Abstract
Nonsense-mediated mRNA decay (NMD) is a eukaryotic quality-control mechanism that recognizes and degrades mRNAs with premature termination codons (PTCs). In yeast, PTC-containing mRNAs are targeted to processing bodies (P-bodies), and yeast strains expressing an ATPase defective Upf1p mutant accumulate P-bodies. Here we show that in human cells, an ATPase-deficient UPF1 mutant and a fraction of UPF2 and UPF3b accumulate in cytoplasmic foci that co-localize with P-bodies. Depletion of the P-body component Ge-1, which prevents formation of microscopically detectable P-bodies, also impairs the localization of mutant UPF1, UPF2, and UPF3b in cytoplasmic foci. However, the accumulation of the ATPase-deficient UPF1 mutant in P-bodies is independent of UPF2, UPF3b, or SMG1, and the ATPase-deficient UPF1 mutant can localize into the P-bodies independent of its phosphorylation status. Most importantly, disruption of P-bodies by depletion of Ge-1 affects neither the mRNA levels of PTC-containing reporter genes nor endogenous NMD substrates. Consistent with the recently reported decapping-independent SMG6-mediated endonucleolytic decay of human nonsense mRNAs, our results imply that microscopically detectable P-bodies are not required for mammalian NMD.
Keywords: mRNA processing body, mRNA surveillance, nonsense-mediated mRNA decay, RNA interference
INTRODUCTION
Eukaryotic cells possess quality control mechanisms that recognize and degrade aberrant mRNAs. A well-studied quality control mechanism is nonsense-mediated mRNA decay (NMD), which recognizes and degrades mRNAs whose open reading frame (ORF) is truncated by a premature translation-termination codon (PTC) (Behm-Ansmant et al. 2007; Chang et al. 2007; Isken and Maquat 2007; Mühlemann et al. 2008). Moreover, NMD also contributes to the post-transcriptional regulation of many physiological mRNAs. Depletion of the core NMD factor UPF1 alters the steady-state level of about 10% of all human mRNAs (Mendell et al. 2004). Furthermore, NMD modulates the phenotype of numerous genetic diseases; it is estimated that one-third of all disease-causing mutations generate a PTC (Holbrook et al. 2004).
The three up-frame shift (UPF) proteins UPF1, UPF2, and UPF3 are conserved from yeast to human and were identified as principal NMD factors in all eukaryotes studied so far. UPF1 is an ATP-dependent RNA helicase (Bhattacharya et al. 2000) that interacts with UPF2, which in turn interacts with UPF3 (Lykke-Andersen et al. 2000; Chamieh et al. 2008). UPF2 contains three conserved middle of eIF4G-like (MIF4G) domains and multiple putative nuclear localization signals (NLSs) and a putative nuclear export signal (NES). Humans encode two different UPF3 proteins, UPF3a and UPF3b, which both contain several NLSs and NESs (Lykke-Andersen et al. 2000; Serin et al. 2001). In metazoans, SMG1, SMG5, SMG6, and SMG7 were identified as additional NMD factors that regulate the phosphorylation status of UPF1 (for review, see Mühlemann et al. 2008).
The molecular mechanism of NMD is not yet fully understood, but recent studies suggest that the basic mechanism of PTC recognition is conserved from yeast to human (Stalder and Mühlemann 2008). According to the current NMD model, a ribosome stalled at a PTC recruits UPF1 and SMG1 through the eukaryotic release factors 1 and 3 (eRF1 and eRF3) (Kashima et al. 2006). UPF2 and UPF3b then associate with UPF1 and SMG1, which is required for the SMG1-mediated phosphorylation of UPF1 in mammals. Phosphorylated UPF1 then interacts with SMG5, SMG6, and/or SMG7, finally leading to the degradation of the mRNA (Stalder and Mühlemann 2008). The exact mechanism by which SMG5, SMG6, and SMG7 participate in the degradation of the PTC-containing (PTC+) mRNA and where in the cells this happens is still unclear. In human cells, SMG7 was found to localize to distinct cytoplasmic foci called processing bodies (P-bodies, also known as “GW182-bodies,” “DCP1-foci,” or “XRN1-foci”), and SMG5 and UPF1 localize to P-bodies when SMG7 is overexpressed (Unterholzner and Izaurralde 2004; Fukuhara et al. 2005; Durand et al. 2007). In contrast, the metazoan-specific NMD factor SMG6, which initiates degradation of PTC+ mRNA by an endonucleolytic cleavage near the PTC in Drosophila and human cells (Huntzinger et al. 2008; Eberle et al. 2009), does not co-localize with P-bodies in HeLa cells (Unterholzner and Izaurralde 2004; L Stalder and O Mühlemann, unpubl.).
