Abstract
The RNA ligase-containing or L-complex is the core complex involved in uridine insertion/deletion RNA editing in trypanosome mitochondria. Blue native gels of glycerol gradient-separated fractions of mitochondrial lysate from cells transfected with the TAP-tagged editing protein, LC-8 (TbMP44/KREPB5), show a ∼1 MDa L-complex band and, in addition, two minor higher molecular weight REL1-containing complexes: one (L*a) co-sedimenting with the L-complex and running in the gel at around 1.2 MDa; the other (L*b) showing a continuous increase in molecular weight from 1 MDa to particles sedimenting over 70S. The L*b-complexes appear to be mainly composed of L-complex components, since polypeptide profiles of L- and L*b-complex gradient fractions were similar in composition and L*b-complex bands often degraded to L-complex bands after manipulation or freeze–thaw cycles. The L*a-complex may be artifactual since this gel shift can be produced by various experimental manipulations. However, the nature of the change and any cellular role remain to be determined. The L*b-complexes from both lysate and TAP pull-down were sensitive to RNase A digestion, suggesting that RNA is involved with the stability of the L*b-complexes. The MRP1/2 RNA binding complex is localized mainly in the L*b-complexes in substoichiometric amounts and this association is RNase sensitive. We suggest that the L*b-complexes may provide a scaffold for dynamic interaction with other editing factors during the editing process to form the active holoenzyme or “editosome.”
Keywords: RNA editing, trypanosomes, mitochondria, editosome, L-complex
INTRODUCTION
The L-complex was first identified as a single band in a native gel of Leishmania tarentolae mitochondrial extract, which sedimented in a glycerol gradient at around 20S–25S (Peris et al. 1997). The term “L-complex” refers to the presence of the auto-adenylatable REL1 and REL2 RNA editing ligases (McManus et al. 2001; Rusche et al. 2001). Identical bands were observed for Trypanosoma brucei, both in terms of sedimentation in glycerol gradients (McManus et al. 2001; Panigrahi et al. 2001; Rusche et al. 2001; Schnaufer et al. 2001) and migration in native gels (Peris et al. 1997). The term, “editosome,” has been used for the homologous complex from T. brucei (Panigrahi et al. 2003a; Stuart et al. 2005; Panigrahi et al. 2006). The L-complex from L. tarentolae and the corresponding complex from T. brucei were shown to consist of ∼18–20 polypeptides, with the functionally identified proteins being the REL1 and REL2 RNA ligases, the REX1 and REX2 3′-5′ U-specific exonucleases, the REN1, REN2, and REN3 RNA editing site-specific endonucleases, and the RET2 3′ uridylyl transferase (Aphasizhev et al. 2003a; Panigrahi et al. 2003a,b; Simpson et al. 2003, 2004; Stuart et al. 2004, 2005). In addition, there are several zinc finger-motif proteins and several additional putative RNA-binding proteins with single-strand binding motifs. Evidence has been presented for a minor L-complex heterogeneity, where the endonucleases are associated with specific subsets of 3-4 proteins (Panigrahi et al. 2006; Carnes et al. 2008). The ∼20S L-complex material purified from L. tarentolae mitochondria by REL1-TAP pull-down showed, in addition, substoichiometric amounts of RET1 3′ TUTase and the MRP1 and MRP2 RNA-binding proteins, both of which are associated with the editing complex by RNase-sensitive linkers (Aphasizhev et al. 2003a,b). The REH1 RNA helicase has also been shown to be an RNA-linked substoichiometric component of the editing complex (F Li and L Simpson, unpubl.).
L-complex gradient fractions were shown to have several partial editing reactions in vitro. These include gRNA-mediated precleaved U-insertion and U-deletion reactions (Igo et al. 2000; Kang et al. 2005) and, at a lower efficiency, full-cycle gRNA-mediated U-insertion and U-deletion reactions (Seiwert and Stuart 1994; Byrne et al. 1996; Kable et al. 1996; Piller et al. 1996; Seiwert et al. 1996). The in vitro editing data were obtained mainly for intrablock single site editing, but a recent report (Alatortsev et al. 2008) has demonstrated an efficient two-site in vitro editing reaction in which the second site editing appears to be coupled to that of the first site. There are no reports of in vitro multi-gRNA-mediated processive editing.
