Abstract
During pre-mRNA splicing, the spliceosome must configure the substrate, catalyze 5′ splice site cleavage, reposition the substrate, and catalyze exon ligation. The highly conserved U2/U6 helix I, which adjoins sequences that define the reactive sites, has been proposed to configure the substrate for 5′ splice site cleavage and promote catalysis. However, a role for this helix at either catalytic step has not been tested rigorously and previous observations question its role at the catalytic steps. Through a comprehensive molecular genetic study of U2/U6 helix I, we found that weakening U2/U6 helix I, but not mutually exclusive structures, compromised splicing of a substrate limited at the catalytic step of 5′ splice site cleavage, providing the first compelling evidence that this helix indeed configures the substrate during 5′ splice site cleavage. Further, mutations that we proved weaken only U2/U6 helix I suppressed a mutation in PRP16, a DEAH-box ATPase required after 5′ splice site cleavage, providing persuasive evidence that helix I is destabilized by Prp16p and suggesting that this structure is unwound between the catalytic steps. Lastly, weakening U2/U6 helix I also compromised splicing of a substrate limited at the catalytic step of exon ligation, providing evidence that U2/U6 helix I reforms and functions during exon ligation. Thus, our data provide evidence for a fundamental and apparently dynamic role for U2/U6 helix I during the catalytic stages of splicing.
Keywords: pre-mRNA splicing, U2/U6 helix I, snRNA, spliceosome; DExD/H-box ATPase, Prp16
INTRODUCTION
Pre-mRNA splicing, the process by which introns are excised from pre-mRNA, is mediated by a large and dynamic ribonucleoprotein complex known as the spliceosome (Nilsen 2003; Matlin and Moore 2007; Valadkhan 2007; Smith et al. 2008). The spliceosome is composed of five highly conserved small nuclear RNAs (snRNAs) (U1, U2, U4, U5, and U6) and over 100 proteins (Stark and Lührmann 2006). The spliceosome identifies a pre-mRNA substrate by recognizing three conserved sequences in the intron: the 5′ splice site, the branch site, and the 3′ splice site (Spingola et al. 1999; Brow 2002). The snRNA components of the spliceosome play a key role in intron recognition. For example, in the catalytically active spliceosome, the snRNAs recognize the pre-mRNA substrate through base-pairing interactions between U6 snRNA and the 5′ splice site and between U2 snRNA and the branch site (Staley and Guthrie 1998). Such interactions indicate that the snRNAs reside at the catalytic core. Further, evidence suggests that the snRNAs promote catalysis. For example, the chemical mechanism utilized by the spliceosome is indistinguishable from the mechanism used by self-splicing group II introns (Villa et al. 2002), and protein-free RNAs corresponding to U2 and U6 can promote chemical reactions that resemble the reaction of pre-mRNA splicing (Valadkhan and Manley 2001; Valadkhan et al. 2007).
The catalytically activated spliceosome promotes the chemistry of splicing through two sequential chemical steps: 5′ splice site cleavage and exon ligation (Staley and Guthrie 1998). In 5′ splice site cleavage, the 2′ hydroxyl of a universally conserved bulged adenosine at the branch site attacks the phosphate at the 5′ splice site, generating a branched lariat intermediate and a free 5′ exon. In exon ligation, the 3′ hydroxyl of the 5′ exon attacks the phosphate at the 3′ splice site, generating the mature mRNA and the excised lariat intron. To permit exon ligation after 5′ splice site cleavage, the spliceosome must reposition the substrate to juxtapose the reactants, which is likely promoted both by disrupting the U6/5′ splice site and U2/branch site interactions (Konarska et al. 2006; Smith et al. 2007).
Notably, the spliceosome itself must also rearrange between the chemical steps; ATP hydrolysis by the DEAH-box ATPase Prp16p (Schwer and Guthrie 1992) destabilizes the 5′ splice site cleavage conformation of the spliceosome (Query and Konarska 2004). Members of the ubiquitous DExD/H-box ATPase family can utilize ATP hydrolysis to unwind RNA duplexes or disrupt RNA–protein interactions (Staley and Guthrie 1998; Pyle 2008), and in the spliceosome these ATPases promote rearrangements throughout the splicing cycle. Prp16p activity has been linked genetically to the destablization of stem IIc of U2 (Hilliker et al. 2007) and less well-defined interactions involving Isy1p, Prp8p, U6 snRNA, and U2 snRNA (Madhani and Guthrie 1994b; McPheeters 1996; Query and Konarska 2004; Villa and Guthrie 2005).
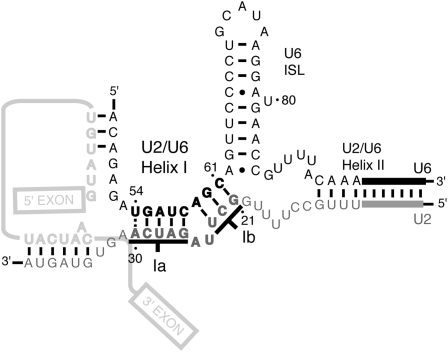
In order to promote chemistry, the spliceosome must position the pre-mRNA reactants within the catalytically active center. This function may be served in part by an intermolecular snRNA structure, U2/U6 helix I, which is composed of two short helices, Ia and Ib, separated by a 2-nucleotide (nt) bulge in U2 (Fig. 1; Madhani and Guthrie 1992). Helix Ia is proximal to both the U6/5′ splice site and U2/branch site interactions, leading Madhani and Guthrie (1992) to propose that helix Ia juxtaposes the substrate for 5′ splice site cleavage, a hypothesis that has remained unproved. Helix Ib contains the invariant AGC triad of U6 that has been suggested to bind a catalytic metal (Fabrizio and Abelson 1992; Yu et al. 1995), like the catalytic triad of group II introns (Toor et al. 2008), but it has not yet been shown that helix Ib functions during either catalytic step. U2/U6 helix I is essential for viability in budding yeast and mutations in either U2 or U6 that would disrupt helix I block spliceosome activation just before 5′ splice site cleavage (Madhani and Guthrie 1992; McPheeters and Abelson 1992; Ryan and Abelson 2002), but these defects have not been definitively linked to U2/U6 helix I. This helix is conserved in both the U2- and U12-dependent spliceosomes of metazoans (Tarn and Steitz 1996) and is required for 5′ splice site cleavage in mammals (Wolff et al. 1994; Sun and Manley 1995), but it is unknown if this reflects a requirement for U2/U6 helix at or before the catalytic stage of 5′ splice site cleavage. While one study suggested that U2/U6 helix Ib promotes aberrant 5′ splice site cleavage (Luukkonen and Séraphin 1998), it remains unclear whether this reflects a role for helix Ib at the catalytic stage of canonical 5′ splice site cleavage. Further, an NMR structure shows an alternative configuration of U2/U6 helix I, where helix Ib is replaced by two intramolecular interactions, a conformation that was hypothesized to promote 5′ splice site cleavage (Sashital et al. 2004).
FIGURE 1.
Model of the snRNA secondary structure of the catalytic core. U6 is black, U2 is gray, and the pre-mRNA is light gray; U2/U6 helix I is bold (Madhani and Guthrie 1992). Complementarity at U2-A30/U6-U54 (dashed line) is neither conserved (Guthrie and Patterson 1988) nor supported by data (Supplemental Data). U2/U6 helix II, the U6 ISL, the U2/branch site interaction, and the U6/5′ splice site interaction are also depicted (Madhani and Guthrie 1994a). U5 is omitted for clarity.