P-bodies have been identified in both yeast and mammalian cells to be sites of mRNA turnover and storage (Sheth and Parker 2003; Cougot et al. 2004; for review, see Eulalio et al. 2007a; Parker and Sheth 2007; Franks and Lykke-Andersen 2008). P-bodies are dynamic structures characterized by a high local concentration of mRNA decapping enzyme (DCP1 and DCP2), activators of decapping (Ge-1, EDC3, Lsm1-7, RAP55, and RCK/p54), the 5′-3′ exonuclease XRN1, the deadenylation-complex CCR4-CAF1-NOT, and factors of the miRNA pathway (GW182, Argonaute proteins) (for review, see Eulalio et al. 2007a; Franks and Lykke-Andersen 2008). Furthermore, different mutants of Upf1p were found to accumulate P-bodies in Saccharomyces cerevisiae (Cheng et al. 2007), and PTC+ mRNAs were found to localize to P-bodies in an Upf1p-dependent manner (Sheth and Parker 2006). This suggests that NMD in S. cerevisiae involves targeting of PTC+ mRNAs to P-bodies. Based on the findings that (1) factors of the mRNA degradation machinery accumulate in P-bodies; (2) SMG5 and UPF1 localize to P-bodies in a SMG7-dependent manner; (3) Upf1p mutants accumulate in P-bodies in S. cerevisiae; and (4) PTC+ mRNAs are targeted to P-bodies in S. cerevisiae, it was suggested that P-bodies might also play an important role in mammalian NMD. On the other hand, it was shown that (1) NMD is not affected by Edc3p (enhancer of decapping 3), which functions in P-body formation in yeast (Kshirsagar and Parker 2004; Decker et al. 2007); (2) general mRNA turnover is not affected in yeast strains lacking microscopically detectable P-bodies (Decker et al. 2007); and (3) the depletion of the P-bodies by knocking down Lsm1 or Lsm3 does not prevent the function of microRNAs (miRNAs), RNA interference (RNAi), or NMD in Drosophila S2 cells (Eulalio et al. 2007b). Furthermore, the depletion of Dcp1, Dcp2, Ge-1, GW182, or other RNAi or miRNA factors does not inhibit NMD in Drosophila S2 cells (Rehwinkel et al. 2005; Eulalio et al. 2007b). In addition, microscopically detectable P-bodies can be depleted in human cells by knockdown of RCK/p54 or Lsm1 without affecting miRNA-mediated repression (Chu and Rana 2006), or by the knockdown of GW182 without affecting the decay of transcripts harboring AU-rich elements (AREs) (Stoecklin et al. 2006).
In the present study, we aimed to elucidate the role of P-bodies for NMD in human cells. We report enrichment of an ATPase-defective UPF1 mutant, but not of wild-type (WT) UPF1, and of a fraction of UPF2 and UPF3b protein in cytoplasmic foci that co-localize with P-bodies in human cells. The co-localization of the ATPase-defective UPF1 protein with P-bodies appears to be independent of UPF2, UPF3b, or SMG1, and the ATPase-deficient UPF1 mutant can co-localize with the P-bodies, independent of its phosphorylation status. This localization of the UPF1 mutant, UPF2, and UPF3b into cytoplasmic foci is lost upon disruption of P-bodies by knockdown of Ge-1. Most importantly, the depletion of P-bodies does neither affect the mRNA levels of PTC+ reporter genes nor the abundance of endogenous NMD substrates. Collectively, this demonstrates that microscopically detectable P-bodies are not required for mammalian NMD.
RESULTS AND DISCUSSION
ATPase-defective UPF1, UPF2, and UPF3b proteins localize to P-bodies
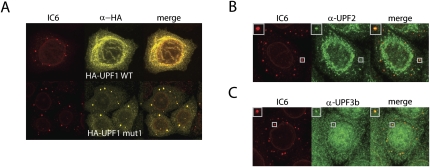
In order to investigate a potential functional relationship between mammalian NMD and P-bodies, we first characterized the cellular localization of the NMD factors UPF1, UPF2, and UPF3b. N-terminally HA-tagged UPF1 was expressed in HeLa cells, and 48 h after transfection the cells were fixed, permeabilized, and incubated with the antibodies. The human reference serum IC6 (Ou et al. 2004; Bloch et al. 2006) was used to visualize the P-bodies. The IC6 serum contains mainly antibodies against Ge-1 (also known as Hedls or EDC4), a major component of P-bodies, and it also stains the nuclear lamina (Fig. 1A, left; Ou et al. 2004; Fenger-Gron et al. 2005; Yu et al. 2005; Bloch et al. 2006). HA-UPF1 WT is distributed quite evenly throughout the cytoplasm, with some tendency to form a fibrillar mesh (Fig. 1A, upper part). In contrast, a large fraction of the HA-tagged UPF1 mutant (Fig. 1A, HA-UPF1 mut1), which bears a K498Q mutation in the ATPase domain (Kashima et al. 2006), accumulated in P-bodies (Fig. 1A, lower part). This is similar to yeast, where expression of a corresponding Upf1 K436A mutant in an upf1Δ strain accumulated large P-bodies (Cheng et al. 2007).
FIGURE 1.
