowing disruption of the endogenous RDR1 locus, but C-terminal tagging was not similarly successful in supporting viability (Lee and Collins 2007). SDS-PAGE analysis of ZZ-Rdr1 purifications (Fig. 1A) suggests four associated polypeptides: a doublet of ∼65-kDa proteins and a doublet of ∼40-kDa proteins. Previously, mass spectrometry of ZZ-Rdr1 associated proteins identified, in addition to Rdr1 and Dcr2, predicted T. thermophila proteins designated 6.m00629, 6.m00633, and 274.m00027 (Lee and Collins 2007). Subsequent ZZ-Rdr1 purifications and mass spectrometry identified a fourth protein, 274.m00028, represented by up to four unique peptides when an isolated ∼40-kDa gel slice from a ZZ-Rdr1 preparation was analyzed.
FIGURE 1.
Rdr1-associated RDRC subunits are two pairs of related proteins with differential mRNA expression profiles. (A) Extracts from starved cells with no tagged protein (Mock) or with ZZ-Rdr1 were used for affinity purification of RDRC. The four proteins recovered specifically in association with Rdr1 are labeled in the enlargements, with assignments based on mass spectrometry, protein tagging, and genetic depletion assays. (B) An alignment of active site residues in Rdn1, Rdn2, and other noncanonical ribonucleotidyl transferases is shown, with consensus underneath. Identical and similar residues are boxed. Asterisks denote residues mutated to alanine to render this class of enzyme catalytically inactive. Nucleotidyl transferase and nucleotide recognition motifs are indicated as previously described (Martin and Keller 2007). Tt, Tetrahymena thermophila; Sp, Schizosaccharomyces pombe; Ce, Caenorhabditis elegans; At, Arabidopsis thaliana; Xt, Xenopus tropicalis; Hs, Homo sapiens. (C) Northern blots for mRNA expression were performed using total RNA isolated from cells in vegetative growth (V), starvation (St), or conjugation (Conj). Time points after the initiation of conjugation are noted in hours (h); where not noted, the 10-h time point of conjugation was used (see Fig. 3B for conjugation stages).
Because gene predictions in T. thermophila are imprecise, we characterized the mRNA transcripts expressed by these four predicted genes using RT-PCR. The largest open reading frames of the experimentally defined mRNAs encode polypeptides with molecular weights matching the RDRC polypeptides (two of ∼65 kDa, two of ∼40 kDa). BLAST and other analyses of primary protein sequences revealed homology of both ∼65 kDa proteins with the poly(A) polymerase/2′-5′ oligo(A) synthetase family of noncanonical ribonucleotidyl transferases (Fig. 1B). This family includes proteins shown to have poly(A) and/or poly(U) polymerase activity (Kwak and Wickens 2007; Martin and Keller 2007; Rissland and Norbury 2008). Following T. thermophila nomenclature rules, the genes encoding these ∼65-kDa Rdr1-associated proteins were designated as the Rdr1-associated nucleotidyl transferases RDN1 and RDN2. The T. thermophila Rdn1 and Rdn2 proteins are highly related, exhibiting 39% identity and 24% additional similarity. Rdn1 and Rdn2 are more similar to each other than either is to other putative nucleotidyl transferases encoded by T. thermophila macronuclear genome, including proteins that we infer by sequence homology to represent the canonical poly(A) polymerase and the noncanonical Trf4-family poly(A) polymerase involved in RNA turnover (Martin and Keller 2007).
In contrast to the Rdn proteins, the smaller Rdr1-associated proteins bear no structural motifs that are readily discernible by either primary sequence analysis or tertiary structure threading methods. Following T. thermophila nomenclature rules, the genes encoding the novel ∼40-kDa Rdr1-associated proteins were designated as the Rdr1-associated factors RDF1 and RDF2. While no homologs of the Rdf proteins were found by BLAST of protein sequences deposited in GenBank, comparison of the Rdf proteins to each other revealed 24% identity and 23% additional similarity. The genes encoding the Rdf proteins are located in tandem in the genome, with no intervening open reading frames, which suggests recent gene duplication. The genes encoding the Rdn proteins are also located in close proximity in the genome, but they are separated by ∼12 kbp with three intervening predicted open reading frames.
We used Northern blot hybridization to examine mRNA expression for each of the RDRC subunits in cells undergoing rapid growth and fission (vegetative growth) or cells starved for nutrients and mixed with an alternate mating type to induce the sexual cycle of conjugation. All of our Northern blot conclusions were recently supported and extended by whole-genome microarray analysis of mRNA expression across highly sampled time courses of T. thermophila growth, starvation, and conjugation (Miao et al. 2009), and are therefore culled to show the most relevant results. DCR2 and RDR1 are robustly expressed in vegetative growth (Fig. 1C, left; Lee and Collins 2006). RDN1 expression parallels that of RDR1 (Fig. 1C, left). Curiously, RDN2 expression instead peaks in conjugation, when expression of RDN1 is relatively low (Fig. 1C, left; additional data not shown). This inverse relationship is consistent with the unequal silver staining intensity of Rdn1 and Rdn2 in ZZ-Rdr1 purifications from growing or starving cells, which suggests generally higher abundance of Rdn1 than Rdn2 in our RDRC preparations (Fig. 1A) (note that the indicated protein assignments are confirmed by additional mass spectrometry and by protein tagging and genetic depletion studies described below).
Like RDN1 and RDN2, RDF1 and RDF2 show differential expression in vegetative growth versus conjugation. RDF2 expression is high in vegetative growth and down-regulated by midconjugation, while RDF1 expression is low in vegetative growth and increases in midconjugation (Fig. 1C, right). Oddly, in silver-stained ZZ-Rdr1 purifications from growing or starving cells, Rdf2 does not stain well and appears of equal or lesser abundance than Rdf1; we suspect that this is due to the acidic isoelectric point of Rdf2, predicted to be 5.46. Together, our observations indicate a discordance in the expression of RDN1 versus RDN2 and a discordance in the expression of RDF1 versus RDF2. These results suggest that T. thermophila Rdr1-associated proteins may form alternate RDRCs rather than the single RDRC of invariant composition suggested for S. pombe (Motamedi et al. 2004).