Several accessory factors have also been characterized, which appear to be involved in general mitochondrial RNA metabolism, but also have some editing-specific roles. The mitochondrial TbRGG2 protein binds preferentially the oligo (U) tail of gRNAs and a small fraction co-immunoprecipitates with editing complexes (Fisk et al. 2008). Down-regulation of expression of TbRGG2 led to a dramatic decrease in pan-edited mRNA and to a moderate stabilization of never-edited and pre-edited mRNAs (Fisk et al. 2008). RBP16 is an oligo (U) binding protein that is associated with ∼30% of the gRNA population. Depletion of this protein led to decrease of the never-edited ND4 and COI mRNAs and severe effects on editing of Cyb mRNA (Pelletier and Read 2003; Miller et al. 2006). RBP16 also stimulated in vitro RNA editing (Miller et al. 2006).
The MRP1 and MRP2 proteins act in vitro as nonspecific RNA matchmakers and catalyze RNA annealing (Koller et al. 1997; Allen et al. 1998; Lambert et al. 1999; Muller et al. 2001; Aphasizhev et al. 2003b; Zikova et al. 2008). Simultaneous down-regulation of expression of both proteins showed a phenotype similar to that of down-regulation of RBP16 (Vondruskova et al. 2005). It has been proposed that MRP1/2 catalyzes the initial annealing of the gRNA and the mRNA to form the anchor duplex, as well as having a general RNA regulatory role, but there is yet no direct evidence for the former model.
Several additional macromolecular complexes have been identified, which are involved with gRNA metabolism, and some of which also interact via RNA linkers with the L-complex. The GRBC1 and GRBC2 proteins (originally AP1 and AP2) (Aphasizhev et al. 2003b) together with eight additional proteins form a 200 kDa heterotetramer, which binds gRNA and acts as a gRNA-stabilizing factor (Panigrahi et al. 2008; Weng et al. 2008; Acestor et al. 2009). This complex also interacts with the KPAP1 complex that adds the poly A tails to mitochondrial mRNAs (Etheridge et al. 2008) and with the MRP1/2 complex (Aphasizhev et al. 2003b). The MERS1 (originally AP3) complex of five proteins appears to control the stability of edited mRNAs. A model for these various interactions has recently been proposed (Weng et al. 2008).
RESULTS
Discovery of the L*-complexes
The L-complex single copy (data not shown) LC-8 (TbMP44/KREPB5) protein was TAP-tagged and used as the target for all TAP pull-downs. LC-8 has a highly diverged RNase III motif (Aphasizhev et al. 2003a), and is required for cell viability and stability of the ∼20S L-complex (Wang et al. 2003). LC-8 was also shown to be a common component of the REN1, REN2, and REN3 TAP pull-downs (Panigrahi et al. 2006; Carnes et al. 2008).