An ATPase-deficient UPF1 mutant, UPF2, and UPF3b localize to P-bodies in HeLa cells. (A) HeLa cells were stained with patient autoimmune serum IC6, which mainly contains α-Ge-1 antibodies, to visualize the P-bodies (shown in red; left column). HA-tagged UPF1 WT or the ATPase-deficient HA-UPF1 mut1 (Kashima et al. 2006) was expressed from transfected plasmids and detected with α-HA antibody (shown in yellow; middle column). The merge of both channels is shown in the right column. Endogenous (B) UPF2 and (C) UPF3b was visualized using rabbit polyclonal antisera against the respective proteins (Lykke-Andersen et al. 2000). Insets in the upper-left corner of the pictures in panels B and C show a single P-body zoomed in.
In a previous study, it was shown that UPF2 localizes perinuclear and that UPF3b is mainly nuclear (Lykke-Andersen et al. 2000). Using the same antibodies as Lykke-Andersen and colleagues, we also find most of the endogenous UPF2 localizing perinuclear, but a minor fraction co-localizes with P-bodies (Fig. 1B). Likewise, a small fraction of the otherwise mainly nuclear UPF3b also co-localizes with P-bodies (Fig. 1C). The finding that at a given time point only a minor fraction of the cell's total UPF2 and UPF3b protein is localized to P-bodies, whereas the major fractions localize perinuclear or nuclear, respectively, suggests a highly dynamic association of UPF2 and UPF3b with P-bodies.
RNAi-mediated depletion of Ge-1 disrupts P-bodies and the localization of mutant UPF1, UPF2, and UPF3b to cytoplasmatic foci
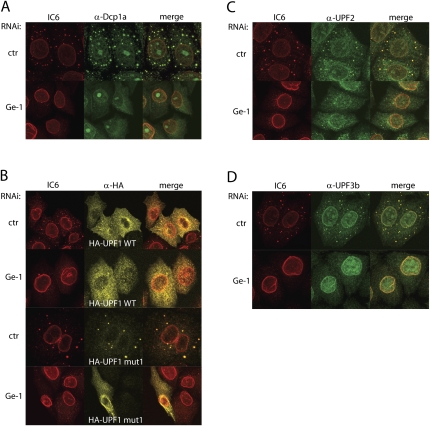
Next we wanted to test if the co-localization of the recombinant UPF1 mutant, and endogenous UPF2 and UPF3b with the IC6-stained cytoplasmatic foci depends on intact P-bodies. We therefore knocked down Ge-1, which was previously shown to be an essential component for the formation of the P-bodies (Yu et al. 2005). Ninety-six hours after transfection with shRNA-expressing plasmids, the cells were fixed, permeabilized, and incubated with the antibodies. To monitor the efficiency of the Ge-1 knockdown microscopically, the cells were stained with the IC6 serum and with an antibody against the decapping protein 1a (DCP1a). As knockdown control (ctr), a plasmid encoding a shRNA with no predicted target in human cells was used (Buhler et al. 2006). In the knockdown control, the cytoplasmic foci stained by IC6 serum and by the α-DCP1a antibody co-localized completely (Fig. 2A, upper part), suggesting that all P-bodies containing Ge-1 also contain DCP1a. In cells with a Ge-1 knockdown, no P-bodies could be observed anymore as judged by the uniform cytoplasmic distribution of DCP1a under these conditions (Fig. 2A, lower part). This demonstrates that knocking down Ge-1 disrupts microscopically detectable Ge-1 and DCP1a-containing P-bodies. Notably, a fraction of DCP1a was always observed in nuclear foci that might represent nucleoli (Fig. 2A). Whether this represents unspecific binding of the antibody or whether some DCP1a indeed resides in the nucleoli remains unclear.
FIGURE 2.
The localization of HA-UPF1 mut1, UPF2, and UPF3b to cytoplasmic foci depends on the integrity of the P-bodies. (A) HeLa cells were transfected with a plasmid encoding a shRNA targeting Ge-1 or encoding a shRNA with no predicted target in human cells (control). The cells were stained with the IC6 serum (red) and α-DCP1a antibodies (green). (B) Ge-1 was knocked down, using the same conditions as in A, and HA-UPF1 WT or HA-UPF1 mut1 were co-transfected. The cells were stained with the IC6 serum (red) to visualize the P-bodies, and with α-HA antibody (yellow). (C,D) Ge-1 knockdown as in A and staining of the cells with the IC6 serum (red), with (C) α-UPF2 or (D) α-UPF3b antibodies (green).