Distinct phenotypes of RDRC subunit depletion in vivo
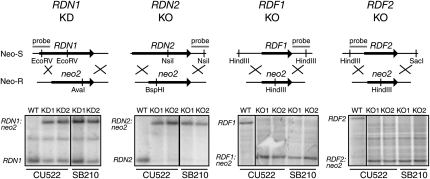
We next tested whether genes encoding each of the newly identified RDRC subunits are essential in T. thermophila, as is the case for RDR1 and DCR2 (Malone et al. 2005; Mochizuki and Gorovsky 2005; Lee and Collins 2006, 2007). We targeted the open reading frames of each of the Rdr1-associated proteins for replacement with a gene cassette conferring resistance to neomycin. Initial targeting replaces only a few of the 45 gene copies in the macronuclear genome, but increased replacement by the neomycin-resistance cassette can be achieved by gradually increasing the selective pressure. Nonessential genes can be eliminated entirely from the macronucleus (knockout [KO] strains), while essential genes reach a functionally limiting extent of gene knockdown (KD strains). Genomic DNA was prepared from several independently selected strains, which were subsequently released from selection to allow back-assortment of any remaining copies of the endogenous locus. The state of each gene locus was assayed using Southern blot hybridization. Probes were designed to distinguish wild-type chromosomes from disrupted chromosomes by hybridization to differently sized DNA fragments in KO or KD strains compared with wild type (Fig. 2). Wild-type chromosomes were missing in strains where RDN2, RDF1, or RDF2 was targeted, indicating that each of the genes had been replaced entirely by the selectable marker. We conclude that RDN2, RDF1, and RDF2 are not essential genes. In contrast, only incomplete loss of the RDN1 locus was attained. Thus, like RDR1 and DCR2, RDN1 is essential in T. thermophila.
FIGURE 2.
Three RDRC subunits are not essential for vegetative growth. Gene disruption for RDN1, RDN2, RDF1, and RDF2 was performed by integration of a selectable marker cassette in two independent strains, CU522 and SB210. Southern blot analysis of genomic DNA was performed separately for each locus, using the restriction enzymes and probes shown in the schematics. Neo-S and Neo-R indicate relative phenotypic sensitivity and resistance to the selective drug. The appropriate wild-type (WT) and disrupted locus fragment sizes are indicated to the right of each blot. Incomplete loss of the endogenous RDN1 locus indicates that the strains are gene knockdown (KD) in the polyploid macronucleus; complete loss of RDN2, RDF1, or RDF2 indicates that these strains are gene knockouts (KO).
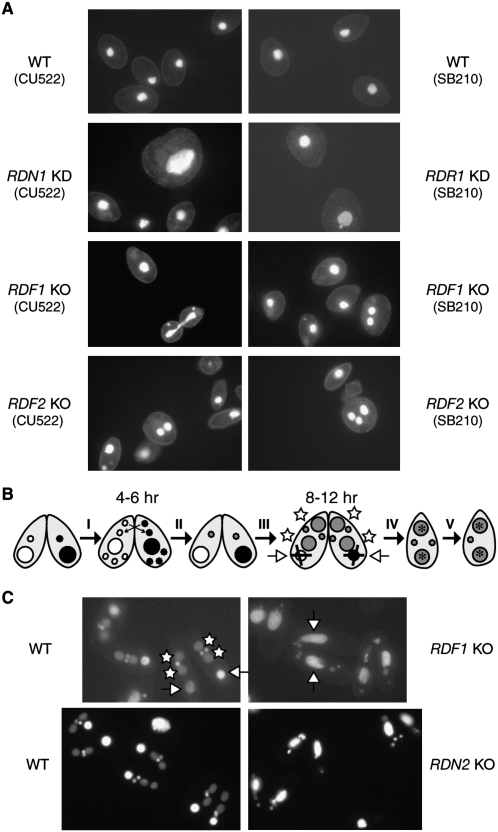
Up to 10% of cells in RDR1 KD strain cultures were enlarged, overly round, and harboring an often larger macronucleus than wild-type cells (Fig. 3A, right upper panels). This characteristic “monster” phenotype has been frequently noted as a consequence of delayed or defective cell division. Cells from RDN1 KD cultures also showed an increased frequency of monsters in comparison to wild-type (Fig. 3A, left upper panels), even when wild-type cells were cultured at a drug concentration that limited culture growth. Cells in RDF1 KO or RDF2 KO cultures displayed phenotypes rarely observed in wild-type cell cultures but occasionally noted in RDR1 KD cultures as well: a minority of cells had more than a one macronucleus per cell and/or incomplete DNA segregation between dividing cells (Fig. 3A, lower four panels). Curiously, cells from RDN2 KO cultures showed no cellular phenotype during vegetative growth. To confirm the generality of these phenotypes, we made each RDRC subunit gene knockdown or knockout in different strain backgrounds. In strain CU522, a mutation in the β-tubulin 1 gene that confers taxol hypersensitivity (Gaertig et al. 1994) also sensitizes cells for division defects (Smith et al. 2004). In the CU522 background even in the absence of taxol, the phenotypes of RDN1 KD, RDF1 KO, and RDF2 KO cells were similar to those described above in SB210 background and were generally more penetrant; again, RDN2 KO cultures displayed no cellular phenotype in vegetative growth.
FIGURE 3.
RDRC subunits have genetic depletion phenotypes in growing and conjugating cells. (A) Growing cells were fixed and stained with DAPI. Cell size and the intensity of nuclear staining vary among cells in a growing population due to changes over the cell cycle, but aberrantly large and rounded monster cells were frequent only in RDR1 and RDN1 KD cultures. All images are shown at the same relative magnification, and a relatively high exposure level was used to reveal faint cell outlines for context. Note that the macronucleus in each cell is strongly stained with DAPI, while the much smaller micronucleus may or may not be visible depending on cell orientation. (B) A schematic of the nuclear events of conjugation is shown. (C) Conjugating cells were fixed and stained with DAPI. Two separate time courses were performed for conjugation of wild-type and RDF1 KO (top panels) or wild-type and RDN2 KO (bottom panels) using SB210 and CU522 strains as mating partners. Matched panels of wild-type and KO cells were taken from the same time point within the interval of new macronuclear differentiation by the wild-type cells; KO cells did not progress to this stage at the expected time or even later (RDN1 KO is shown at 12 h; RDN2 KO is shown at 9 h). Note that cells of mated pairs are somewhat smaller than growing cells due to starvation prior to conjugation. A few nonpartnered cells stain brightly for the large macronucleus and adjacent small micronucleus. In B and C, differentiating zygotic macronuclei (white stars) and degenerating parental macronuclei (open arrows) are present at the same time within a cell pair; KO cell pairs retain parental macronuclei (open arrows) and do not differentiate new macronuclei.