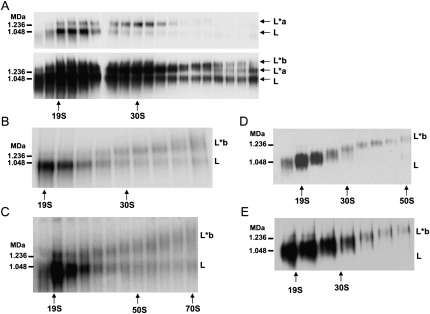
LC-8 whole cell TAP pull-down (purified through IgG Sepharose and Calmodulin agarose) was fractionated in glycerol gradients. The gradient fractions were electrophoresed in 3%–12% blue native gradient gels, which were blotted and probed with anti-REL1 monoclonal antibody to visualize REL1-containing complexes. The L-complex band, which sedimented around 20S–25S in the gradient, migrated in the blue native gels at ∼1 MDa (Fig. 1A, upper panel; Fig. 1B). There was, in addition, a minor REL1-containing band that co-sedimented in the gradient with the L-complex, but migrated in the blue native gel separate from the L-complex at ∼1.2 MDa (labeled L*a in Fig. 1, upper panel). The same overexposed gel (Fig. 1A, lower panel) shows the presence of a second minor band of continuously increasing molecular weight (as observed in Fig. 1D,E), which we labeled L*b. The progressive increase in size of the L*b-complex can be more clearly seen in the blue native gel of gradient fractions of whole cell LC-8-TAP pull-down in Figure 1B. Gradient fractions of mitochondrial lysate of LC-8-TAP cells gave a similar blue native gel pattern (Fig. 1C). The L*b-complex sedimented from ∼20S to over 70S and became progressively more heterodisperse in the native gel. We attribute the presence of variable amounts of the 1 MDa L-complex band in gradient fractions sedimenting greater than 30S to the breakdown of L*b-complex material after gradient fractionation, perhaps during the electrophoresis process (Fig. 1A,B,C). Some preparations from both mitochondrial TAP pull-down and mitochondrial lysate (Fig. 1D) showed a single continuously increasing L*b-complex profile with no indication of breakdown to the 1 MDa L-complex. The variable profiles in different preparations probably reflect the degree of breakdown of the L*b-complex due to the lengthy experimental manipulations involved in TAP isolation. A similar gel profile was obtained using wild-type L. tarentolae UC cells, showing that L*b-complex bands were not produced by incorporation of the TAP-tagged Leishmania major LC-8 protein into the editing complex (Fig. 1E).
FIGURE 1.
Blue native Western blots with glycerol gradient fractions of whole cell and mitochondrial LC-8-TAP pull-downs. (A) Western blot of whole cell LC-8-TAP pull-down sedimented in a 10%–30% glycerol gradient. Anti-REL1 antibody used to visualize REL1-containing complexes. (Upper panel) Ten second film exposure and (lower panel) 2 min exposure to visualize the two minor upper bands labeled L*a and L*b. (B) Auto-adenylation of whole cell LC-8-TAP pull-down with [α-32P]ATP. (C) Auto-adenylation of mitochondrial lysate from LC-8-TAP cells. (D) Western blot of an LC-8-TAP pull-down from isolated mitochondria. Anti-REL1 antibody used to visualize REL1-containing complexes. (E) Western blot of wild-type mitochondrial lysate. Anti-REL1 antibody used to visualize REL1-containing complexes. High molecular weight native marker sizes are indicated. L, L-complex; L*a, 1.2 MDa REL1-containing complex; L*b, REL1-containing complex of increasing size.
A gel shift to the 1.2 MDa L*a-complex could be produced with gel-isolated 1 MDa L-complex by several experimental manipulations (data not shown). This suggests that the L*a-complex is an experimental artifact, which occurs during or after glycerol gradient sedimentation. However, the nature of the change and any possible cellular role remain to be determined.
The L*b-complexes are RNase sensitive and are associated by RNA linkers with the MRP1/2 RNA-binding proteins
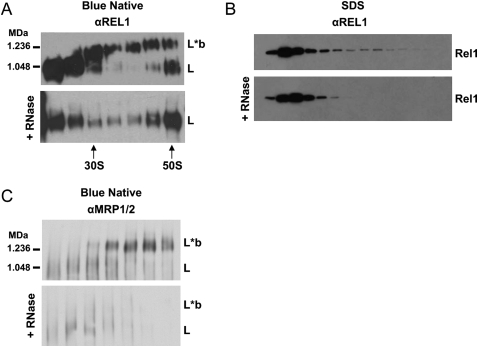
RNase A treatment of gradient fractions from either crude mitochondrial lysate or LC-8-TAP pull-down caused a breakdown of the L*b-complex and an appearance of L-complex bands (Fig. 2A). There was no apparent effect of RNase A on the L-complex itself (Fig. 2A). This sensitivity of L*b-complexes to RNase A can also be seen in the Western blot of the SDS gel in Figure 2B, in which the TAP pull-down was pretreated with RNase prior to running the gradient. There is a complete disappearance of REL1-containing material in the L*b-complex region. These data suggest that the L*b-complex material consists mainly of RNA-linked L-complexes or L-complexes with additional L-complex proteins associated by RNA linkers.