Next we examined if intact P-bodies are required for the NMD factors UPF1, UPF2, and UPF3b to localize in cytoplasmic foci, or if these factors are sufficient to form such foci on their own. First, we knocked down Ge-1 in HeLa cells that were co-transfected with HA-UPF1 WT or HA-UPF1 mut1, respectively. As expected, neither the control nor the Ge-1 knockdown did affect the localization of HA-UPF1 WT (Fig. 2B, upper two rows). Similar to Figure 1A, HA-UPF1 WT was distributed throughout the cytoplasm with a slight fibrillar and dotty enrichment around the nucleus. In contrast, the P-body localization of HA-UPF1 mut1 was lost upon knockdown of Ge-1, and HA-UPF1 mut1 now also distributed throughout the cytoplasm in a similar pattern as HA-UPF1 WT (Fig. 2B, third and fourth rows). Furthermore, the co-localization of endogenous UPF2 and UPF3b with the P-bodies was also impaired in cells depleted of Ge-1 (Fig. 2C,D). During the course of these experiments, we noticed that for unknown reasons the proportion of UPF2 and UPF3b co-localizing with P-bodies seemed generally higher in cells with a control knockdown compared to untreated HeLa cells (cf. Fig. 1B,C and Fig. 2C,D). Collectively, these results show that the localization of the exogenously expressed HA-UPF1 mut1, as well as the endogenous UPF2 and UPF3b in cytoplasmic foci depend on the integrity of the P-bodies, suggesting that UPF1, UPF2, and UPF3b are not sufficient to form such foci on their own.
Disruption of P-bodies does not stabilize NMD substrates
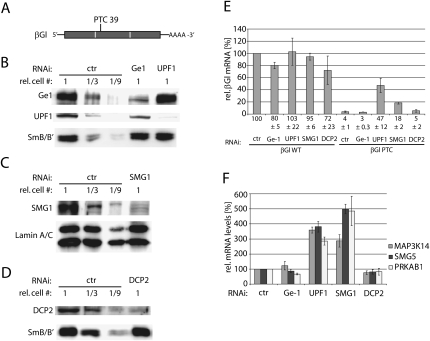
Next we wanted to test if the disruption of the P-bodies by knocking down Ge-1 influences the mRNA levels of PTC+ transcripts. Knockdown of the NMD core factor UPF1 served as a positive control. A β-globin (βGl) minigene was used as NMD reporter gene (Fig. 3A; Thermann et al. 1998; Mohn et al. 2005). Judged from the Western blots shown in Figures 3B–D, the amount of Ge-1 and UPF1 protein was reduced more than threefold, and the amount of SMG1 and DCP2 protein was reduced at least ninefold in the respective knockdowns. The βGl WT mRNA levels were reduced to 80% in the Ge-1 knockdown and to 72% in the DCP2 knockdown, and remained constant under knockdown conditions of UPF1 or SMG1, respectively (Fig. 3E). The mRNA level of the PTC+ βGl construct was reduced to 4% of the βGl WT construct in the control knockdown, reflecting the efficiency of NMD. Surprisingly, the mRNA of the PTC+ βGl was not stabilized upon the Ge-1 or DCP2 knockdowns, whereas the mRNA level increased about 15-fold to 47% of βGl WT upon the knockdown of UPF1 and about 4.5-fold to 18% of βGl WT upon the knockdown of SMG1 (Fig. 3E). Additionally, we also measured the effect of Ge-1, UPF1, SMG1, and DCP2 knockdowns on the mRNA levels of several known endogenous NMD targets. In a microarray study, MAP3K14 (for “mitogen-activated protein kinase kinase kinase 14”), SMG5 (for “suppressor with morphological effect on genitalia”), and PRKAB1 (for “5′-AMP-activated protein kinase subunit beta-1”) showed increased mRNA levels upon UPF1 knockdown (Mendell et al. 2004). In accordance with the microarray data, the mRNA steady-state levels of MAP3K14, SMG5, and PRKAB1 increased three- to fivefold upon depletion of UPF1 or SMG1. In contrast, knockdown of Ge-1 or DCP2 did not change the relative abundance of MAP3K14, SMG5, and PRKAB1 mRNA (Fig. 3F). Given that (1) an ATPase defective UPF1 mutant, UPF2 and UPF3b localize to P-bodies in a Ge-1-dependent manner (Figs. 1, 2B–D); (2) and that microscopically detectable P-bodies are disrupted by Ge-1 knockdown (Fig. 2A); (3) but that the transcripts of neither NMD reporter genes nor endogenous NMD targets are stabilized by a Ge-1 knockdown (Fig. 3E,F), these results suggest that the localization of NMD factors to P-bodies is not required for NMD in human cells. These results are consistent with previous findings in Drosophila S2 cells, where disruption of P-bodies by depletion of GW182 or Ge-1 also did not affect NMD (Rehwinkel et al. 2005; Eulalio et al. 2007b), and may reveal a metazoan-specific feature of NMD.
FIGURE 3.