While a requirement to form U2/U6 helix I during spliceosome activation provides a rationalization for the requirement to unwind the mutually exclusive U4/U6 stem I (Madhani and Guthrie 1992), several observations in the literature are difficult to reconcile with a role for U2/U6 helix I at the catalytic step of 5′ splice site cleavage. First, a mutation, U6-U57C, that would disrupt base pairing in U2/U6 helix I nonetheless strongly suppresses a mutation of the branch site adenosine to cytosine (brC) that leads to a defect at the catalytic step of 5′ splice site cleavage (McPheeters 1996). Further, U6-U57C appears to hyperstabilize the 5′ splice site cleavage conformation of the spliceosome (McPheeters 1996; Query and Konarska 2004). Second, mutations in the U2 and U6 strands of U2/U6 helix I destabilize the 5′ splice site cleavage confirmation, and thereby suppress a cold-sensitive mutation in PRP16; but attempts to attribute this suppression to the disruption of U2/U6 helix I failed (Madhani and Guthrie 1994b), raising the possibility that the mutations suppress by disrupting alternative structures at the 5′ splice site cleavage stage. Finally, although some mutations in U2/U6 helix I impair exon ligation, they permit 5′ splice site cleavage (Fabrizio and Abelson 1992; McPheeters and Abelson 1992; Wolff et al. 1994; Hilliker and Staley 2004; McGrail et al. 2006). These puzzling observations raise the possibility that alternative structures function during catalysis.
Given that a role for U2/U6 helix I at the stage of 5′ splice site cleavage is compelling, but definitive data in support of the model have been lacking, we have reinvestigated the role of U2/U6 helix I to determine whether this highly conserved and essential structure functions at either catalytic step. Through a systematic molecular genetic analysis of U2/U6 helix I in the yeast Saccharomyces cerevisiae, we found that U2/U6 helix I, but not alternative structures, promotes the catalytic step of 5′ splice site cleavage, providing the first compelling support for the model that U2/U6 helix I configures the substrate for 5′ splice site cleavage. We also found persuasive evidence that U2/U6 helix I is destabilized after 5′ splice site cleavage during the Prp16p-dependent rearrangement. Finally, we found evidence that U2/U6 helix I reforms to function during exon ligation. Together, these data provide evidence that U2/U6 helix I forms a key component of the catalytically active spliceosome during both chemical steps of splicing, and suggests that unwinding of U2/U6 helix I facilitates the transition of the spliceosome from the 5′ splice site cleavage stage to the exon ligation stage.
RESULTS
Evidence that U2/U6 helix I promotes 5′ splice site cleavage
Previous studies of U2/U6 helix I, using wild-type splicing substrates, were unable to attribute a role for this structure at a defined stage. To test precisely for a role of U2/U6 helix I at the catalytic stage of 5′ splice site cleavage, rather than a prior stage, we sensitized splicing to the catalytic step of 5′ splice site cleavage using a pre-mRNA substrate limited at this step. This substrate contains a mutation of the conserved branch site adenosine to cytosine (brC), which has been shown to destabilize the 5′ splice site cleavage conformation of the spliceosome (Query and Konarska 2004; Liu et al. 2007). If U2/U6 helix I promotes 5′ splice site cleavage, then mutations that disrupt this structure should worsen the 5′ splice site cleavage defect of the brC mutation. Consistent with this idea, the mutation U6-U57A, which effectively destabilizes the 5′ splice site cleavage conformation, worsens the defect of the brC substrate (Liu et al. 2007).
To test for such genetic interactions, we utilized the ACT1-CUP1 splicing reporter, in which excision of the ACT1 intron is required for expression of the CUP1 gene, which confers copper resistance to budding yeast in a dosage-dependent manner and thereby allows a quantitative assay of splicing efficiency (Lesser and Guthrie 1993). In this reporter, the brC mutation reduces the growth of yeast from 0.8 mM to 0.2 mM copper (Lesser and Guthrie 1993; McPheeters 1996). While genetic interactions between the brC substrate and mutations at U6-U57 have been reported (McPheeters 1996; Query and Konarska 2004; Liu et al. 2007), genetic interactions between other mutations in U2/U6 helix I have not been reported. Given the potential importance of U2/U6 helix I at the catalytic center of the spliceosome, we analyzed all possible combinations of alleles at each position in U2/U6 helix I. We show only a subset of the data to illustrate trends, but have based our conclusions on the full data set (available online as Supplemental Data).
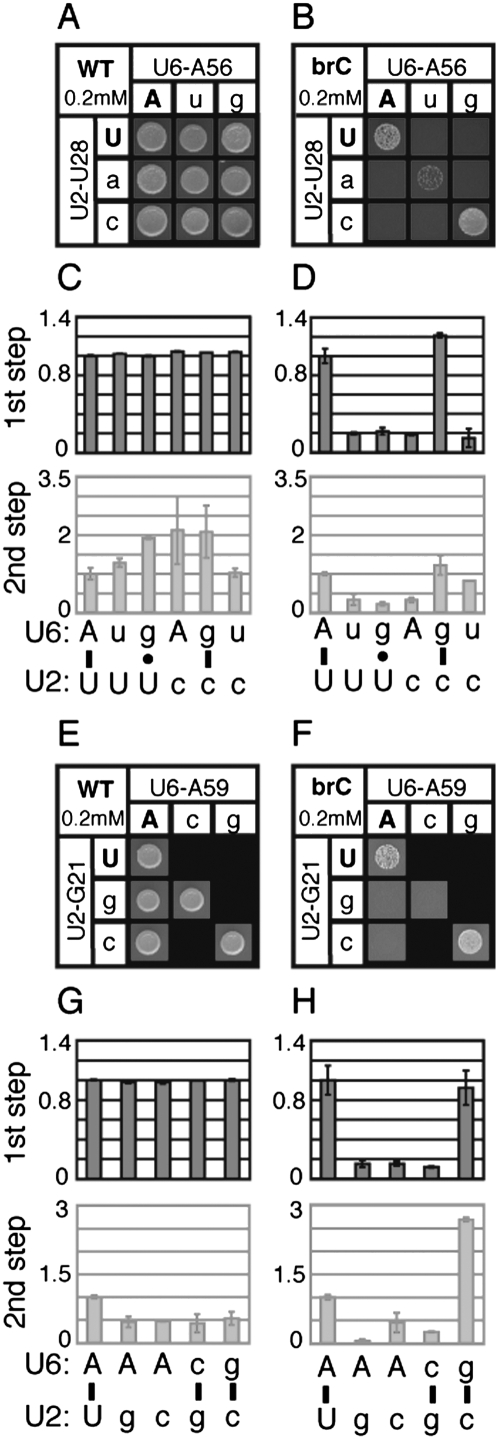
We found that growth of yeast containing the brC mutated substrate was exacerbated by most of the viable point mutations in U2/U6 helix I (Fig. 2B,F; Supplemental Data). In contrast, none of the point mutations compromised growth conferred by the wild-type reporter at 0.2 or 0.8 mM copper, where growth becomes limited in the isogenic wild-type strain (Fig. 2A,E; data not shown). In addition, with the brC reporter, none of the snRNA point mutations showed a growth defect in the absence of copper (data not shown). Thus, in the snRNA mutants, the growth conferred by the brC reporter decreased because of a genetic interaction between the brC mutation and the snRNA mutations.