FIGURE 2.
Effect of RNase A on blue native and SDS-PAGE Western blot profiles. (A) Western blot of blue native gels of LC-8-TAP pull-down gradient fractions with and without RNase A (0.1 mg/mL). Blots were probed with anti-REL1 antiserum. (B) Western blot of SDS-PAGE of gradient fractions with and without treatment with RNase A (0.1 mg/mL) prior to glycerol gradient sedimentation. The blot was probed with anti-REL1 antiserum. (C) Blue native Western blot of LC-8-TAP mitochondrial lysate gradient fractions with and without treatment with RNase A (0.1 mg/mL). The blot was probed with anti-MRP1/2 antiserum.
We showed previously by co-IP experiments that the MRP1/2 complex (and RET1 complex) associated in substoichiometric amounts with the L-complex via RNA linkers (Aphasizhev et al. 2003a,b). This work employed standard native gels which did not resolve the L*b-complexes. We examined the localization of the MRP1/2 complex in blue native gels of gradient fractions of LC-8-TAP mitochondrial lysate. In addition to the 100–400 kDa MRP1/2 complexes described previously (Zikova et al. 2008), which sedimented between 5S and 20S, MRP also sedimented above 30S in the L*b-complexes (Fig. 2C, upper panel). Incubation of each gradient fraction with RNase A resulted in the loss of the bound MRP1/2 material (Fig. 2C, lower panel). We attribute the minor signal in the L-complex region remaining after RNase treatment to be due to the presence of the protein A tag, which was recognized by the secondary antibody.
The L*b-complexes contain a similar set of major proteins as the L-complex
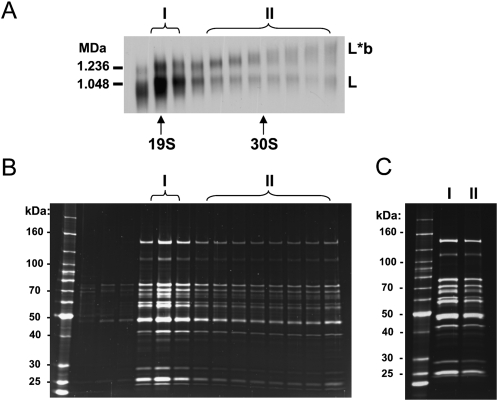
LC-8-TAP pull-down material was fractionated in a glycerol gradient. The fractions were run in a blue native gel, which was blotted and probed with anti-REL1 antibody (Fig. 3A). The L- and L*b-complexes are indicated. The gradient fractions were also run in an SDS gel, which was stained with Sypro Ruby (Fig. 3B). The gradient fractions indicated by I and II in Figure 3A were pooled, concentrated, and run side-by-side in an SDS gel (Fig. 3C). The polypeptide profile of the pooled L*b-complex fractions was very similar to that of the L-complex fractions (Aphasizhev et al. 2003a), except for some minor quantitative variations in the polypeptide bands and the absence of an endogenous LC-8 band in the LC-8-TAP pull-down (data not shown). This is consistent with the above conclusion that the L*b-complex material consists mainly of L-complex material. It should be noted that the presence of substoichiometric amounts of auxiliary polypeptides, such as MRP1/2 in the L*b-complex fractions would not be visualized by this method.
FIGURE 3.