The disruption of P-bodies does not influence NMD. (A) Schematic depiction of the previously characterized NMD reporter transcript β-globin NS39 (Thermann et al. 1998). The open reading frame is shown as gray bar, exon–exon junctions are depicted by white lines, and the amino acid position of the PTC is indicated. (B–D) The efficacy of the RNAi-mediated Ge-1, UPF1, SMG1, and DCP2 knock-downs is monitored by Western blot analysis. Detection of SmB/B′ or Lamin A/C served as loading control. (E) A Ge-1 knockdown was performed as in Figure 2A, and additionally, the NMD reporter gene β-globin was co-transfected. The steady-state mRNA levels were quantified by RT-PCR and normalized to the miniμ WT mRNA from a co-transfected plasmid. (F) Relative mRNA levels of the three endogenous NMD substrates MAP3K14, SMG5, and PRKAB1 were measured after knockdown of Ge-1, UPF1, SMG1, or DCP2. Endogenous GAPDH mRNA levels were used for normalization. Average values from one representative experiment with three real-time PCR runs each are shown.
Although the knockdowns of Ge-1 or DCP2 are efficient (Fig. 3B,D) we cannot exclude residual decapping activity. However, the observations that the mRNA levels from NMD substrates are not affected by the depletion of Ge-1 or DCP2 are consistent with recently published studies, which document decapping-independent NMD in human and Drosophila S2 cells (Rehwinkel et al. 2005; Huntzinger et al. 2008; Eberle et al. 2009). Given that in human and Drosophila S2 cells, nonsense mRNAs are efficiently degraded via SMG6-mediated endonucleolytic cleavage near the PTC (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009), one would not a priori expect a requirement for decapping activity in metazoan NMD. On the other hand, evidence for a role of decapping in human NMD has also been reported. First, UPF1 was shown to interact with the decapping complex via PNRC2 (Cho et al. 2009). Moreover, more decapped PTC+ β-globin transcripts were detected compared WT β-globin by using a circularization real-time polymerase chain reaction (RT-PCR) assay (Couttet and Grange 2004), and an increase of PTC+ (and to a lesser extent WT) β-globin and GPx1 mRNA levels upon knockdown of DCP2 has been observed (Lejeune et al. 2003). Collectively, the currently available data suggest that in mammals both decay pathways may be active in NMD, and we do not know yet if indeed the SMG6-dependent, decapping-independent endocleavage pathway prevails, or if the conventional decapping-dependent exonucleolytic pathway is also important.
The localization of mutant UPF1 to P-bodies is independent of its phosphorylation status and does not require UPF2, UPF3b, and SMG1
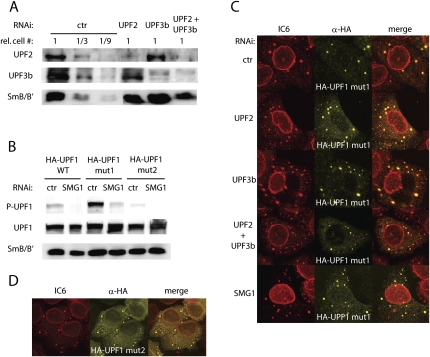
With regard to the findings in yeast (Sheth and Parker 2006), it was unexpected to find that UPF1 mut1, UPF2, and UPF3b can localize to P-bodies in a Ge-1-dependent manner, but that a knockdown of Ge-1 or DCP2 did not stabilize PTC+ mRNAs. Although alternative explanations are also possible, this could indicate that the localization of UPF1, UPF2, and UPF3b to the P-bodies might not be linked to NMD, but to another cellular process instead. An essential step in NMD is the phosphorylation of UPF1, which is mediated by SMG1, and it has been shown that SMG1, UPF2, and UPF3b are required for this phosphorylation step (Yamashita et al. 2001; Kashima et al. 2006). SMG7 binds phosphorylated UPF1 through its N-terminal 14-3-3 like domain, and it has been shown that SMG7 can target UPF1 to P-bodies (Unterholzner and Izaurralde 2004; Fukuhara et al. 2005). Thus, if UPF1 localizes to the P-bodies during the process of NMD, one would predict that this localization requires the SMG1-, UPF2-, and UPF3b-dependent phosphorylation of UPF1. To test this hypothesis, we knocked down SMG1, UPF2, or UPF3b in HeLa cells, and co-transfected the HA-UPF1 mut1. The efficiency of the knockdowns was monitored by Western blotting (Figs. 3C, 4A). Consistent with previous publications (Yamashita et al. 2001; Kashima et al. 2006), phosphorylation of the otherwise hyperphosphorylated HA-UPF1 mut1 was strongly reduced in SMG1-depleted cells (Fig. 4B, lanes 3,4). As already seen before, HA-UPF1 mut1 accumulated in P-bodies in the cells with the control knockdown (Fig. 4C, upper row). Upon knockdown of SMG1, UPF2, or UPF3b, HA-UPF1 mut1 still localized to the P-bodies, indistinguishable from the control knockdown (Fig. 4C). Even in cells with a combined knockdown of UPF2 and UPF3b together, HA-UPF1 mut1 still accumulated in P-bodies. This SMG1-, UPF2-, and UPF3b-independent localization of HA-UPF1 mut1 to P-bodies suggests that the UPF1 protein residing in P-bodies does not need to be phosphorylated, at least not in a SMG1-, UPF2-, or UPF3b-dependent manner. To test this hypothesis, we used a double mutant (HA-UPF1 mut2), which in addition to the ATPase deficiency has mutated the four preferentially by SMG1 phosphorylated S/Q-motifs in the C-terminus (S1073A, S1078A, S1096A, S1116A) (from Kashima et al. 2006). The phosphorylation state of HA-UPF1 mut2 is strongly reduced compared to HA-UPF1 mut1 and HA-UPF1 WT (Fig. 4B, lanes 1,3,5; the residual signal stems mainly from the endogenous UPF1 in the cells). Nevertheless, a large fraction of HA-UPF1 mut2 still accumulated in P-bodies (Fig. 4D), demonstrating that the ATPase deficient UPF1-mutant can localize to P-bodies independent of its phosphorylation status. The fact that the ATPase deficient UPF1 can localize to P-bodies in a nonphosphorylated state and in a SMG1-, UPF2-, and UPF3b independent manner suggests that the fraction of the P-body confined UPF1-mutant is not active in NMD, which is consistent with the conclusion that P-bodies are not required for mammalian NMD.