SB210 and CU522 strains with macronuclear gene knockouts of the nonessential RDRC subunits were mated to test for conjugation phenotypes. RDN2 KO or RDF1 KO cells paired and appeared to complete meiosis with an efficiency and timing comparable to wild-type cells (Fig. 3B, step I), but few knockout pairs examined after these events appeared normal. Representative images from the interval when wild-type cells have differentiating new macronuclei (Fig. 3B, 8-12-h post-mixing) are shown in Figure 3C. The number and arrangement of nuclei suggest that knockout cells could not progress through zygotic mitoses and differentiation of two new macronuclei (Fig. 3B, step III). Progression through conjugation was halted rather than delayed, because paired KO cells examined at later time points (up to 19 h) still harbored a parental macronucleus and several small nuclei rather than the two macronuclei plus one micronucleus that result from successful conjugation (Fig. 3B, step V).
In contrast with RDN2 KO or RDF1 KO cells, RDF2 KO cells that paired did not exhibit a defect in conjugation at the cellular level. The finding of conjugation defects resulting from loss of Rdn2 or Rdf1 but not Rdf2 is consistent with the differential mRNA expression profiles of these RDRC subunits (Fig. 1C). The use of macronuclear gene knockout strains for conjugation has the potential to underestimate the importance of a gene product if it is needed late in conjugation, because the zygotic nuclei may supply some mRNA prior to the completion of their differentiation. Zygotic rescue is unlikely to account for the lack of conjugation phenotype for RDF2 KO, however, because this locus is down-regulated in mRNA expression by midconjugation (Fig. 1C). Overall, this phenotypic analysis of cells depleted or eliminated for expression of individual RDRC subunits suggests different biological roles for each Rdn or Rdf protein in vivo.
Mutually exclusive assembly of Rdn1 and Rdn2 into separate RDRCs
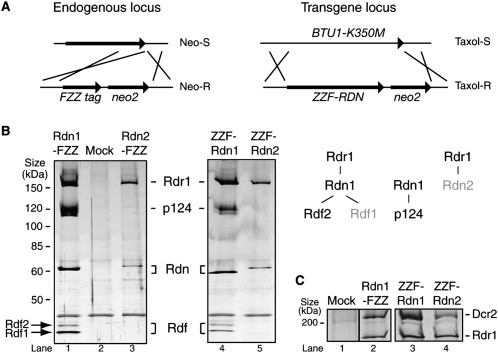
The differential mRNA expression profiles of RDRC subunits and their distinct genetic depletion phenotypes suggested separation of function among RDRC assemblies. We focused subsequent studies on the T. thermophila Rdn subunits, due in part to their disparate loss-of-function phenotypes and in part to the presence of a potentially similar subunit with noncanonical ribonucleotidyl transferase motifs in the S. pombe RDRC (Motamedi et al. 2004). To resolve RDRCs harboring Rdn1 versus Rdn2, we created two types of strains expressing tagged Rdn proteins. At RDN1 and RND2 endogenous loci, we integrated a C-terminal tag with FLAG epitopes, a TEV protease cleavage site and tandem Protein A domains (the FZZ tag) by selecting for cointegration of a downstream neomycin resistance cassette (Fig. 4A, left). Because complete replacement of the endogenous RDN1 locus with the tag cassette was achieved (data not shown), we infer that Rdn function is not perturbed by tag fusion to the C terminus. We also expressed N-terminally tagged Rdn proteins with FLAG and ZZ modules placed in the opposite orientation of the FZZ tag (the ZZF tag) by selecting for transgene replacement of the nonessential, taxol-hypersensitive β-tubulin encoded at BTU1 in strain CU522 (Fig. 4A, right).
FIGURE 4.
Rdn1 and Rdn2 assemble as distinct RDRCs that share association with Dcr2. (A) Strains expressing tagged Rdn1 or Rdn2 were created using two strategies: introduction of a C-terminal tag cassette at the endogenous locus or integration of an N-terminally tagged protein transgene at the nonessential BTU1 locus. Neo-S and Neo-R or Taxol-S and Taxol-R indicate relative phenotypic sensitivity and resistance to the selective drug. (B) Protein complexes were examined following purification of the indicated tagged proteins from extracts of starved cells (lanes 1,4–5) or cells at the 9-h time point of conjugation (lanes 2,3). Parallel mock purifications were performed using wild-type cell extracts, only one of which is shown but all of which were similar in nonspecific background. The composition of the multisubunit complexes identified here is summarized on the right. Lighter text denotes the RDRC subunits that exhibit increased expression during conjugation. (C) Dcr2 association with Rdr1 was examined following purification of the indicated tagged proteins from extracts of starved cells using the gentle washing conditions that preserve RDRC-Dcr2 interaction.
Using strains that express the Rdn proteins tagged at their endogenous loci, optimal purification of Rdn1-FZZ was accomplished using cell extracts from growing or starved cells whereas optimal purification of Rdn2-FZZ was accomplished using extracts from conjugating cells (Fig. 4B, lanes 1–3), as predicted by RDN1 and RDN2 mRNA expression profiles. Using strains that express tagged Rdn proteins from the BTU1 promoter of the transgene locus, both tagged proteins could be readily purified from extracts of cells grown and starved in parallel (Fig. 4B, lanes 4–5). Independent of tag location and whether the tagged protein was expressed from an endogenous or transgene locus, Rdn1 copurified Rdr1 and both Rdf subunits (Fig. 4B, lanes 1,4). In contrast, tagged Rdn2 copurified only Rdr1 (Fig. 4B, lanes 3,5). Equal loadings of tagged Rdn1 and Rdn2 confirmed this disparity in associated Rdf subunits (data not shown) along with additional studies described below. Under the less stringent wash conditions used to stabilize Dcr2 association with RDRC (Lee and Collins 2007), RDRCs with tagged Rdn1 or Rdn2 each copurified Dcr2 (Fig. 4C; additional data not shown).
