Abstract
In Drosophila, miRNA is processed by Dicer-1 (DCR-1) from its precursor and loaded onto Argonaute1 (AGO1). AGO1 recognizes target mRNAs based on the miRNA sequence and suppresses the expression at post-transcriptional levels. GW182, a P-body component, localizes the AGO1 complex to processing bodies (P-bodies) where mRNA targets are decayed or stored. However, the details of the pathway remain elusive. In this study, two distinct types of AGO1-containing complexes from Drosophila Schneider2 (S2) cells were isolated and compared at the molecular level. The AGO1 complex with DCR-1 contained neither mature miRNA nor GW182 but exhibited pre-miRNA processing activity. The resultant mature RNA was loaded onto AGO1 within the complex. The AGO1 complex with GW182 excluded DCR-1, but possessed mature miRNA and showed no pre-miRNA processing activity. Thus, the AGO1-DCR-1 and AGO1-GW182 complexes correspond to miRLC (miRISC loading complex) and miRISC, respectively. The requirement for various domains of AGO1 in miRNA-loading and DCR-1/GW182 interaction was also examined. The Mid domain mutant (F2V2) interacted with DCR-1 but not with mature miRNA and GW182. The AGO1-PAZ mutant lacks the mature miRNA-binding ability but associates with either DCR-1 or GW182. The AGO1-PIWI mutant showed no Slicer activity but associates with mature miRNA. These results indicate that these domains are required differently for miRLC and miRISC formation in the miRNA pathway.
Keywords: Argonaute1, GW182, Dicer-1, miRLC, miRISC, Drosophila
INTRODUCTION
RNA silencing is an evolutionarily conserved physiological process regulating gene expression (Zaratiegui et al. 2007). RNA interference (RNAi), the prototype of RNA silencing, negatively regulates gene expression by cleaving target mRNAs in a sequence-dependent manner (Tolia and Joshua-Tor 2007). A molecule that renders sequence specificity to RNAi is a short interfering RNA (siRNA) of ∼21 nucleotides (nt) (Bernstein et al. 2001). siRNAs are produced from long double-stranded (ds) RNA precursors that are exogenously introduced into, or expressed within, cells. The RNase III enzyme required for dicing long dsRNA precursors into siRNAs in Drosophila is Dicer-2 (DCR-2) (Bernstein et al. 2001; Lee et al. 2004a). DCR-2 associates with R2D2, a dsRNA-binding domain protein during the processing step (Liu et al. 2003). DCR-2-R2D2 is also necessary for the siRNA loading step onto the RNA-induced silencing complex (RISC) (Liu et al. 2003); thus, it is considered as the RISC-loading complex (RLC) (Lee et al. 2004a; Tomari et al. 2004a). Other components of the RLC remain unclear. The siRNA duplex processed by DCR-2-R2D2 is passed onto Argonaute2 (AGO2), an endo-nuclease required for cleaving target mRNAs in RNAi (Tolia and Joshua-Tor 2007). AGO2 cleaves a particular strand of the siRNA duplex (called “passenger”; this strand is not loaded onto RISC), which then disappears in the environment (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005; Leuschner et al. 2006; Kim et al. 2007). Through these processes, RISC is finally activated.
miRNAs are an endogenous subset of small RNAs of 21–23 nt that function in RNA silencing by repressing translation or by facilitating degradation of target mRNAs (Bushati and Cohen 2007; Farazi et al. 2008). miRNA target genes are implicated in vital biological processes such as development, apoptosis, neural patterning, and viral infection (Bushati and Cohen 2007). To date, about 150 miRNAs have been identified in Drosophila. Each miRNA is thought to target multiple genes, suggesting that the vast majority of protein-coding genes are regulated by miRNAs in vivo (Bushati and Cohen 2007).