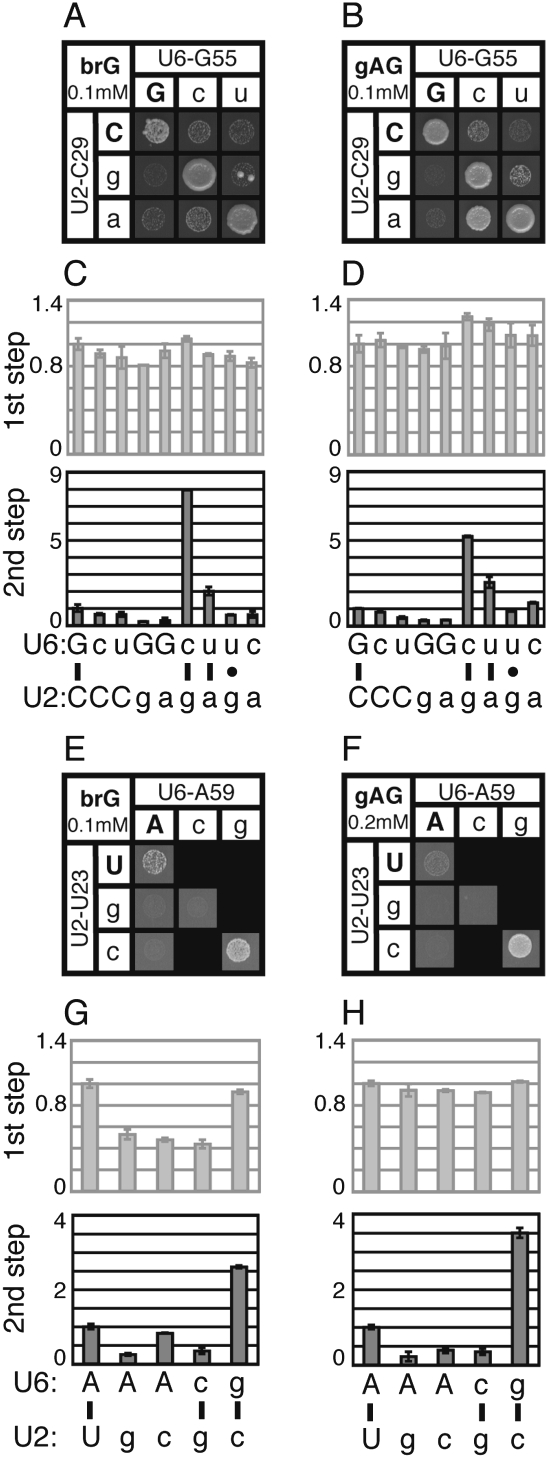
FIGURE 2.
Disrupting U2/U6 helix I complementarity exacerbates the 5′ splice-site cleavage defect of a splicing reporter mutated at the branchpoint (brA→brC). Yeast strains containing the indicated alleles at the U2/U6 positions U28/A56 (A–D) and U23/A59 (E–H) and expressing the ACT1-CUP1 splicing reporter (Lesser and Guthrie 1993) were assayed for growth and splicing. (A,B,E,F) Yeast growth on media containing 0.2 mM copper sulfate for 3 d at 30°C. The yeast express either a wild-type (WT, A,E) or brC mutated (B,F) reporter. The identity of the U6 or U2 alleles are shown in each column or row, respectively. Wild-type alleles are capitalized; mutants are lower case. Watson–Crick combinations fall on the diagonal (upper left to lower right). Lethal allele combinations are indicated by black boxes. (C,D,G,H) Quantitation of primer extension reactions of whole-cell RNA isolated from selected strains containing the WT (C,G) or brC (D,H) ACT1-CUP1 reporter. The selected allele combinations are indicated below the bar graphs and correspond to the base pair analyzed directly above by growth. Throughout, 5′ splice site cleavage efficiency (1st step) is calculated as (LI+M)/(P+LI+M) and exon ligation efficiency (2nd step) is calculated as M/LI (see Materials and Methods). Bars reflect the average value for duplicate samples; error bars reflect the range of duplicates. Values in each case were normalized to the strain expressing wild-type U2 and U6. In C and G, variation in exon ligation efficiency is due to the low signal for WT lariat intermediate. The exon ligation efficiency is faded to emphasize the 5′ splice site cleavage efficiency.
Many previous investigations of U2/U6 helix I found defects in the regions of U2 and U6, but failed to repair U2/U6 helix I to test whether the defects were indeed due to disruption of this structure. In order to test rigorously for a role for helix I in 5′ splice site cleavage, we combined U2 and U6 mutations to restore base pairing in helix I and assayed for loss of exacerbation of the brC defect. Significantly, in approximately half of the cases tested, compensatory mutations that restored base pairing indeed restored growth on copper (Fig. 2B,F; Supplemental Data); for instance, the conservative U2/U6 double mutations U28C/A56G and U23C/A59G reversed the exacerbation of brC conferred by the corresponding point mutations (Fig. 2B,F). In contrast, noncompensatory mutations that failed to repair U2/U6 helix I generally failed to reverse the exacerbation of brC (Fig. 2B; Supplemental Data). We also tested mutations that would repair alternative mutually exclusive structures, such as the intramolecular U6 central stem (Vidaver et al. 1999), U2 stem I (Guthrie and Patterson 1988), a possible intramolecular U2 stem–loop (Yan and Ares 1996), or the four-helix junction structure (Sashital et al. 2004); however, repair in each case failed to reverse the exacerbation of brC (Supplemental Data). Thus, these data suggest that U2/U6 helix I, and not alternative structures, are especially important for splicing the brC substrate. Because this substrate is limited at the step of 5′ splice site cleavage, these data suggest that U2/U6 helix I functions at this catalytic step.
To test for a role for U2/U6 helix I in splicing the brC substrate at the 5′ splice site cleavage stage, we directly assayed splicing of the brC substrate in the snRNA mutants. Primer extension analysis of whole cell RNA revealed that point mutations that disrupted U2/U6 helix I did not decrease the efficiency of 5′ splice site cleavage for the wild-type substrate, but did decrease the efficiency of 5′ splice site cleavage for the brC substrate by 10%–80% (Fig. 2D,H; Supplemental Data). In nearly all cases tested, compensatory mutations that restored Watson–Crick base pairing in U2/U6 helix I restored the efficiency of 5′ splice site cleavage (Fig. 2D,H; Supplemental Data). In contrast, noncompensatory mutations did not restore the efficiency of 5′ splice site cleavage. While it is possible that U2/U6 helix I is only required for splicing of aberrant substrates or that U2/U6 helix I disruption exacerbates an earlier silent defect of the brC substrate, we ruled out these caveats (see below). In total, at five of the seven conserved base pairs in U2/U6 helix I, we found evidence that this helix promotes 5′ splice site cleavage (Supplemental Data).
Evidence that U2/U6 helix I is destabilized after 5′ splice site cleavage by Prp16p
Because the spliceosome appears to disrupt the U6/5′ splice site interaction and the U2/branch site interaction after 5′ splice site cleavage (Konarska et al. 2006; Smith et al. 2007), and because these structures adjoin U2/U6 helix I (Fig. 1), we hypothesized that the spliceosome destabilizes U2/U6 helix I in parallel to facilitate repositioning of the substrate between the chemical steps. We further hypothesized that U2/U6 helix I is destabilized by the DEAH-box ATPase Prp16, either directly or indirectly, given the requirement for Prp16p after 5′ splice site cleavage (Schwer and Guthrie 1992). Consistent with this hypothesis, previous work showed that double mutations in both U2 and U6 suppressed the cold-sensitive phenotype of prp16-302, but compensatory mutations that restored helix I did not abolish suppression, challenging the hypothesis (Madhani and Guthrie 1994b). However, these mutations, while restoring base pairing in U2/U6 helix I, may have failed to restore the stability of the helix due to the severity of quadruple mutants and the disruption of tertiary interactions.