Tagged Rdn1 also copurified a protein of ∼120 kDa, which was identified by mass spectrometry as the predicted protein TTHERM_00782040. The putative ∼124 kDa protein has no obvious primary sequence motifs or homology with other proteins. No evidence of p124 was detected in any purification of Rdn2 or in any previous affinity purification of tagged Rdr1 or Dcr2 (Lee and Collins 2007). In summary (Fig. 4B, right), these findings suggest that distinct RDRCs harbor Rdn1 or Rdn2 and that only RDRC harboring Rdn1 can recruit Rdf proteins; also, Rdn1 assembles a putative non-RDRC complex with p124.
We next examined the interdependence of RDRC subunit assembly. We created strains that express ZZ-Rdr1 in the background of RDN2 KO, RDF2 KO, or RDF1 KO. Cell extracts were used for affinity purification of RDRC under the stringent wash conditions that release Dcr2 (Fig. 5A) or the gentle wash conditions that retain Dcr2–RDRC association (Fig. 5B). Purifications of ZZ-Rdr1 in the absence of Rdn2, Rdf1, or Rdf2 confirmed a lack of association of each genetically eliminated protein, but no additional subunits were depleted (Fig. 5A). Thus, the loss of each nonessential RDRC subunit did not perturb the association of the remainder of the subunits with Rdr1. Dcr2 association with RDRC was also unaffected by the absence of Rdn2, Rdf2, or Rdf1 (Fig. 5B). These observations contrast with the interdependence of subunits in the assembly of the S. pombe RDRC (Motamedi et al. 2004). Based on independent assembly the two T. thermophila Rdf proteins with RDRC and their reciprocal mRNA expression profiles, we suggest that the RDRC harboring Rdr1, Rdn1, and Rdf1 is distinct from the RDRC harboring Rdr1, Rdn1, and Rdf2. However, the full complexity of Rdn1 association with the novel Rdf1, Rdf2, and p124 polypeptides remains to be explored in future extensions of this work.
FIGURE 5.
RDRC subunit interactions occur in the absence of Rdn2, Rdf1, or Rdf2. Protein complexes were examined following purification of ZZ-Rdr1 from extracts of starved cells lacking tagged protein (Mock) or the indicated gene knockout (KO) cells. Affinity purifications employed either robust (A) or gentle (B) washing conditions.
Nucleotidyl transferase activity of Rdn1 and Rdn2
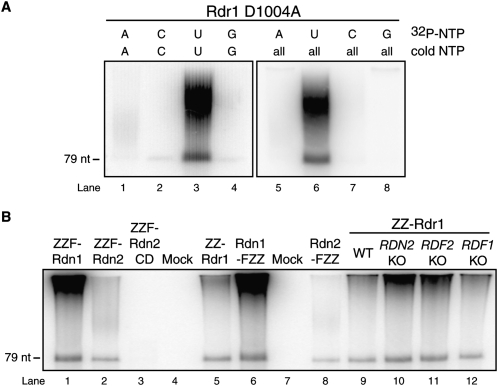
To characterize the biochemical activities of Rdn1 and Rdn2 as potential nucleotidyl transferases, we used RDRC complexes harboring the D1004A catalytic-dead (CD) variant of Rdr1 (Lee and Collins 2007). We performed separate reactions with each radiolabeled NTP and a 79 nt ssRNA template used in previous studies. Each radiolabeled NTP was supplemented with the same NTP unlabeled or with the full set of unlabeled NTPs (Fig. 6A). Robust elongation of the 79 nt ssRNA was detected only in reactions with 32P-UTP. Notably, product signal in reactions with 32P-UTP was not diminished by inclusion of other NTPs in the reaction (Fig. 6A, cf. lane 3 and lane 6), suggesting that the activity is specific for incorporation of 32P-UTP. Treatment of the reaction products with ssRNA-specific nucleases entirely eliminated product signal (see Lee and Collins 2007; data not shown) confirming that the product did not represent residual dsRNA synthesis activity by Rdr1 D1004A. Some ssRNA products were extended by one or a few nucleotides, while others gained a longer polynucleotide tail. Because the specific activity of product RNA varies with the number of nucleotides added, it is not possible to use radiolabel intensity to infer which product is most abundant in vitro. However, we note that both short and long products were generated under all in vitro reaction conditions tested. Similar results were obtained in assays performed using input ssRNA templates of different lengths and sequence compositions (data not shown).
FIGURE 6.
Rdn1 and Rdn2 are uridine-specific nucleotidyl transferases. (A) Products of nucleotidyl transferase assays using a 79-nt input ssRNA and purified RDRCs containing catalytic-dead Rdr1–D1004A. (B) Uridylation of a 79-nt input ssRNA by specific RDRCs. RDRCs were purified by the indicated tagged protein from extracts of cells that were starved (all lanes except 7,8) or at the 9-h time point of conjugation (lanes 7,8). The intensity of reaction product correlates with the level of Rdn present in the preparation (not shown). ZZ-Rdr1 was also purified from extracts of starved wild-type or gene knockout (KO) cells as noted.
We next compared the dependence of nucleotidyl transferase activity on RDRC composition using a panel of complexes purified by tagged Rdr1, Rdn1, or Rdn2. ZZ-Rdr1 purification enriched nucleotidyl transferase activity relative to mock purifications from control cells lacking tagged protein (Fig. 6B, lanes 4–5). Importantly, ZZF-tagged Rdn1 and Rdn2 each enriched nucleotidyl transferase activity in proportion to the amount of Rdn protein recovered by affinity purification (Fig. 6B, lanes 1–2; additional data not shown). Rdn1-FZZ and Rdn2-FZZ tagged at their endogenous loci also copurified nucleotidyl transferase activity in proportion to Rdn protein (Fig. 6B, lanes 5,8). Because Rdn1 and Rdn2 do not copurify each other (Fig. 4B), these results suggest that each Rdn protein catalyzes ssRNA uridylation. Finally, we found that RDRC purified by ZZ-Rdr1 from extract of any RDRC subunit knockout strain retained comparable nucleotidyl transferase activity (Fig. 6B, lanes 9–12).