FIGURE 4.
The ATPase-deficient UPF1 localizes to P-bodies independent of SMG1, UPF2, and UPF3b. (A) The efficacy of the knockdowns was monitored by Western blot analysis. Detection of SmB/B′ served as loading control. (B) Western blot of SMG1-depleted cells expressing HA-UPF1 WT, HA-UPF1 mut1, or HA-UPF1 mut2. Detection of SmB/B′ served as loading control. (C) Cells expressing HA-UPF1 mut1 and with a knockdown of UPF2 (second row), UPF3b (third row), UPF2 and UPF3b together (fourth row), or with SMG1 (bottom row) were stained with the IC6 serum (red) and with α-HA antibodies (yellow). (D) HeLa cells transfected with HA-UPF1 mut2 were stained with the IC6 serum (red) and with α-HA antibodies (yellow).
CONCLUSIONS
In this study, we show that an exogenously expressed ATPase-deficient UPF1 mutant and a minor fraction of the endogenous UPF2 and UPF3b accumulate in cytoplasmic foci that co-localize with P-bodies in HeLa cells, whereas wild-type UPF1 was not detected in P-bodies. RNAi-mediated depletion of Ge-1 prevented the formation of microscopically detectable P-bodies and also abrogated localization of the ATPase-defective UPF1 mutant, UPF2, and UPF3b in cytoplasmic foci. However, under these conditions, NMD was not affected, demonstrating that the microscopically detectable P-bodies are not required for mammalian NMD. P-bodies have also been found to be dispensable for NMD in Drosophila cells (Rehwinkel et al. 2005; Eulalio et al. 2007b), whereas in S. cerevisiae, Upf1p was shown to target PTC+ transcripts to P-bodies (Sheth and Parker 2006). This apparent difference between yeast and metazoan cells might reflect mechanistic differences in the degradation of NMD substrates. In Drosophila and human cells, SMG6 can initiate degradation of PTC+ transcripts by an endonucleolytic cleavage (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009), and SMG6 resides outside of P-bodies in human cells (Unterholzner and Izaurralde 2004). It is conceivable that such an endonucleolytic degradation pathway operates independently of decapping activity (consistent with our negative result in the DCP2 knockdown) (Fig. 3) and hence also independently of P-bodies. In yeast NMD, which lacks a SMG6 homolog, NMD relies on decapping followed by exonucleolytic decay, as evidenced by accumulation of PTC+ transcripts in P-bodies of dcp1Δ strains (Sheth and Parker 2006).
Even if microscopically visible P-bodies are not required for the function of NMD in human cells, our experiments do not exclude the possibility that some steps of NMD occur within submicroscopic P-bodies under physiological conditions. In this case, SMG6-mediated endonucleolytic cleavage and the localization of the mRNP destined for degradation into the P-bodies would have to occur prior to UPF1 phosphorylation and ATP hydrolysis in mammalian NMD. According to this view, the accumulation of the ATPase-deficient UPF1 in P-bodies would be the consequence of inhibition of a late step in NMD that takes place in P-bodies. This scenario is, however, inconsistent with the current mechanistic model of NMD, according to which SMG1-mediated UPF1 phosphorylation precedes the initiation of RNA degradation (Behm-Ansmant and Izaurralde 2006; Stalder and Mühlemann 2008). Moreover, because the ATPase-deficient UPF1 also co-localizes with P-bodies independently of its phosphorylation status (Fig. 4), this mutant UPF1 protein accumulating in P-bodies most likely does not represent a trapped functional intermediate of the NMD pathway, but rather results from a cellular process unrelated to NMD. This interpretation is consistent with the conclusion derived from the Ge-1 knockdown, namely, that P-bodies are not required for mammalian NMD.
Collectively, our results are also consistent with the emerging view that P-bodies are not cellular structures that recruit RNPs destined for degradation, but rather represent microscopically visible aggregates of mRNPs forming as a consequence of a temporarily high load on mRNA silencing and decay pathways in cells (Eulalio et al. 2007a; Franks and Lykke-Andersen 2008). Undoubtedly, more work is required to clarify if P-bodies are involved in NMD in human cells under physiological conditions.