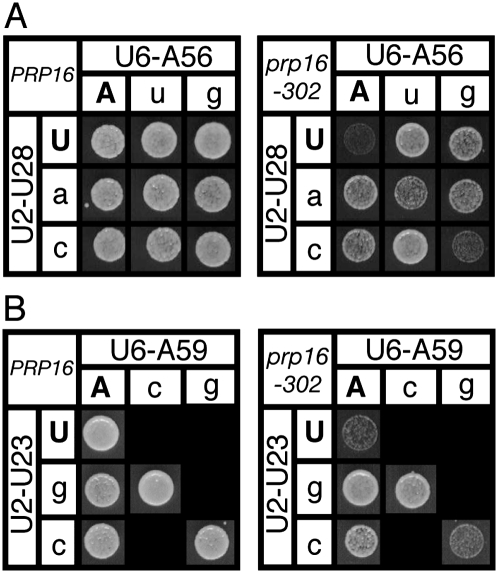
To evaluate the hypothesis with more subtle changes, we first tested all point mutations in U2/U6 helix I for suppression of the cold-sensitive phenotype of prp16-302 (Madhani and Guthrie 1994b), and found that the majority of point mutants in either U2 or U6 suppressed prp16-302; none compromised growth of wild-type PRP16 (Fig. 3; Supplemental Data). Unlike the previous analysis with double mutations, in almost all cases tested with point mutations, repairing U2/U6 helix I restored the cold-sensitive phenotype of prp16-302 (Fig. 3; Supplemental Data). For instance, the U2/U6 double mutants U28C/A56G and U23C/A59G, which both restored base pairing, abolished the suppression of prp16-302 by the corresponding point mutations (Fig. 3). All double mutants that restored base pairing but failed to abolish suppression of prp16-302 also failed to abolish enhancement of brC, suggesting that these double mutants failed to restore the stability of either U2/U6 helix I or tertiary interactions. In contrast to the double mutations that restored base pairing and abolished suppression, double mutations that did not restore helix I did not abolish suppression of prp16-302 (Fig. 3A; Supplemental Data). Further, suppression was not abolished by repairing alternative structures such as the U6 central stem, U2 stem I, or the U2/U6 four-helix junction structure (Supplemental Data; see below). In total, at five of the seven conserved base pairs in U2/U6 helix I, we found evidence that U2/U6 helix I, but not alternative structures, is destabilized during the Prp16p-dependent remodeling step. Importantly, the mutations that suppress prp16-302 correlate almost exactly with the mutations that exacerbate the brC substrate. Thus, even with all wild-type pre-mRNA substrates in the prp16-302 mutant, the prp16-302 data corroborate the brC data and thereby provide further evidence for a role for U2/U6 helix I during 5′ splice site cleavage.
FIGURE 3.
Disruption of U2/U6 helix I complementarity suppresses the cold-sensitive growth defect of the DEAH-box ATPase mutant prp16-302. (A,B) Wild-type PRP16 (left) or mutant prp16-302 (right) containing mutated alleles at U2/U6 helix I positions U28/A56 (A) and U23/A59 (B) were grown on media containing 5-FOA at the restrictive temperature (20°C) for 3 (PRP16) or 5 (prp16-302) d. Matrices are arranged as described in the legend for Figure 2.
A mutually exclusive four-helix junction does not promote 5′ splice site cleavage
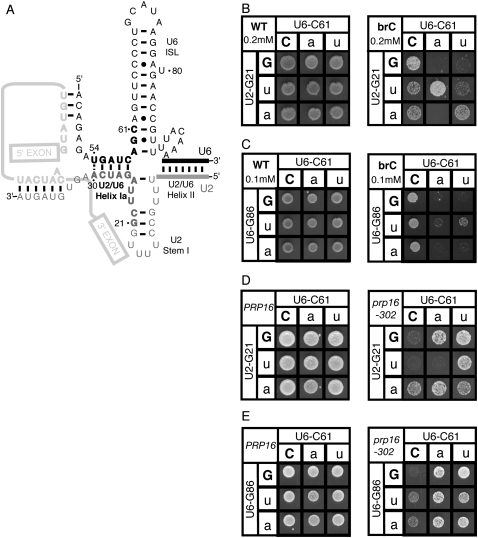
As evidenced by NMR studies, portions of U2 and U6 snRNA anneal to form an alternative conformation of U2/U6 helix I. The U2/U6 helix Ia portion forms as predicted; however, the U2/U6 helix Ib portion does not (Sashital et al. 2004). Instead, the U6 strand of helix Ib pairs to a downstream region of U6, extending the U6 ISL, while the U2 strand of helix Ib pairs to an upstream region of U2, forming part of U2 stem I. Together with U2/U6 helix II and U2/U6 helix Ia, these alternative structures form a four-helix junction (Fig. 4A). Given the previously described role for U2/U6 helix Ib before or during exon ligation (Hilliker and Staley 2004), this alternative conformation was proposed to function during 5′ splice site cleavage (Sashital et al. 2004).
FIGURE 4.
Repair of the putative mutually exclusive four-helix junction failed to provide evidence for this model at the stage of 5′ splice site cleavage. (A) Model of four-helix junction as proposed by Sashital et al. (2004). Regions of U2 and U6 that participate in U2/U6 helix I are in bold black and gray, as in Figure 1. U2/U6 helix Ia remains formed in this model, but residues in U2/U6 helix Ib are involved in mutually exclusive intramolecular interactions. (B,D) Repair of U2/U6 helix Ib at G21/C61 or (C,E) repair of the mutually exclusive intramolecular U6 stem at C61/G86. (B,C) Yeast containing the WT (left) or brC mutated (right) ACT1-CUP1 reporter were grown at 30°C for 3 d on media containing copper sulfate. (D,E) Wild-type PRP16 or mutant prp16-302 strains were grown on media containing 5-FOA at the restrictive temperature, 20°C, for 3 (PRP16) or 5 (prp16-302) d. Matrices are labeled as in Figure 2, except that in C and E the U6-G86 alleles are shown in each row.
We tested this alternative model for a role during 5′ splice site cleavage. Unlike most mutations in U2/U6 helix Ib, most mutations in the downstream region of U6 or upstream region of U2 failed to exacerbate brC or suppress prp16-302 (Fig. 4; data not shown). Where point mutations that disrupted both U2/U6 helix Ib and the four helix junction exacerbated brC or suppressed prp16-302, repair of the four-helix junction failed to alleviate exacerbation of brC or suppression of prp16-302 (Fig. 4C,E; data not shown), while repair of U2/U6 helix Ib did alleviate exacerbation of brC or suppression of prp16-302 (Figs. 2, 3, 4B,D; data not shown). For example, although U6-C61A exacerbated brC, this exacerbation was not alleviated by repairing the U6 ISL, but was alleviated by repairing helix Ib (Fig. 4, cf. B and C); by primer extension analysis, we confirmed directly that the exacerbation of the brC 5′ splice site cleavage defect conferred by C61 mutations was not alleviated by repair of the U6 ISL (Supplemental Data). Similarly, although the U6 mutant C61A suppressed prp16-302, this suppression was not diminished by repair of the U6 ISL, but was diminished by repair of helix Ib (Fig. 4, cf. D and E). These data fail to support the alternative hypothesis that the U2/U6 four-helix junction, rather than U2/U6 helix Ib, promotes 5′ splice site cleavage.