We attempted to create strains expressing tagged CD versions of Rdn1 or Rdn2, using the same transgene approach employed for expression of wild-type ZZF-Rdn1 and ZZF-Rdn2 described above (Fig. 4A). Aspartic acids in the putative active site (Fig. 1B) were substituted by alanines. Strains with complete replacement of BTU1 by the ZZF-Rdn2 CD expression cassette were obtained, but expression of ZZF-Rdn1 CD was toxic enough to induce loss of viability and prevent the establishment of strains fully replaced at the BTU1 locus by the ZZF-Rdn1 CD cassette. Affinity purification of ZZF-Rdn2 CD failed to enrich for nucleotidyl transferase activity (Fig. 6B, lane 3), despite enriching for the dsRNA synthesis and dicing activities of Rdr1 and Dcr2 (see below). The disruption of RDRC nucleotidyl transferase activity by substitution of two conserved aspartic acids in Rdn2 suggests that the T. thermophila Rdn polypeptides rely on the same active site as other noncanonical poly(A) and poly(U) polymerases to generate the nucleotidyl transferase activity detected in preparations of T. thermophila RDRC. We observed no difference in nucleotidyl transferase activity associated with wild-type versus CD Rdr1, consistent with the requirement for the Rdn active site. In contrast, studies of partially purified recombinant A. thaliana RDR6 suggest that this Rdr by itself may act as a nucleotidyl transferase to extend ssRNA or ssDNA, with some preference for use of UTP as the nucleotide substrate (Curaba and Chen 2008).
RDRCs share general properties of dsRNA synthesis and dicing in vitro
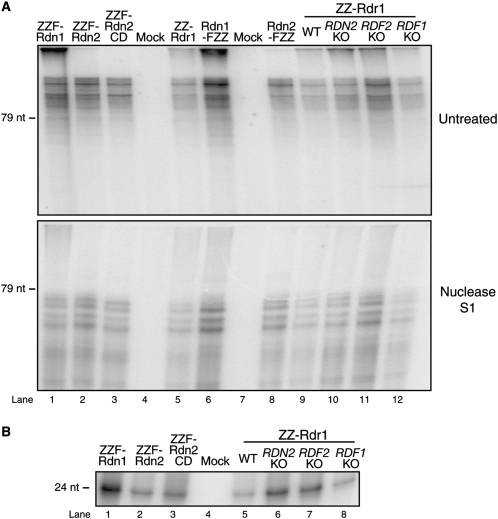
We next investigated whether Rdr1 catalytic activity was affected by RDRC composition. Rdr reactions were performed using the same 79-nt ssRNA substrate tailed by Rdn proteins in studies above, in reactions with all four unlabeled NTPs and 32P-CTP. Reaction products were divided for mock treatment or treatment with the single-strand specific Nuclease S1 prior to electrophoresis (Fig. 7A). We found that this post-reaction processing resolves the nuclease-resistant dsRNA portion of product from product region(s) with some ssRNA nature (Lee and Collins 2007). As shown previously, ZZ-Rdr1 purification enriched for synthesis of products that migrated larger than the input ssRNA template without Nuclease S1 treatment but resolved into a series of products of slightly less than input template length with Nuclease S1 treatment (Fig. 7A, lane 5). Mock purifications from various cell extracts lacking a tagged RDRC subunit did not enrich for this activity (Fig. 7A, lanes 4,7). RDRC complexes isolated by purification of ZZF-Rdn1, ZZF-Rdn2, or ZZF-Rdn2 CD all harbored similar Rdr1 activity (Fig. 7A, lanes 1–3). Thus, under the conditions used here, dsRNA synthesis is not influenced differentially by the two Rdn proteins. Moreover, our results indicate that the transferase activity of an Rdn is not required for dsRNA synthesis by Rdr1. RDRC complexes isolated by purification of Rdn1-FZZ or Rdn2-FZZ, the tagged Rdn proteins expressed from endogenous loci, also did not display differential dsRNA synthesis activity in vitro (Fig. 7A, lanes 6,8).
FIGURE 7.
Synthesis and dicing of dsRNA do not require a specific RDRC composition in vitro. (A) Products of dsRNA synthesis untreated or treated with Nuclease S1 prior to electrophoresis. Assays were performed using a 79-nt ssRNA template and radiolabeled CTP. The intensity of reaction product correlates with the level of Rdr1 present in the preparation (not shown). (B) Small RNA products of coupled dsRNA synthesis and dicing assays using the same 79-nt ssRNA template as in A and RDRC complexes that were gently washed to preserve Dcr2 association. The intensity of reaction product correlates with the level of Dcr2 protein present in the preparation (not shown).
We also tested the activity of RDRCs isolated by purification of ZZ-Rdr1 from strains lacking the nonessential RDRC subunits Rdn2, Rdf1, and Rdf2. Wild-type and gene-knockout strains all yielded RDRCs with similar Rdr1 product synthesis activity (Fig. 7A, lanes 9–12). Together, these Rdr1 activity assays demonstrate that synthesis of dsRNA is independent of the presence of any individual RDRC subunit other than Rdr1, because normal Rdr1 activity was retained in preparations of (1) tagged Rdn2, which does not copurify Rdn1, Rdf1, or Rdf2; (2) tagged Rdn1, which does not copurify Rdn2; and (3) ZZ-Rdr1 from RDN2 KO, RDF1 KO, or RDF2 KO cell extracts. Furthermore, Rdr1 activity in vitro does not require the catalytic activity of a Rdn subunit: although ZZF-Rdn2 CD lacks nucleotidyl transferase activity (Fig. 6B, lane 3), RDRC harboring ZZF-Rdn2 CD still carries out normal dsRNA synthesis (Fig. 7A, lane 3).
T. thermophila Rdr1 complexes containing Dcr2 are capable of coupled dsRNA synthesis and dicing, such that input ssRNA generates ∼24-nt sRNA products in short duplexes (Lee and Collins 2007). We tested whether RDRC composition had an influence on Dcr2 activity in the coupled reaction system in vitro. None of the changes in RDRC subunit composition affected copurification of Dcr2 under gentle wash conditions (Fig. 4C, and additional data not shown). Likewise, none of the changes in RDRC subunit composition reduced the generation of ∼24-nt Dcr2 products from dsRNA synthesized by RDRC (Fig. 7B). Therefore, coupled dsRNA synthesis and dicing in vitro is independent of the presence of any specific subunit other than Rdr1 and Dcr2.