miRNAs are mostly transcribed by RNA polymerase II (Cai et al. 2004; Lee et al. 2004b). The primary transcripts, pri-miRNAs, are processed to pre-miRNAs (miRNA precursors) in the nucleus by an RNase III enzyme, Drosha, in Drosophila (Lee et al. 2003). Upon export to the cytoplasm, pre-miRNAs are further processed by another RNase III enzyme, DCR-1 (Lee et al. 2004a). Loquacious (LOQS; also known as R3D1), a cytoplasmic double-stranded (ds) RNA-binding domain protein, stimulates and directs the specific pre-miRNA processing activity of DCR-1 by tightly associating with DCR-1 (Forstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). Upon processing, miRNAs are still in a duplex form (miRNA/miRNA*) and thus should be unwound prior to miRISC (miRNA-containing RISC) formation. In contrast to siRNA duplexes, strand rejection of miRNA* is not accompanied by miRNA* strand cleavage by AGO1 (Matranga et al. 2005). Factor(s) responsible for miRNA/miRNA* unwinding remain unknown. Within miRISC, AGO1, another member of the Argonaute family in Drosophila, directly associates with mature, single-stranded miRNA (Miyoshi et al. 2005).
Human GW182 was shown to be involved in post-transcriptional regulation of a subset of mRNAs by localizing them to cytoplasmic foci termed P-bodies (Eystathioy et al. 2002, 2003; Liu et al. 2005; Meister et al. 2005). P-bodies contain enzymes required for mRNA decay, for example, CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. In Caenorhabditis elegans, AIN-1, a homolog of human GW182, interacts with ALG-1, one of the nematode Argonautes, and targets ALG-1 to P-bodies (Ding et al. 2005). GW182 in Drosophila (also known as Gawky) (Schneider et al. 2006) was also shown to interact with AGO1 and accumulate in P-bodies (Behm-Ansmant et al. 2006; Schneider et al. 2006). Recent studies suggested a functional involvement of GW182 in the miRNA-mediated silencing pathway. Loss of GW182 changes mRNA expression profiles in a similar way to those observed in AGO1 depletion (Behm-Ansmant et al. 2006). It was also demonstrated that GW182 interaction with AGO1 is essential for miRNA-mediated translational repression and mRNA decay (Liu et al. 2005; Jakymiw et al. 2007; Eulalio et al. 2008). Taken together, it is thought that DCR-1-LOQS, AGO1, and GW182 are indispensable in the Drosophila miRNA pathway. However, in contrast to the RNAi pathway, the RLC-like complex and miRISC, which should be formed in the miRNA pathway, have not yet been isolated and characterized.
To gain further insights into the miRNA-mediated gene silencing mechanism, we immunopurified AGO1-containing complexes from S2 cells and found two such complexes. The AGO1 complex that involves DCR-1 excludes GW182 and mature miRNAs but exhibits pre-miRNA activity. miRNAs processed by the complex were loaded onto AGO1 within the complex. The other AGO1 complex contained GW182 and mature miRNAs but excluded DCR-1 and pre-miRNA activity. These results suggest that the association of AGO1 with DCR-1 and GW182 occurs in a mutually exclusive manner and that the AGO1-DCR-1 and the AGO1-GW182 complexes correspond to miRLC and miRISC, respectively. The requirements of the various domains in AGO1 for binding with miRNA, DCR-1, and GW182 were also examined. The Mid domain mutant (F2V2) interacted with DCR-1. However, it does not associate with mature miRNA and GW182; thus, it did not localize to P-bodies (Eulalio et al. 2008; this study). The PAZ mutant lacks the mature miRNA-binding activity, but associates with either DCR-1 or GW182. The Piwi mutant showed no Slicer activity but still associates with mature miRNA. These results indicate that these domains are required for different AGO1 functions and for miRLC and miRISC formation in the miRNA pathway.