Evidence that U2/U6 helix I reforms to promote exon ligation
Although U2/U6 helix Ib is required for exon ligation (Hilliker and Staley 2004) and some helix Ia residues are required for exon ligation (Fabrizio and Abelson 1992; McPheeters and Abelson 1992; Wolff et al. 1994), it has remained unclear whether these data reflect a role for U2/U6 helix I during, rather than before, exon ligation. To determine whether U2/U6 helix I functions during exon ligation, we sensitized splicing to this catalytic step using ACT1-CUP1 pre-mRNA substrates limited at this step. These substrates contain a mutation of the branch site adenosine to guanosine (brG) or a mutation of the 3′ splice site UAG to gAG. These mutations reduce splicing and copper resistance by destabilizing the exon ligation conformation of the spliceosome (Query and Konarska 2004; Liu et al. 2007). If U2/U6 helix I promotes exon ligation, then point mutations in either U2 or U6 that would disrupt helix I would exacerbate the defects of these substrate mutations, just as mutations in prp8 that effectively destabilize the exon ligation conformation worsen these defects (Liu et al. 2007).
Indeed, by copper resistance, most point mutations in the U2 or U6 strands of helix I exacerbated the growth defect conferred by the brG mutation or the gAG mutation (Fig. 5A,B,E,F; Supplemental Data). Significantly, in almost all cases, restoring base pairing restored copper resistance. For instance, within U2/U6 helix Ia, the repaired double mutants C29G/G55C and U23C/A59G completely eliminated the exacerbation of both the brG and gAG mutations conferred by the corresponding point mutants (Fig. 5A,B,E,F). In contrast, double mutant combinations that failed to restore base pairing failed to restore copper resistance, with a few exceptions (Fig. 5; Supplemental Data; see below). Importantly, repair of other mutually exclusive structures such as the U6 central stem, U2 stem I, or the four-helix junction structure failed to eliminate exacerbation (Supplemental Data), arguing against a role for these alternative structures during exon ligation. These data provide evidence that disrupting U2/U6 helix I per se exacerbates the brG and gAG substrate mutations and, consequently, that U2/U6 helix I is required to efficiently splice these substrates.
FIGURE 5.
Disrupting U2/U6 helix I complementarity exacerbates the exon ligation defect of a branch site (brG) and a 3′ splice site (gAG) mutated substrate. (A–H) Yeast strains containing the indicated alleles at the U2/U6 helix I positions C29/G55 (A–D) and U23/A59 (E–H) and expressing a mutated ACT1-CUP1 splicing reporter were assayed for growth and splicing. Strains expressing the brG (A,E) and gAG (B,F) mutated splicing reporters were grown on media containing 0.1 mM copper sulfate for 3 d at 30°C. Alleles are arranged as in Figure 2. (C,D,G,H) Quantitation of primer extension reactions from selected strains containing the indicated alleles of U2 and U6, corresponding to the positions tested by growth directly above. Strains were tested with the brG (C,G) or gAG (D,H) mutated ACT1-CUP1 reporter. Quantitation was performed as described in Figure 2. The 5′ splice site efficiency is faded to emphasize the exon ligation efficiency.
Importantly, disrupting U2/U6 helix I not only exacerbated the copper resistance of the brG and gAG reporters, but also exacerbated the exon ligation defects of these mutated substrates. Specifically, by primer extension analysis, disrupting U2/U6 helix I decreased the efficiency of exon ligation for the brG and gAG substrates by 10%–80% in all but one case tested (Fig. 5C,D,G,H; Supplemental Data); in contrast, the efficiency of 5′ splice-site cleavage was not compromised for these substrates, with a few exceptions in the case of brG, which has a mild defect in 5′ splice site cleavage (McPheeters 1996). Importantly, restoring base pairing at all positions tested abolished the exacerbation of exon ligation by the corresponding point mutants (Fig. 5; Supplemental Data; see below). In contrast, noncompensatory mutations generally failed to abolish exacerbation (Fig. 5; Supplemental Data). In total, at five of the seven conserved base pairs in U2/U6 helix I, we found evidence that U2/U6 helix I promotes exon ligation for these substrates.
Because we were unable to corroborate these findings using prp22 suppressors (data not shown), as we could above for 5′ splice site cleavage using prp16 suppressors, these data do not formally rule out that U2/U6 helix I functions only on aberrant substrates. However, U2/U6 helix I promotes splicing of all three mutated substrates (Figs. 2, 5; Supplemental Data), and U2/U6 helix Ib promotes splicing of a wild-type substrate after 5′ splice site cleavage (Hilliker and Staley 2004), supporting a general role for U2/U6 helix I during exon ligation. Additionally, we cannot rule out that exacerbation occurs at a stage prior to exon ligation, but since the 3′ splice site is not recognized in yeast prior to exon ligation, this possibility is unlikely. Therefore, the simplest interpretation of our data is that U2/U6 helix I functions during exon ligation. Considering the evidence for a parallel role for U2/U6 helix I during 5′ splice site cleavage, these conclusions imply that U2/U6 helix I plays a fundamental role in shaping the structure of the catalytically active spliceosome.
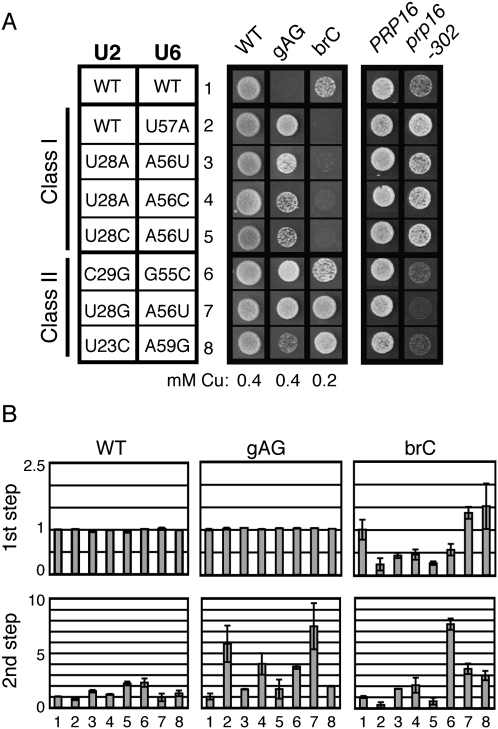
Several combinations of alleles promote splicing of aberrant substrates
Significantly, a few combinations of alleles in U2/U6 helix I improved rather than exacerbated the defects of the brG and gAG mutated substrates. These suppressors increased the copper resistance of brG and/or gAG by up to fourfold and increased the efficiency of exon ligation by two- to 10-fold, as measured by primer extension (Fig. 6; Supplemental Data). These suppressors can be grouped into two classes: those that also suppress prp16-302 and those that fail to suppress prp16-302. The first class includes two mismatches, a wobble, and two Watson–Crick combinations that reverse the purine/pyrimidine orientation of a base pair (Fig. 6; Supplemental Data; see Discussion). Each of these suppressors also decreased the efficiency of 5′ splice site cleavage for the brC substrate by 50%–80% (Fig. 6B), indicating that these suppressors act by destabilizing the 5′ splice site cleavage conformation, similar to U6-U57A and other previously characterized suppressors of exon ligation defective mutations (McPheeters 1996; Query and Konarska 2004). The second class includes a U2/U6 wobble combination and two Watson–Crick combinations. In contrast to the first class of suppressors, two of the three members in this class increased the efficiency of 5′ splice site cleavage of the brC substrate, and they all failed to suppress prp16-302 (Fig. 6), indicating that they do not simply suppress the exon ligation defect by destabilizing the 5′ splice site cleavage conformation. This second class of suppressors could suppress by either generally stabilizing both catalytic conformations or by destabilizing a competing conformation, such as the intermediate state that appears to occur after 5′ splice site cleavage (see Discussion).