RDRC subunit requirements for 23–24-nt sRNA accumulation in vivo
Our previous finding of physical and functional coupling of Rdr1 and Dcr2 in generating ∼24-nt sRNA in vitro implicated these enzymes in the biogenesis of constitutively accumulated 23–24-nt sRNAs (Lee and Collins 2007). Before investigating the role of Rdn and Rdf RDRC subunits in 23–24-nt sRNA biogenesis in vivo, we first wanted to verify dependence of the in vivo process on Rdr1. Because RDR1 and DCR2 are both essential genes (Malone et al. 2005; Mochizuki and Gorovsky 2005; Lee and Collins 2006, 2007), it was not possible to use gene knockout strains to test whether their loss of function also resulted in loss of 23–24-nt sRNA. Furthermore, although gene knockdown strains often yield phenotypic insights, the genotypic variation in any growing cell population will obscure molecular phenotypes by “averaging” them across the culture. To escape these limitations, we tested for potential reduction of 23–24-nt sRNA following short-term overexpression of CD Rdr1 D1004A. Because Rdr1-D1004A still assembles with all of the RDRC-associated proteins including Dcr2 (Lee and Collins 2007), it could have a dominant-negative impact by competing with wild-type Rdr1 for biological templates and by inhibiting RDRC-associated activities that are coupled to dsRNA synthesis.
Expression of ZZ-Rdr1-D1004A was placed under the control of the cadmium-inducible MTT1 promoter integrated at BTU1. Selection for transgene integration was performed without cadmium in the medium, allowing complete replacement of BTU1 with the transgene (data not shown). After release from selection, cell cultures were expanded by vegetative growth. Protein expression was induced by cadmium addition to cells either in vegetative growth or after transfer of growing cells to starvation medium to halt cell growth. Cadmium addition induced similar levels of ZZ-Rdr1-D1004A protein accumulation in growing and starving cells (data not shown). Wild-type cells were cultured and induced with cadmium in parallel as a control.
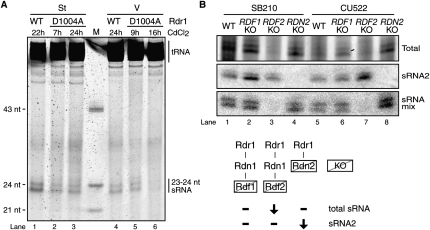
At various time points within 24 h after cadmium addition, cells were harvested for total RNA purification. Total RNA was normalized for recovery, size-enriched for sRNA, and then examined by denaturing gel electrophoresis and direct staining. Expression of CD Rdr1 in starved cells reduced the level of 23–24-nt sRNA (Fig. 8A, lanes 1–3). In continuously growing cells, the impact of CD Rdr1 expression was more dramatic: 23–24-nt sRNA became almost undetectable after 16 h (Fig. 8A, lanes 4–6). The greater impact of CD Rdr1 expression on 23–24-nt sRNA accumulation in growing cells could reflect greater dilution of the sRNA present prior to cadmium addition or greater sRNA turnover. No cellular phenotypes of CD Rdr1 expression were detected in the 24-h interval of cell culture employed for these studies.
FIGURE 8.
RDRC subunits differentially impact 23–24-nt sRNA accumulation in vivo. (A) Total RNA isolated from starving (St) or growing (V) wild-type cells or cells expressing Rdr1–D1004A was size-enriched for small RNA, resolved by denaturing gel electrophoresis and stained directly. Starved or growing cell cultures were treated with 0.1 μg/mL CdCl2 or 1 μg/mL CdCl2, respectively, for the times indicated to induce catalytic-dead Rdr1 overexpression. Wild-type cells were similarly treated with CdCl2 as a control. (B) Aliquots of enriched sRNA from growing wild-type or gene-knockout cells were used for direct staining (top panel) or Northern blots. Representative sRNA expression results are shown for an individual sRNA (sRNA2) or 14 other sRNAs (sRNA mix) of known sequence used for Northern blot assays in previous work (Lee and Collins 2006). A summary of sRNA phenotypes is also provided.
We next investigated the accumulation of 23–24-nt sRNA in strains lacking Rdn2, Rdf1, or Rdf2. Total RNA was size-enriched and used to visualize 23–24-nt sRNA by direct staining. In both SB210 and CU522 backgrounds, RDF2 KO strains had extremely low levels of 23–24-nt sRNA (Fig. 8B, top panel). This result was reproduced over many repetitions of sRNA purification. We also examined the accumulation of 27–30-nt sRNA in conjugating RDN2 KO, RDF1 KO, or RDF2 KO cells sampled across the normal time course of conjugation (data not shown). While 27–30-nt sRNA levels were largely unaffected, RDN2 KO cells exhibited a slight reduction in sRNA that could reflect a direct contribution of Rdn2 to production of scan RNAs or more likely an indirect impact of disrupted progression through conjugation (see Fig. 3C).
The genomic loci from which sequenced T. thermophila 23–24-nt sRNA originate harbor predicted protein-coding genes antisense to the sRNA; these genes can be classified into several homology groups or families (Lee and Collins 2006). In strains lacking Rdn2, Rdf1, or Rdf2, the presence of known sRNAs was probed by Northern blot hybridization with end-labeled oligonucleotides. This approach revealed approximately wild-type levels of a specific sRNA, sRNA2, in the RDF2 KO 23–24-nt sRNA population despite the much lower accumulation of 23–24-nt sRNA overall (Fig. 8B, middle panel). Remarkably, sRNA2 was missing from the 23–24-nt sRNA pool in the RDN2 KO strain, despite an abundance of 23–24-nt sRNA similar to wild type. Additional oligonucleotide probes complementary to known T. thermophila 23–24-nt sRNA were used individually and as mixtures, with some sRNA found to be missing in the RDN2 KO strain (i.e., sRNA2, and other sRNA from this sequence family) and others found to be missing in the RDF2 KO strain (Fig. 8B, bottom panel).
Together, these results suggest that accumulation of most 23–24-nt sRNA is dependent on Rdf2 but not Rdf1 or Rdn2, consistent with preferential expression of Rdf2 in growing cells. However, because specific subsets of 23–24-nt sRNA require the presence of Rdn2, distinct forms of RDRC have unique roles in sRNA biogenesis during growth. These findings indicate that compositionally different RDRCs play functionally different roles in sRNA biogenesis in vivo.