Polypeptide profiles of L- and L*b-complexes are similar. (A) Blue native gel of gradient fractions of whole cell LC-8-TAP pull-down. Blot was probed with anti-REL1 antibody. (B) Sypro-stained SDS gel of same fractions as in A. (C) Sypro-stained SDS-PAGE of pooled and concentrated L- and L*b-complex gradient fractions indicated by I and II in A and B.
LC-8-TAP pull-down contains kinetoplast DNA and kinetoplast RNA
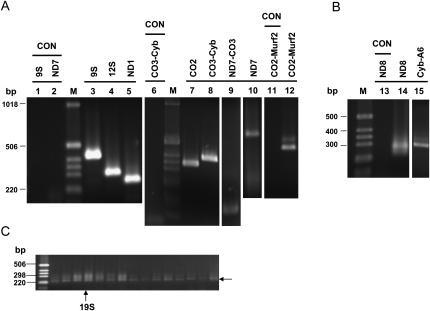
RNA was isolated from total TAP pull-down and subjected to RT-PCR using primer sets specific for various maxicircle transcripts, both never-edited, pre-edited, and edited (see Supplemental Fig. S3). Several extensive DNase I treatments were required to remove contaminating kinetoplast DNA molecules. RT-PCR cDNA products from the purified RNAs were obtained for several never-edited mRNAs and for pre-edited ND7 mRNA (Fig. 4A). Partially edited cDNA products were obtained with two pre-edited primer sets (see Supplemental Fig. S3) that were located on adjacent maxicircle genes (ND7-CO3, CO3-Cyb, and CO2-Murf2), indicating the presence of unprocessed maxicircle transcripts in the TAP pull-down, as was previously reported (Koslowsky and Yahampath 1997; Bessolitsyna et al. 2005). The presence of partially edited maxicircle transcripts was also examined by RT-PCR using downstream edited and upstream unedited primer sets for the pan-edited ND8 transcripts and for the Cyb-A6 unprocessed transcripts (Fig. 4B). Interestingly, these cDNA sequences showed extensive misediting at the junction regions between fully edited and unedited regions (Sturm et al. 1992; see Supplemental Figs. S1, S2).
FIGURE 4.
RT-PCR of maxicircle mRNA transcripts. (A) Never-edited (9S, 12S, ND1) and pre-edited (CO2, ND7) mRNA transcripts were detected in the LC-8-TAP pull-down. (B) RT-PCR of partially edited ND8 and Cyb-A6 junction regions. (C) RT-PCR of ND8 mRNA from glycerol gradient fractions of LC-8-TAP pull-down. A 5′-unedited and 3′-edited primer set was used. The 230 bp band and the smear above this band were eluted from a 20S fraction, cloned and sequenced. The sequences confirmed the 230 bp band as unedited ND8 mRNA and the upper smear as partially edited ND8 RNAs (data not shown). RNA templates were treated multiple times with DNase I. PCR control lanes are indicated. Products visualized by ethidium bromide. See Supplemental Figure S3 for primer sets used and Supplemental Tables S1, S2 for primer sequences.
To analyze the distribution of maxicircle transcripts in LC-8-TAP gradient fractions, RNA was isolated from each fraction and subjected to RT-PCR. The primer set consisted of ND8-unedited (5′) and ND8-edited (3′). Products of the correct size for unedited RNA and a smear of several larger bands were obtained with all fractions (Fig. 4C). The cDNA products of the 20S fractions were cloned and sequenced to confirm the identity of the 230 base-pair (bp) band as unedited ND8 mRNA and the upper bands as partially edited ND8 mRNAs (Fig. 4C; data not shown).