FIGURE 6.
Combinations of alleles in U2/U6 helix I that compromise the fidelity of splicing. (A) Examples of combinations of U2 and U6 alleles that suppress brG and/or gAG reporters. The two classes of suppressors are indicated to the left. Class I suppressors exacerbate the brC reporter and suppress prp16-302, while Class II suppressors fail to exacerbate brC and fail to suppress prp16-302. Yeast were grown with the WT, gAG, or brC ACT-CUP1 reporter, as labeled, on media containing the indicated concentration of copper sulfate at 30°C for 3 d. Yeast were grown with either PRP16 or prp16-302, as labeled, on media containing 5-FOA at 20°C for 3 (PRP16) or 5 (prp16-302) d. (B) The efficiency of 5′ splice-site cleavage (1st step) and exon ligation (2nd step) for each reporter are shown and were calculated as in Figure 2.
DISCUSSION
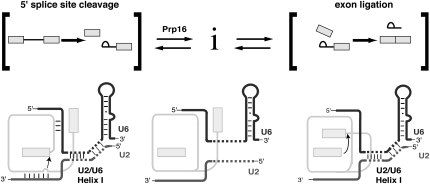
Previous investigations of mutations in the strands of U2/U6 helix I have demonstrated that these residues are important at several stages in the splicing cycle, but the studies have failed to convincingly attribute these defects to a role for U2/U6 helix I or to define a specific stage when U2/U6 helix I is required (Fabrizio and Abelson 1990; McPheeters and Abelson 1992; McPheeters 1996; Ryan and Abelson 2002; Hilliker and Staley 2004; McGrail et al. 2006). We provide the first compelling evidence that U2/U6 helix I plays an important role in promoting the catalytic step of 5′ splice site cleavage, as we show that disrupting U2/U6 helix I exacerbates a pre-mRNA substrate limited at this catalytic step. We also present persuasive evidence that Prp16p remodels the spliceosome after 5′ splice site cleavage, in part by destabilizing U2/U6 helix I, as we show that disrupting U2/U6 helix I per se suppresses the cold-sensitive mutation prp16-302 (Schwer and Guthrie 1991; Madhani and Guthrie 1994b), an observation that corroborates a role for U2/U6 helix I at the catalytic step of 5′ splice site cleavage. Finally, disrupting U2/U6 helix I also exacerbated pre-mRNA substrates that were limited at the catalytic step of exon ligation, suggesting that U2/U6 helix I also promotes this second chemical step of splicing. Together, our data extend previous studies of U2/U6 helix I by establishing evidence for the importance of U2/U6 helix I during both catalytic steps and by implicating U2/U6 helix I dynamics in between these steps (Fig. 7).
FIGURE 7.
Model for U2/U6 helix I function and rearrangement during the catalytic phase of splicing. U2/U6 helix I forms during 5′ splice site cleavage, where it could juxtapose the substrate for 5′ splice site cleavage and position a catalytically important metal ion. After 5′ splice site cleavage, U2/U6 helix I is destabilized during the Prp16p-dependent rearrangement, potentially allowing formation of undefined mutually exclusive interactions, which could aid in repositioning the substrate. U2/U6 helix I also promotes exon ligation, where it likely functions again to position a catalytically important metal ion and possibly the substrate.
U2/U6 helix I appears to be a key element of the spliceosomal catalytic core
Previous work has highlighted differences between the conformations that promote the two chemical steps of splicing. For instance, the Query and Konarska laboratories found evidence for a “two-state” model for the spliceosome, in which one state promotes 5′ splice site cleavage, the other state promotes exon ligation, and each state equilibrates with the other (Query and Konarska 2004; Liu et al. 2007). This model was inspired by observations that a number of spliceosomal mutations interact genetically in opposite ways with substrates limited for either 5′ splice site cleavage or exon ligation. In contrast to these distinct features of the two catalytic states, more recent work provided evidence that the two catalytic conformations also share an important structure, U2 stem IIc (Hilliker et al. 2007). Similarly, we found that mutations that disrupted U2/U6 helix I exacerbated both a substrate limited for 5′ splice site cleavage and a substrate limited for exon ligation, providing evidence that U2/U6 helix I functions as a general component of the catalytically active spliceosome. Thus, although the spliceosome shows differences between the two catalytic conformations, it appears to show remarkable similarity in the snRNA structure of each conformation. This similarity may reflect a general catalytic conformation of the snRNAs that efficiently promotes the phosphoryl transfer reactions of splicing.
Role of U2/U6 helix Ib and the catalytic AGC triad
The helix Ib component of U2/U6 contains the invariant AGC triad of U6, which likely binds a catalytic metal ion in analogy with the catalytic AGC triad of group II introns (Fabrizio and Abelson 1992; Yu et al. 1995; Toor et al. 2008). Given the importance of the AGC triad, it is crucial to understand what positions, and consequently, what activates this triad within the catalytically active spliceosome. However, two different strands have been proposed to bind and position the AGC triad: a strand in U2 that forms U2/U6 helix Ib (Madhani and Guthrie 1992; Tarn and Steitz 1996; Frilander and Steitz 2001; Hilliker and Staley 2004), and a strand in U6 that forms an extension of the U6 ISL (Sun and Manley 1995; Sashital et al. 2004). Here, we provide the first evidence to resolve these two alternative models in yeast. Specifically, we found evidence that U2/U6 helix Ib promotes splicing at the stage of 5′ splice site cleavage and exon ligation (Figs. 2–5) and failed to find any evidence to support a role for the U6 ISL extension in promoting splicing at the stage of 5′ splice site cleavage (Fig. 4), although our data do not rule out a role for U6 in binding to the AGC triad at other stages of splicing. Given evidence for a role for helix Ib at both catalytic stages of splicing, our data suggest that one important role for U2 is to position the AGC triad within the catalytic core to bind a catalytic metal as in group II introns (Toor et al. 2008). By using a separate RNA to position the catalytic triad, the spliceosome may more readily regulate the formation and dynamics of the catalytic core.
Interactions beyond U2/U6 helix I base pairing
While the majority of the data are consistent with a role for U2/U6 helix I at the catalytic steps, the entire data set is nonetheless complex. For example, some non-native Waston–Crick combinations fail to reveal evidence for helix I and a few rare non-Watson–Crick combinations appear inconsistent with helix I. Such observations could, in principle, indicate a function for an alternative, mutually exclusive structure at the catalytic steps. However, we have found no evidence for five alternative structures that we have tested. Furthermore, an alternative structure would be difficult to reconcile with the majority of data consistent with helix I formation. Thus, we favor a model in which U2/U6 helix I functions at both catalytic steps and we propose that the complexity of the data reflects both the importance of a mutually exclusive structure at a noncatalytic step in splicing (see below) and the importance of U2/U6 helix I residues beyond simple Watson–Crick base pairing.