DISCUSSION
Roles for Rdr polypeptides have been established at the levels of transcriptional and post-transcriptional regulation (Wassenegger and Krczal 2006). Endogenous synthesis of dsRNA complicates the necessary cellular repertoire of response to nucleic acids, because dsRNA is also a hallmark of invasion by selfish foreign genomes. Much remains to be learned about how Rdr activity is recruited to and/or restrained from acting on potential ssRNA targets in vivo.
The Rdr polypeptide itself has conserved N- and C-terminal extensions from the active site motifs, so some functional specialization may be conferred by these accessory domains. In addition, because Rdr proteins isolated from their endogenous sources form RDRCs, tightly associated subunits are likely to be a general solution for increasing biological specificity. Here we show that there is yet more gain in Rdr specificity by assembly of distinct RDRCs responsible for separate sRNA biogenesis pathways. Mechanisms for RDRC subunit function in the biogenesis of T. thermophila sRNAs remain to be addressed. The subunits may govern specificity for recruitment to ssRNA templates, determine the synthesis of product structures with endogenous rather than foreign dsRNA hallmarks, and/or direct the fate of product dsRNA to siRNA generation or other currently unknown end points.
In comparing the activities and in vivo functions of the T. thermophila RDRCs resolved here, it is clear that some features are shared while others are distinct. By in vitro assays of dsRNA synthesis, T. thermophila RDRCs differing in the presence of Rdn1 or Rdn2 have similar activity. Likewise, both types of RDRC interact with Dcr2 and promote Dcr2 cleavage of RDRC products to generate ∼24-nt sRNA in vitro. In addition, both types of RDRC support equivalent nucleotidyl transferase activity on purified ssRNA templates. Beyond these similarities in Rdr1, Dcr2, and Rdn catalytic activities, differences are apparent. Curiously, only RDRC with Rdn1 can assemble the Rdf1 and Rdf2 subunits. Furthermore, loss of Rdn2 precludes in vivo accumulation of some sRNAs while loss of Rdf2 reduces accumulation of other sRNAs. Cellular phenotypes also distinguish loss of function by the different RDRCs: loss of Rdf1 or Rdf2 induces DNA segregation phenotypes, while loss of Rdn2 or Rdf1 halts conjugation. In addition to revealing a division of labor among RDRCs sharing the same Rdr1 catalytic core, our results demonstrate conclusively that T. thermophila Rdr1 and its associated proteins play important roles in accumulation of 23–24-nt sRNA in vivo.
In the simplest model for the biogenesis of strand-asymmetric T. thermophila 23–24-nt sRNAs in vivo, Rdr1 acts at the top of a pathway that selects RNA targets to yield sRNAs in a manner specified by RDRC context. In other organisms, Rdr family members are proposed to act downstream of initial sRNA generation. C. elegans RRF-1 acts downstream from primary siRNA to generate secondary siRNA bearing a 5′ triphosphate (Aoki et al. 2007; Pak and Fire 2007; Sijen et al. 2007). The S. pombe RDRC functions in a positive feedback loop that integrates transcription by DNA-dependent RNA polymerase, chromatin modification enzymes, an Argonaute-containing RITS complex and Dicer (Bühler and Moazed 2007). A. thaliana RDR6 generates endogenous siRNA from transcripts targeted by miRNA (Allen et al. 2005; Yoshikawa et al. 2005). In all these cases, whether dsRNA synthesis by the Rdr is considered initiating or amplifying, RDRCs share the common need to recognize a non-mRNA target transcript, such as one that may originate from a degenerate gene no longer encoding a functional protein (as for T. thermophila Rdr1) or a nascent RNA (as for S. pombe Rdp1) or a transcript otherwise compromised in its integrity (as for A. thaliana RDR6 or C. elegans RRF-1). Our findings suggest that this specificity for transcripts potentially recognized as aberrant mRNAs may be influenced by RDRC context.
Our finding that biochemically active poly(U) polymerases are subunits of T. thermophila RDRCs is intriguing in light of the recent recognition that these noncanonical nucleotidyl transferase proteins have wide conservation in diverse eukaryotes (Kwak and Wickens 2007; Martin and Keller 2007; Rissland and Norbury 2008). Some members of the family have been implicated to have function(s) in RNA silencing pathways in other organisms. Substitution of putative active site residues of S. pombe Cid12 disrupts RNAi-dependent heterochromatin formation and accumulation of centromeric siRNAs (Win et al. 2006). S. pombe Cid14, the ortholog of Saccharomyces cerevisiae Trf4/5, also functions in heterochromatic gene silencing (Bühler et al. 2007). For C. elegans RDE-3, substitutions predicted to inhibit nucleotidyl transferase activity abrogate protein function in RNAi (Chen et al. 2005).
How do T. thermophila Rdn1 and Rdn2 contribute to RDRC function in vivo? One plausible model is that they catalyze uridylation of ssRNA templates for Rdr1. Uridylation could enhance target RNA recognition by stabilization of the RNA 3′ end (Song and Kiledjian 2007; Wilusz and Wilusz 2008). On the other hand, uridylation has been linked to enhanced 5′ decapping and/or decay (Shen and Goodman 2004; Song and Kiledjian 2007; Heo et al. 2008; Mullen and Marzluff 2008; Wilusz and Wilusz 2008) and decapped transcripts may be preferential Rdr targets for dsRNA synthesis (Gazzani et al. 2004). Alternately, Rdn1 and Rdn2 may act on the sRNA duplexes produced by Dcr2 to impact Piwi loading or sRNA turnover. Indeed, approximately half of the cloned 23–24-nt sRNAs from T. thermophila include a nontemplated 3′ nucleotide that is most often uridine (Lee and Collins 2006). Terminal uridylation of miRNAs has been reported in C. elegans (Ruby et al. 2006), and destabilized sRNA in A. thaliana hen1 mutants can gain several terminal uridines (Li et al. 2005). More recently, 3′ adenylation of miR-122 in mice and human cells by the noncanonical poly(A) polymerase GLD-2 was implicated in miR-122 stabilization (Katoh et al. 2009). Much remains to be uncovered about the biochemical and biological specificity of noncanonical nucleotidyl transferases, which will be fascinating to dissect in future studies of T. thermophila as a favorable model system.