DISCUSSION
The use of blue native gel electrophoresis has uncovered several minor REL1-containing bands, which we have termed the L*a- and L*b-complexes. The 1.2 MDa L*a-complex appears to represent an experimental artifact since it appears de novo after glycerol gradient sedimentation and can be produced by several experimental manipulations with gel-isolated 1 MDa L-complex (data not shown). The L*b-complex migrated in the blue native gel from around 1 MDa at 20S to an undetermined higher molecular weight sedimenting at over 70S. L*b-complex material from mitochondrial lysate and from TAP pull-down of mitochondrial lysate contained substoichiometric amounts of RNA-linked MRP1/2 complexes. RNase A digestion of gradient fractions led to the breakdown of the L*b-complex bands and the appearance of the 1 MDa L-complex band, as well as to the removal of the bound MRP1/2 material. This breakdown of L*b-complexes also occurred spontaneously in different preparations, especially in TAP pull-downs from whole cells. The L-complex band was unaffected by RNase treatment except for a slight decrease in S value, as reported previously (Aphasizhev et al. 2003a). The breakdown of L*b-complexes to L-complex, as well as the similarity of the polypeptide profiles of the L-complex and the L*b-complex material, suggest that the L*b-complexes consist of RNA-linked L-complex components.
We found that unedited, edited, and partially edited maxicircle RNA transcripts could be detected in the purified whole cell LC-8-TAP pull-down and that these RNAs were distributed across all gradient fractions. The fact that edited and partially edited transcripts could be obtained using primers in the adjacent Cyb and A6 genes suggests that editing is initiated prior to completion of RNA processing, as has been shown previously (Read et al. 1992).
It is interesting that there was extensive kinetoplast DNA contamination in the TAP pull-down in spite of the double affinity purification. This could suggest that RNA editing reactions are closely linked to sites of transcription. In this regard, it is interesting to note that we showed recently by immunofluorescence, that several editing enzymes are concentrated in the kinetoplast DNA portion of the mitochondrion (F Li and L Simpson, unpubl.).
It is likely that minor amounts of L*b-complexes were responsible for the previously observed RNA-linked association of the MRP1/2 and RET1 proteins with L-complex proteins (Aphasizhev et al. 2003b), and probably also for the reported in vitro partial and full-cycle editing activities of both column-purified and TAP-isolated editing complex material. It was, in fact, shown previously that gel-isolated RNase-treated L-complex was inactive in partial editing reactions in vitro (Aphasizhev et al. 2003a)
The total substoichiometric RNA-linked protein components of L*b-complex are probably not limited to the MRP1/2 complex, but also to other editing factors, including the RET1 TUTase complex that adds U's to the 3′ end of gRNAs (Aphasizhev et al. 2003c), the GRBC1/2 complex that controls the stability of gRNAs (Weng et al. 2008), and the REH1 RNA helicase, which is probably involved in multi-gRNA mediated editing (F Li and L Simpson, unpubl.). A complete inventory of the RNA-linked substoichiometric polypeptide composition of the L*b-complexes remains to be done and it indeed may be dynamic. We suggest that the L*b-complexes may represent a scaffold for interaction with other editing factors during the editing process to form the active holoenzyme or “editosome,” but this remains to be confirmed.
MATERIAL AND METHODS
Cell culture and mitochondria isolation
L. tarentolae UC strain cells were grown in brain heart infusion supplemented with 10 μg/mL hemin at 27°C. For large-scale mitochondria isolation, cells were grown in 22 L quantities in a New Brunswick Bioflow 4500 Fermenter to late log phase and harvested using a Cepa Model Z-41 continuous flow centrifuge. Cells were washed in SET and stored as pellets at -80°C for whole cell TAP isolation. Mitochondria were isolated from late log phase cells using Renografin density gradients as described (Braly et al. 1974) and stored as pellets at ∼80°C.
Plasmid construction
The LC-8 gene from L. major was cloned into a TAP vector containing a neomycin-resistance marker in frame with the calmodulin binding and Protein A tags (Aphasizhev et al. 2003a). L. tarentolae UC strain cells were transfected by electroporation as described. Transformants were selected by plating on agarose containing G418 and grown at 27°C with gentle agitation in BHI-hemin with 100 μg/mL G418.