Surprisingly, the optimal pair of bases for stabilizing the catalytic state did not always correspond to Watson–Crick pairs; in these cases, the most stabilizing combinations could form wobbles with a purine in U2 and a pyrimidine in U6. In particular, the base combination U2/U6-U28G/A56U improved both 5′ splice site cleavage and exon ligation, strongly suppressing the brC, brG, and gAG substrate mutations (Fig. 6; Supplemental Data). Additionally, the base combination U2/U6-G26/C58U improved 5′ splice site cleavage for the brC substrate (Supplemental Data). Interestingly, uracil is found at the equivalent of position U6-58 in most other species (Guthrie and Patterson 1988), suggesting that a wobble at this position promotes 5′ splice site cleavage in other organisms and that a Watson–Crick base pair is selected in budding yeast to stabilize exon ligation. These wobble combinations parallel the U2/U6 wobble combination A27/U57C, which improves 5′ splice site cleavage for the brC substrate (McPheeters 1996; Query and Konarska 2004). Because each of these wobbles also exacerbated the prp16-302 mutation (McPheeters 1996), these data imply that the wobbles stabilize the 5′ splice site cleavage conformation, even with wild-type substrates. Consistent with this trend, three of the four noncompensatory combinations that alleviated exacerbation of brC corresponded to wobbles (Supplemental Data). Wobble combinations efficiently coordinate metal ions (Varani and McClain 2000), suggesting a possible mechanism by which wobbles could stabilize the catalytic core. Wobbles may be disfavored during natural selection, because catalysis may not be rate-limiting or because highly efficient splicing could compromise fidelity.
Sequence constraints on helix I were also reflected by some Watson–Crick repair combinations that failed to restabilize the spliceosome, as indicated by their failure to reverse genetic interactions with substrate mutations and prp16-302. For instance, while all but one of the conservative transition combinations did restabilize the spliceosome and revert suppression of prp16-302, of the five double-mutant, Watson–Crick repaired combinations that failed to revert suppression of prp16-302, four corresponded to the more extreme transversion base-pair combinations (Fig. 4; Supplemental Data), suggesting a role for the orientation of the Watson–Crick base pairs. These sequence constraints could reflect the importance of tertiary interactions between U2/U6 helix I and RNA or protein binding partners. Thus, our results provide a foundation to test for tertiary interactions and, in addition, provide a framework for designing stable and catalytically active models of U2/U6 helix I.
Implications for a role for Prp16 in destabilizing U2/U6 helix I and remodeling the spliceosome
Our data suggest a function for the DEAH-box ATPase Prp16p in promoting exon ligation. Previous studies found that mutations in helix I suppressed prp16 mutants, but these studies failed to attribute suppression to the disruption of U2/U6 helix I because they were unable to show that suppression was lost upon repair of U2/U6 helix I (Madhani and Guthrie 1994b), likely because the severe mutations precluded restoration of wild-type stability to helix I (see above). In contrast, by using subtle mutations that allowed restoration of wild-type stability to helix I, we found evidence that repair of U2/U6 helix I abolished suppression of prp16-302, implying that prp16-302 is suppressed by disruption of helix I per se. Thus, we have found the first evidence that Prp16p destabilizes U2/U6 helix I explicitly. Although destabilization does not necessarily equate with unwinding, Prp16p-mediated unwinding of this key catalytic component, directly or indirectly, could facilitate disruption of the neighboring U6/5′ splice site and U2/branch-site interactions (Fig. 1) and thereby promote repositioning of the substrate for exon ligation after 5′ splice site cleavage. Since simply changing the identity of some helix I base pairs suppressed prp16-302, Prp16p, rather than unwinding helix I, may instead dissociate U2/U6 helix I from an RNA or protein binding partner, as proposed by Madhani and Guthrie (1994b). Finally, if U2/U6 helix I is thermodynamically coupled to dynamic substrates of Prp16p, then Prp16p may destabilize U2/U6 helix I, without disrupting or dissociating the helix, as a downstream consequence of disrupting energetically coupled structures. However, given the implication of an intermediate mutually exclusive with U2/U6 helix I (see below), we favor a role for Prp16p in unwinding U2/U6 helix I, directly or indirectly. Remarkably, prp16 is also suppressed by mutations in U2 stem IIc, U6, Prp8p, and Isy1p (McPheeters 1996; Query and Konarska 2004; Villa and Guthrie 2005; Hilliker et al. 2007). Consequently, Prp16p may disrupt multiple interactions, disrupt a single interaction that in turn leads to the disruption of other elements, or disrupt a single interaction whose stability is dependent on neighboring structures that are not dynamic.
Consistent with a role for Prp16p in promoting unwinding of U2/U6 helix I, directly or indirectly, our analysis suggests that the spliceosome assumes a mutually exclusive intermediate state after 5′ splice site cleavage. Prp16p has previously been implicated in unwinding U2 stem IIc, directly or indirectly, to allow formation of a mutually exclusive intermediate state containing U2 stem–loop IIa, in part because mutations that disrupt U2 stem–loop IIa suppress a 3′ splice site mutant without destabilizing the 5′ splice site cleavage conformation (Hilliker et al. 2007). Similarly, a number of mutant combinations in the U2/U6 helix I region suppressed the exon ligation defects of the brG or gAG 3′ splice site mutations, but without destabilizing the 5′ splice site cleavage conformation. Such suppressors did not worsen the growth defect of brC or suppress prp16-302 (Fig. 6). All three suppressor combinations suppressed substrate defects regardless of the substrate mutation, suggesting that the suppressors act via an indirect mechanism. Although these mutations could conceivably suppress by generally stabilizing the exon ligation conformation, we favor an indirect mechanism for suppression in which the mutations destabilize a mutually exclusive on-pathway intermediate, an intermediate that is also implied by the direct or indirect destabilization of U2/U6 helix I by Prp16p after 5′ splice site cleavage. Although our efforts, to date, to define alternative RNA–RNA interactions, such as the U6 central stem or U2 stem I, have failed (data not shown), such an alternative conformation would likely cooperate with the intermediate stem–loop IIa state of U2 to promote an open state of the spliceosome that permits rearrangement of the substrate.
Implications for splicing fidelity
Our work offers insights into how fidelity in splicing may be achieved. First, a role for Prp16p in antagonizing the catalytically important U2/U6 helix I would provide a simple rationale for how Prp16p antagonizes splicing of an aberrant substrate to promote the fidelity of splicing. Second, the formation of a catalytically inactive intermediate lacking U2/U6 helix I after 5′ splice site cleavage would provide a sink to sequester the spliceosome from the catalytically active exon ligation conformation when the substrate is inappropriate for exon ligation. This mechanism would complement the 5′ splice site cleavage conformation as a sink for sequestering substrates inappropriate for exon ligation. Such mechanisms for fidelity would be surprising, given that they appear to involve unwinding of the active core of a large and complex ribonucleoprotein enzyme.