MATERIALS AND METHODS
RNA and DNA manipulations
Total RNA was isolated using Trizol (Invitrogen). Northern blot hybridization for mRNAs and Southern blot hybridizations used hexamer-labeled probes; sRNA blots were probed using end-labeled oligonucleotides as described previously (Lee and Collins 2006). Direct staining of RNA resolved on denaturing acrylamide gels (7 M urea) was performed using SYBR Gold (Molecular Probes). Open reading frames of RDN1, RDN2, RDF1, and RDF2 were sequenced from cDNA amplified by RT-PCR from total RNA. GenBank accession numbers for RDN1, RDN2, RDF1, and RDF2 sequences are EU009112, EU009113, EU009114, and EU009115, respectively. These revise predicted TTHERM protein numbers 00094094 (6.m00633), 00094000 (6.m00629), 01207560 (274.m00027), and 01207570 (274.m00028). Rdn1-associated p124 was identified as TTHERM_00782040 in the T. thermophila Genome Database (www.ciliate.org). The catalytically inactive Rdn2 variant (D267A, D269A) was created by site-specific mutagenesis.
Cell culture and strain construction
Cell cultures were grown shaking at 30°C in 2% proteose peptone, 0.2% yeast extract, 10 μM FeCl3 supplemented with 150 μg/mL ampicillin and streptomycin and 1.25 μg/mL amphotericin B (Fungizone). Cultures were starved by harvesting and transfer into 10 mM Tris-HCl at pH 7.5 with shaking for up to 24 h at 30°C. Conjugation was initiated through the mixing of an equal number of starved cells of each mating type, followed by incubation without shaking at 30°C.
To create gene knockdown or knockout strains, integration cassettes were designed to replace an ∼1.5-kbp region encompassing the catalytic motifs of RDN1 or RDN2 or the entire coding region of RDF1 or RDF2 with a standard T. thermophila expression cassette encoding resistance to neomycin (neo2). Cells were transformed by particle bombardment and selected for gene replacement using paromomycin as described previously (Witkin et al. 2007). To create strains expressing epitope-tagged Rdn1 and Rdn2, two sets of integration cassettes were constructed. For C-terminus tagging at endogenous loci, the tag elements were followed by the poly(A) signal of RPL29 and then the neo2 cassette. This integration unit was flanked by RDN1 or RDN2 genomic regions immediately prior to and following the translation stop codon. For N terminus tagging, the tag elements were fused to the start of the PCR-amplified protein coding region, and its translation stop codon was followed by the BTU1 poly(A) signal and neo2 cassette; this integration unit was targeted for integration by flanking BTU1 genomic regions upstream of the endogenous start codon and downstream of the poly(A) signal. Selection was performed using paromomycin. CD Rdr1 D1004A was expressed from a transgene integrated in substitution of the BTU1 locus under transcription control of a transplanted ∼1-kbp promoter region of MTT1 (Shang et al. 2002).
Cell staining and microscopy
Cells were washed once with 10 mM Tris-HCl and fixed for 1 h at room temperature in 2% paraformaldehyde prepared in PHEM buffer (60 mM PIPES at pH 6.9; 25 mM HEPES; 10 mM EGTA; 2 mM MgCl2). Cells were then washed for 5 min with modified PBS at pH 7.2 (130 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4, 10 mM EGTA, 2 mM MgCl2) and incubated in 0.1–1 μg/mL DAPI in PBS for 10 min with end-over-end rotation. Following three washes in PBS, cells were resuspended in PBS and mounted on a slide with 90% glycerol containing antifade. Cells were imaged using the 40× objective of an Olympus BX61 fluorescent microscope. Images were captured using Metamorph software.
Affinity purification, mass spectrometry, and activity assays
One-step affinity purifications of ZZ-tagged proteins using IgG agarose and mass spectrometry were performed as described previously (Lee and Collins 2007). Silver staining was used to detect proteins following SDS-PAGE. Rdr-mediated dsRNA synthesis, NTP transferase and coupled Rdr/Dicer assays were performed as described previously (Lee and Collins 2007), except that the unlabeled NTPs used were at 20 μM final concentration. Formamide denaturing acrylamide gels containing 7 M urea and 45% formamide were used in the analysis of Rdr and coupled Rdr/Dicer reaction products to eliminate dsRNA structure (Lee and Collins 2007).
ACKNOWLEDGMENTS
This work was supported by a Howard Hughes Medical Institute Predoctoral Fellowship to S.R.L and a National Science Foundation Predoctoral Fellowship to K.B.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1630309.
REFERENCES
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- Bühler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;18:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim Biophys Acta. 2008;1783:2130–2136. doi: 10.1016/j.bbamcr.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, Mello CC. A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr Biol. 2005;15:378–383. doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Curaba J, Chen X. Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem. 2008;283:3059–3066. doi: 10.1074/jbc.M708983200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte DJ, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, Gu L, Gorovsky MA. Electroporation-mediated replacement of a positively and negatively selectable β-tubulin gene in Tetrahymena thermophila. Proc Natl Acad Sci. 1994;91:4549–4553. doi: 10.1073/pnas.91.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003;28:1–23. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes & Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 2007;14:604–610. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes & Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. The Cid1 poly(U) polymerase. Biochim Biophys Acta. 2008;1779:286–294. doi: 10.1016/j.bbagrm.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell M, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Shiu PK, Zickler D, Raju NB, Ruprich-Robert G, Metzenberg RL. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc Natl Acad Sci. 2006;103:2243–2248. doi: 10.1073/pnas.0508896103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Yakisich JS, Kapler GM, Cole ES, Romero DP. A β-tubulin mutation selectively uncouples nuclear division and cytokinesis in Tetrahymena thermophila. Eukaryot Cell. 2004;3:1217–1226. doi: 10.1128/EC.3.5.1217-1226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes & Dev. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win TZ, Stevenson AL, Wang SW. Fission yeast Cid12 has dual functions in chromosome segregation and checkpoint control. Mol Cell Biol. 2006;26:4435–4447. doi: 10.1128/MCB.02205-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Prathapam R, Collins K. Positive and negative regulation of Tetrahymena telomerase holoenzyme. Mol Cell Biol. 2007;27:2074–2083. doi: 10.1128/MCB.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]