TAP isolation
A modified TAP isolation procedure was used (Aphasizhev et al. 2003a), with either whole cells or isolated mitochondria. The lysis was performed in TKM buffer (10 mM Tris-HCl at pH 7.4, 60 mM KCl, 10 mM MgCl2). The lysate was sonicated using a Biologics Model 150 V/T sonicator for 3 × 30 s. For whole cell TAP isolation, the lysate was clarified by centrifugation in the SS34 rotor for 15 min at 19,000 rpm or the SW24 rotor at 20,000 rpm for 15 min, followed by filtration through a 2.7 μm Whatman glass fiber syringe filter. TAP isolation from mitochondrial lysate or whole cells yielded pull-downs of equivalent purity.
Gradient centrifugation
Gradients were 10%–30% glycerol in TKM buffer. Mitochondrial pellets were lysed in TKM with 0.5% Triton X-100, and the lysate clarified by centrifugation. The mitochondrial lysate or TAP pull-down was layered on top and the gradients centrifuged for 2–4 h at 50,000 rpm in a Beckman SW55 rotor, or for 20 h at 30,000 rpm in an SW41 rotor. Gradients were fractionated from the top using the ISCO Model UA-6 fractionator.
Gel electrophoresis
Novex (Invitrogen) 3%–12% gradient acrylamide gels in Bis-Tris buffer were used. Molecular weight standards from Invitrogen were used to calibrate the gel, with the largest being 1.236 MDa and 1.048 MDa. Electrophoresis was for 1 h at 150 V followed by 2 h at 250 V at 4°C. Blue native gels were electro-blotted onto Immobilon-PVDF filters (Millipore), which were processed for Western blot analysis using anti-REL1 monoclonal antibody. The Pierce West Pico system was used to visualize the bands. SDS gel electrophoresis was performed using Novex (Invitrogen) 4%–12% or 8%–16% Tris-Glycine gradient gels. Gels were stained with Sypro Ruby (Invitrogen), and visualized using a Biochimi System (UVP) with a First Light 300 nm UV transilluminator (UVP), and analyzed using LabWorks software (UVP).
Antibody production
The mouse monoclonal antibody against L. tarentolae REL1 was generated by the Cal Tech Monoclonal Facility. Polyclonal antibody against the MRP1/2 proteins were obtained from Covance Research (Aphasizhev et al. 2003b).
PCR and RT-PCR analysis
Specific oligonucleotides for PCR and RT-PCR were commercially synthesized (IDT). Reverse transcription reactions were performed with Superscript II (Invitrogen) using gene specific primers. PCR products were visualized by ethidium bromide staining using agarose gel electrophoresis.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We acknowledge the assistance of Martina Nebohacova and Agda Simpson in screening the mouse monoclonal sera for anti-REL1 antibody. We thank Robert E. Nelson for cloning the LC-8 gene into the TAP vector. This research was supported in part by a research grant (AI09102) from the National Institutes of Health to L.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1642809.