MATERIALS AND METHODS
Plasmids
The plasmids SR-HA-UPF1 WT and SR-HA-UPF1 K498Q (HA-UPF1 mut1) are described elsewhere (Kashima et al. 2006). SR-HA-UPF1 K498Q_4SA (HA-UPF1 mut2) was generated by insertion of the Kpn I–Xho I fragment of SR-HA-UPF1 4SA (Kashima et al. 2006) into Kpn I–Xho I digested SR-HA-UPF1 K498Q. pSUPERpuro-scrambled for control knockdowns, pSUPERpuro-hUPF1, pSUPERpuro-hUPF2, and pSUPERpuro-hUPF3b are described elsewhere (Paillusson et al. 2005; Buhler et al. 2006; Eberle et al. 2008). pSUPERpuro-Ge-1A, pSUPERpuro-Ge-1B, pSUPERpuro-Ge-1C, pSUPERpuro-SMG1, and pSUPERpuro-DCP2 were generated by insertion of double-stranded oligonucleotides encoding for shRNAs into pSUPERpuro between the Bgl II and Hin dIII sites as described previously (Brummelkamp et al. 2002). The three target sequences for Ge-1 were 5′-AGGCCATGTTCCAGCAAAT-3′ (pSUPERpuro-Ge-1A), 5′-GGAATGGCCTTCAGGAAAA-3′ (pSUPERpuro-Ge-1B), and 5′-GCATACGGGATGAGATCAA-3′ (pSUPERpuro-Ge-1C), the target sequence for SMG1 was 5′-GATGAATGGTGGAGAGTTA-3′ (pSUPERpuro-SMG1), and the target sequence for DCP2 was 5′-CACGGAAACTTCAGGATAA-3′ (pSUPERpuro-DCP2).
Cell culture, transient transfections, and RNAi
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen), supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin (Amimed). For all transfections, 2 × 105 HeLa cells were seeded into six-well plates and transfected the next day. For Figure 1, 500 ng SR-HA-UPF1 WT or SR-HA-UPF1 K498Q were transfected with 4 μL DreamFect (OZ Biosciences) according to the manufacturer's protocol. The cells were then fixed, permeabilized, and incubated with the antibodies 48 h after the transfection. For Figures 2 and 4, the cells were transfected with 400–800 ng SR-HA-UPF1 and with 400 ng pSUPERpuro using 4 μL DreamFect. For Figure 3, the cells were transfected with 100 ng reporter plasmid, 100 ng plasmid encoding gene for normalization, 400 ng pSUPERpuro, and 4 μL DreamFect. For normalization, pβ μwt containing an iron-responsive element (IRE) in the 5′UTR was used (Stalder and Mühlemann 2007). Twenty-four hours after transfection, untransfected cells were eliminated by culturing the cells in the presence of 1.5 μg/mL puromycin for 48 h. For Figure 3, the medium was replaced by fresh DMEM and the cells were harvested the day after. For Figures 2 and 4, the cells were then washed in PBS, trypsinized and 4–8 × 105 cells were seeded onto coverslips in six-well plates. The cells were fixed, permeabilized, and incubated with the antibodies the day after. The knock-down control was obtained by transfection of pSUPERpuro scrambled, which endcodes a shRNA with no predicted target in human cells (Buhler et al. 2006). The knockdown of Ge-1, UPF1, UPF2, and UPF3b was induced by transfection of pSUPERpuro plasmids targeting three different sequences in Ge-1 (see above), two different sequences in UPF1 (Paillusson et al. 2005), one sequence in UPF2 (Wittmann et al. 2006), or two different sequences in UPF3b (Eberle et al. 2008), respectively. In all RNAi experiments, the efficiency of the knockdown was assessed on the mRNA level by quantitative RT-PCR (data not shown).
Quantitative RT-PCR
Total cellular RNA was isolated using “Absolutely RNA RT-PCR Miniprep Kit” (Stratagene) and 1 μg RNA was reverse transcribed in 20 μL StrataScript 6.0 RT buffer in the presence of 1 mM dNTPs, 300 ng random hexamers, 40 U RNasin (Promega), and 50 U StrataScript 6.0 reverse transcriptase (Stratagene) according to the manufacturer's protocol. RT-PCR was performed essentially as previously described (Buhler et al. 2006). Ge-1, MAP3K14, SMG5, and PRKAB1 mRNA was measured using assay-on-demand reagents Hs00204913_m1, Hs01089749_m1, Hs00383399_m1, and Hs00272166_m1 from Applied Biosystems.