MATERIALS AND METHODS
Yeast strains and plasmids
For copper growth assays, we used strain yJPS1035, containing the plasmid pU2U6U (Hilliker et al. 2007). For the Prp16p growth assays, we used strains YHM187, containing the prp16-302 mutation, and the wild-type YHM118 (Madhani and Guthrie 1994b,c). These three shuffle strains allow analysis of U2 and U6 on plasmids. pJPS1920, a LEU2-marked vector, encodes ACT-CUP1 (Hilliker et al. 2007); pJPS216, a HIS3 marked plasmid, encodes U2 (Shuster and Guthrie 1988); and pSX6, a TRP1 marked plasmid, encodes U6 (Madhani and Guthrie 1992). All mutations in ACT1-CUP1, U2, and U6 were generated by QuikChange (Stratagene) and verified by sequencing or obtained from previous studies (Hilliker and Staley 2004; Hilliker et al. 2007).
Copper growth assays
yJPS1035 was first transformed with pJPS1920 variants and then cotransformed with pJPS216 and pSX6 variants. Cotransformants were streaked to media lacking leucine and containing 5-flouroorotic acid (5-FOA) (Sikorski and Boeke 1991) and grown for 3 d at 30°C. Colonies were then purified on minimal media lacking leucine for subsequent analysis by growth on copper (Lesser and Guthrie 1993) or by primer extension (see below). To assess the growth of yeast strains on media containing copper, yeast were grown overnight in liquid media lacking leucine to an OD600 of 0.8–1.0, then spotted onto plates lacking leucine with the following concentrations of copper sulfate (mM): 0, 0.013, 0.05, 0.1, 0.2, 0.4, and 0.8, where growth becomes limiting in the wild-type strain. Yeast were grown at 30°C for 3–5 d for phenotype analysis. In each figure, growth is shown at a single concentration of copper to facilitate a comparison of the fundamental, qualitative phenotypes through a matrix. Quantitative analysis of copper resistance or splicing efficiency (see below) leads to observations that support the significance of the qualitative phenotypes. A standard lithium acetate method was used for all transformations (Ito et al. 1983).
Primer extension assays
The same yeast that were generated for the copper growth assays were grown in liquid media lacking leucine at 30°C and harvested at a final OD600 of 0.8. Whole cell RNA was isolated by hot phenol extraction. Primer extensions were performed as described (Stevens and Abelson 2002) using the 32P-labeled oligos oJPS233 and oJPS234 (Hilliker et al. 2007). Primer extension products were separated on a 6% polyacrylamide/7 M urea gel and imaged by PhosphorImager (Molecular Dynamics). Gels were quantitated using ImageQuant software (Molecular Dynamics). The efficiency of 5′ splice-site cleavage was calculated as the ratio of mRNA and lariat intermediate to total RNA [(M+LI)/(M+LI+P)], and exon ligation was calculated as the ratio of mRNA to lariat intermediate (M/LI) or [M/(M+LI)]. In the figures, we have shown M/LI, rather than M/(M+LI), because the M/LI measure of exon ligation is sensitive to changes in the levels of lariat intermediate regardless of the levels of mature mRNA; nonetheless, the trends were similar regardless of which calculation was used. These ratios are self-normalizing; yet U14 was primer extended along with the substrate to provide a visual control for loading. All ratios were expressed relative to the efficiency of a mutated substrate in a strain expressing wild-type U2 and U6, a substrate whose efficiency was arbitrarily set to 1. For low-intensity bands less than twofold above background, we set the value of these bands to twice the local background to establish a conservative upper limit.
Plasmid shuffle growth assays
Variants of pJPS216 and pSX6 were cotransformed into YHM118 and YHM187. Transformants were grown in liquid media lacking histidine and tryptophan to an OD600 of ∼0.8, and then spotted onto media containing 5-FOA to counter select for the pU2U6U plasmid (Sikorski and Boeke 1991). Cells were grown at the permissive temperature of 30°C for 3–5 d and the restrictive temperature of 20°C for 5–7 d for phenotypic analysis.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank current members of the Staley laboratory, Angela Hilliker, Eliza Small, and Sam Butcher for critical reading of this manuscript; Christine Guthrie for strains and plasmids; and Channon Jordan and Martha Norman for technical assistance. This work was supported by a grant from NIH (GM62264) to J.P.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1582609.
REFERENCES
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990;250:404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res. 1992;20:3659–3664. doi: 10.1093/nar/20.14.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander MJ, Steitz JA. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol Cell. 2001;7:217–226. doi: 10.1016/s1097-2765(01)00169-1. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes & Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- Luukkonen BG, Séraphin B. A role for U2/U6 helix Ib in 5′ splice site selection. RNA. 1998;4:915–927. doi: 10.1017/s1355838298980591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Dynamic RNA–RNA interactions in the spliceosome. Annu Rev Genet. 1994a;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Genetic interactions between the yeast RNA helicase homolog Prp16 and spliceosomal snRNAs identify candidate ligands for the Prp16 RNA-dependent ATPase. Genetics. 1994b;137:677–687. doi: 10.1093/genetics/137.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Randomization-selection analysis of snRNAs in vivo: Evidence for a tertiary interaction in the spliceosome. Genes & Dev. 1994c;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Moore MJ. Spliceosome assembly and composition. Adv Exp Med Biol. 2007;623:14–35. doi: 10.1007/978-0-387-77374-2_2. [DOI] [PubMed] [Google Scholar]
- McGrail JC, Tatum EM, O'Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–3822. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS. Interactions of the yeast U6 RNA with the pre-mRNA branch site. RNA. 1996;2:1110–1123. [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Abelson J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell. 1992;71:819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. The spliceosome: The most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- Ryan DE, Abelson J. The conserved central domain of yeast U6 snRNA: Importance of U2-U6 helix Ia in spliceosome assembly. RNA. 2002;8:997–1010. doi: 10.1017/s1355838202025013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE. U2-U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster EO, Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988;55:41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: From cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing. Mol Cell. 2007;26:883–890. doi: 10.1016/j.molcel.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: Spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stark H, Lührmann R. Cryo-electron microscopy of spliceosomal components. Annu Rev Biophys Biomol Struct. 2006;35:435–457. doi: 10.1146/annurev.biophys.35.040405.101953. [DOI] [PubMed] [Google Scholar]
- Stevens SW, Abelson J. Yeast pre-mRNA splicing: Methods, mechanisms, and machinery. Methods Enzymol. 2002;351:200–220. doi: 10.1016/s0076-6879(02)51849-8. [DOI] [PubMed] [Google Scholar]
- Sun JS, Manley JL. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes & Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S. The spliceosome: Caught in a web of shifting interactions. Curr Opin Struct Biol. 2007;17:310–315. doi: 10.1016/j.sbi.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Wachtel C, Manley JL. Protein-free spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing. RNA. 2007;13:2300–2311. doi: 10.1261/rna.626207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, McClain WH. The G•U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaver RM, Fortner DM, Loos-Austin LS, Brow DA. Multiple functions of Saccharomyces cerevisiae splicing protein Prp24 in U6 RNA structural rearrangements. Genetics. 1999;153:1205–1218. doi: 10.1093/genetics/153.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes & Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T, Pleiss JA, Guthrie C. Spliceosomal snRNAs: Mg2+-dependent chemistry at the catalytic core? Cell. 2002;109:149–152. doi: 10.1016/s0092-8674(02)00726-2. [DOI] [PubMed] [Google Scholar]
- Wolff T, Menssen R, Hammel J, Bindereif A. Splicing function of mammalian U6 small nuclear RNA: Conserved positions in central domain and helix I are essential during the first and second step of pre-mRNA splicing. Proc Natl Acad Sci. 1994;91:903–907. doi: 10.1073/pnas.91.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW. U6 snRNA function in nuclear pre-mRNA splicing: A phosphorothioate interference analysis of the U6 phosphate backbone. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]