REFERENCES
- Acestor N, Panigrahi AK, Carnes J, Zikova A, Stuart KD. The MRB1 complex functions in kinetoplastid RNA processing. RNA. 2009;15:277–286. doi: 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatortsev VS, Cruz-Reyes J, Zhelonkina AG, Sollner-Webb B. Trypanosoma brucei RNA editing: Coupled cycles of U deletion reveal processive activity of the editing complex. Mol Cell Biol. 2008;28:2437–2445. doi: 10.1128/MCB.01886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TE, Heidmann S, Reed R, Myler PJ, Goringer HU, Stuart KD. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol Cell Biol. 1998;18:6014–6022. doi: 10.1128/mcb.18.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003a;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kDa complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003b;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad Sci. 2003c;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessolitsyna EA, Fediakov AV, Merzliak EM, Kolesnikov AA. ATPase subunit 6 gene of Leptomonas seymouri (Trypanosomatidae) is transcribed and edited as a polycistronic transcript. Mol Biol (Mosk) 2005;39:61–66. [PubMed] [Google Scholar]
- Braly P, Simpson L, Kretzer F. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J Protozool. 1974;21:782–790. doi: 10.1111/j.1550-7408.1974.tb03752.x. [DOI] [PubMed] [Google Scholar]
- Byrne EM, Connell GJ, Simpson L. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 1996;15:6758–6765. [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ Adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Ammerman ML, Presnyak V, Read LK. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem. 2008;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo RP, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kable ML, Seiwert SD, Heidmann S, Stuart K. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [see comments] [DOI] [PubMed] [Google Scholar]
- Kang X, Rogers K, Gao G, Falick AM, Zhou SL, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant proteins. Proc Natl Acad Sci. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller J, Muller UF, Schmid B, Missel A, Kruft V, Stuart K, Goringer HU. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J Biol Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- Koslowsky DJ, Yahampath G. Mitochondrial mRNA 3′ cleavage polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol Biochem Parasitol. 1997;90:81–94. doi: 10.1016/s0166-6851(97)00133-3. [DOI] [PubMed] [Google Scholar]
- Lambert L, Muller UF, Souza AE, Goringer HU. The involvement of gRNA-binding protein gBP21 in RNA editing-an in vitro and in vivo analysis. Nucleic Acids Res. 1999;27:1429–1436. doi: 10.1093/nar/27.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, Shimamura M, Grams J, Hajduk SL. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MM, Halbig K, Cruz-Reyes J, Read LK. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA. 2006;12:1292–1303. doi: 10.1261/rna.2331506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller UF, Lambert L, Goringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Gygi SP, Ernst NL, Igo RP, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol Cell Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J Am Soc Mass Spectrom. 2003a;14:728–735. doi: 10.1016/S1044-0305(03)00126-0. [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003b;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in Trypanosoma brucei: A novel complex and a unique oxidoreductase complex. Mol Cell Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- Pelletier M, Read LK. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA. 2003;9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris M, Simpson AM, Grunstein J, Liliental JE, Frech GC, Simpson L. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol Biochem Parasitol. 1997;85:9–24. doi: 10.1016/s0166-6851(96)02795-8. [DOI] [PubMed] [Google Scholar]
- Piller KJ, Rusch LN, Sollner-Webb B. Trypanosoma brucei RNA editing a full round of uridylate insertional editing in vitro mediated by endonuclease and RNA ligase. J Biol Chem. 1996;271:4613–4619. doi: 10.1074/jbc.271.9.4613. [DOI] [PubMed] [Google Scholar]
- Read LK, Myler PJ, Stuart K. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J Biol Chem. 1992;267:1123–1128. [PubMed] [Google Scholar]
- Rusche LN, Huang CE, Piller KJ, Hemann M, Wirtz E, Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential band IV gene and identifying the band V gene. Mol Cell Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2161. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Seiwert SD, Stuart K. RNA editing: Transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K, Panigrahi AK, Schnaufer A. Identification and characterization of trypanosome RNA-editing complex components. Methods Mol Biol. 2004;265:273–291. doi: 10.1385/1-59259-775-0:273. [DOI] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Sturm NR, Maslov DA, Blum B, Simpson L. Generation of unexpected editing patterns in Leishmania tarentolae mitochondrial mRNAs: Misediting produced by misguiding. Cell. 1992;70:469–476. doi: 10.1016/0092-8674(92)90171-8. [DOI] [PubMed] [Google Scholar]
- Vondruskova E, van den Burg J, Zikova A, Ernst NL, Stuart K, Benne R, Lukes J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- Wang B, Ernst NL, Palazzo SS, Panigrahi AK, Salavati R, Stuart K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:1–12. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikova A, Kopecna J, Schumacher MA, Stuart K, Trantirek L, Lukes J. Structure and function of the native and recombinant mitochondrial MRP1/MRP2 complex from Trypanosoma brucei. Int J Parasitol. 2008;38:901–912. doi: 10.1016/j.ijpara.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]