Immunohistochemistry
Cells numbering 4–8 × 105 were seeded onto coverslips in six-well plates. The day after, the cells were washed with PBS, fixed for 30 min at 37°C in DSP-IF-buffer (PBS containing 2 mM MgCl2, 10% glycerol, 0.5 mM Dithiobis [succinimidyl propionate; DSP, Pierce]), and subsequently washed five times with DSP-IF-buffer without DSP and permeabilized for 20 min with PBS containing 0.2% Triton X-100 and 0.2 M glycine. The cells were incubated 30 min with Signal-iT FX Signal Enhancer (Invitrogen), washed with PBS and then incubated with polyclonal rabbit anti-UPF2, anti-UPF3b, or anti-DCP1a antiserum (Lykke-Andersen et al. 2000; Lykke-Andersen and Wagner 2005), with rabbit anti-HA antibody (Santa Cruz Biotechnology), or with human IC6 serum. Anti-rabbit Alexa-Fluor 488, anti-rabbit Alexa-Fluor 647 (Molecular Probes), or anti-human Alexa-Fluor 546 (Molecular Probes) were used as secondary antibodies. All antibodies and antisera were used at 1:500 dilutions. The antibodies were incubated in PBS containing 10% fetal calf serum, and the coverslips were mounted with mowiol (Calbiochem). Images were collected on a Leica TCS SP2 AOBS laser scanning confocal microscope equipped with a HCX PL APOlbd.BL 63.0 1.2W objective (Leica Microsystems, Inc.).
Immunoblotting
Whole cell lysates corresponding to 0.11 × 104–1 × 105 cells per lane were electrophoresed on a 10% SDS-PAGE. Proteins were transferred to Optitran BA-S 85 reinforced nitrocellulose (Schleicher and Schuell) and probed with 1:1000 diluted polyclonal rabbit anti-UPF2, anti-UPF3b, or anti-Ge-1 antiserum (Lykke-Andersen et al. 2000; Fenger-Gron et al. 2005), 1:1000 diluted rabbit anti-UPF1 antibody (Santa Cruz Biotechnology), 1:2000 diluted rabbit anti-DCP2 antibody (Fenger-Gron et al. 2005), 1:500 diluted mouse anti-SMG1 (Abnova; M02), 1:1000 diluted rabbit anti-phospho-(Ser/Thr) antibody (Cell Signaling technology), 1:1000 mouse anti-Lamin A/C (Santa Cruz Biotechnology), or 1:400 diluted supernatant of the mouse hybridoma cell line Y12, which produces a monoclonal antibody against the human Sm B/B′ proteins (Lerner et al. 1981). A 1:2500 dilution of HRP-conjugated anti-rabbit IgG or HRP-conjugated anti-mouse IgG (Promega) was used as secondary antibody. ECL+ Western blotting detection system (Amersham) was used for detection and signals were visualized on a luminescent image analyzer LAS-1000 (Fujifilm).
ACKNOWLEDGMENTS
We are grateful to J. Lykke-Andersen for providing anti-UPF2, anti-UPF3b, anti-DCP2, and anti-Ge-1 antibodies;M. Fritzler for providing the IC6 autoimmune serum; D. Schümperli for providing Y12 supernatant; D. Zünd and M. Ruepp for technical assistance; and A. Yamashita for providing UPF1 expression plasmids. L.S. was supported by a fellowship of the Roche Research Foundation and O.M. is a fellow of the Max Cloëtta Foundation. This work was supported by the Kanton Bern and by grants to O.M. from the European Research Council, the Swiss National Science Foundation, the Novartis Foundation for Biomedical Research, and the Helmut Horten Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1672509.
REFERENCES
- Behm-Ansmant I, Izaurralde E. Quality control of gene expression: A stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes & Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch DB, Gulick T, Bloch KD, Yang WH. Processing body autoantibodies reconsidered. RNA. 2006;12:707–709. doi: 10.1261/rna.17406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. EJC-independent degradation of nonsense immunoglobulin-μ mRNA depends on 3′ UTR length. Nat Struct Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Grange T. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res. 2004;32:488–494. doi: 10.1093/nar/gkh218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes & Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Mohn F, Buhler M, Mühlemann O. Nonsense-associated alternative splicing of T-cell receptor β genes: No evidence for frame dependence. RNA. 2005;11:147–156. doi: 10.1261/rna.7182905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O, Eberle AB, Stalder L, Orozco RZ. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Ou Y, Enarson P, Rattner JB, Barr SG, Fritzler MJ. The nuclear pore complex protein Tpr is a common autoantigen in sera that demonstrate nuclear envelope staining by indirect immunofluorescence. Clin Exp Immunol. 2004;136:379–387. doi: 10.1111/j.1365-2249.2004.02432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson A, Hirschi N, Vallan C, Azzalin CM, Mühlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Mühlemann O. Transcriptional silencing of nonsense codon-containing immunoglobulin μ genes requires translation of its mRNA. J Biol Chem. 2007;282:16079–16085. doi: 10.1074/jbc.M610595200. [DOI] [PubMed] [Google Scholar]
- Stalder L, Mühlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes & Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Yang W-H, Gulick TOD, Bloch KD, Bloch DB. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA. 2005;11:1795–1802. doi: 10.1261/rna.2142405. [DOI] [PMC free article] [PubMed] [Google Scholar]