RESULTS
AGO1 association with DCR-1 and GW182 is mutually exclusive
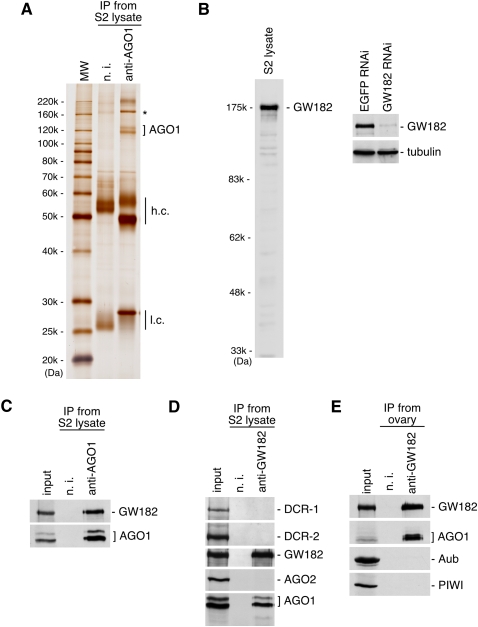
The identification of AGO1-associating proteins in S2 cells by immunoprecipitation with anti-AGO1 antibodies revealed that an ∼175-kDa protein is a tight binder (Fig. 1A, “*”). Mass spectrometric analysis identified the protein as GW182, a factor known to localize the AGO1 complex to P-bodies (Behm-Ansmant et al. 2006). Western blotting using a newly produced anti-GW182 antibody (Fig. 1B) on anti-AGO1 immunoprecipitates confirmed the association (Fig. 1C). Anti-GW182 immunoprecipitates were reciprocally probed with anti-AGO1. Bands corresponding to AGO1 were detected (Fig. 1D; Supplemental Fig. 1A). However, under these circumstances, DCR-1, which was previously shown to interact with AGO1, was not detected (Fig. 1D; the specificity of anti-DCR-1 monoclonal antibody is shown in Supplemental Fig. 1B). These results led us to hypothesize that AGO1 associates with DCR-1 and GW182 in a mutually exclusive manner. AGO2 did not interact with GW182 (Fig. 1D) as reported previously (Behm-Ansmant et al. 2006). DCR-2 was also undetected in the complex (Fig. 1D; the specificity of anti-DCR-2 monoclonal antibody is indicated in Supplemental Fig. 1B). Western blotting on anti-GW182 immunoprecipitates from ovaries revealed no association of GW182 with Aub, Piwi (Fig. 1E), or AGO3 (data not shown), the germline-specific Argonaute. Thus, in Drosophila, GW182 is dedicated to the function of AGO1 in the miRNA pathway and is most likely unrelated to the piRNA pathway.
FIGURE 1.
AGO1 interacts in a mutually exclusive manner with DCR-1 and GW182. (A) AGO1 coimmunoprecipitates with an ∼175-kDa protein, GW182, from S2 cells. The immunoprecipitated proteins were stained with silver. (n.i.) Non-immune IgG used as negative control; (h.c. and l.c.) heavy and light chains of the antibodies, respectively. (B) Western blotting analyses show that anti-GW182 antibodies produced in our laboratory specifically recognize GW182 in S2 cells. (C) Anti-AGO1 immunoprecipitates from S2 cells contain both AGO1 and GW182. (D) AGO1, but not AGO2, DCR-1, and DCR-2, is detected in the immunoprecipitated complex with anti-GW182 from S2 cells. (E) The PIWI proteins, Aub and Piwi, are not detected in anti-GW182 immunoprecipitates obtained from fly ovaries.
AGO1 with GW182, but not with DCR-1, is associated with endogenous mature miRNA
To confirm the exclusion of GW182 from the AGO1-DCR-1 complex, immunoprecipitates with anti-LOQS were probed with anti-GW182. The immunoprecipitation efficiency of anti-DCR-1 was very low (data not shown); thus, hereinafter, we decided to use anti-LOQS to obtain the AGO1-DCR-1 complexes. As expected, GW182 was undetected in the AGO1-DCR-1-LOQS complex (Fig. 2A). These results further suggest that two distinct AGO1-containing complexes exist in S2 cells; one with DCR-1-LOQS and the other with GW182. Glycerol gradient sedimentation experiments further supported the model that the AGO1-DCR-1 and AGO1-GW182 complexes are distinct in S2 cells (Supplemental Fig. 2). Note that GW182 and DCR-1 were detected simultaneously in anti-AGO1 immunoprecipitates, although the DCR-1 and LOQS signals were very faint compared with those of GW182 (Fig. 2A). This suggests that the AGO1-GW182 complex is more abundant than the AGO1-DCR-1-LOQS complex in S2 cells. Thus, to equalize the amount of AGO1 in the immunoprecipitated samples, we employed 10-fold more (by volume; the protein concentration was the same) S2 lysate for anti-LOQS immunoprecipitation (see Fig. 2A).
FIGURE 2.
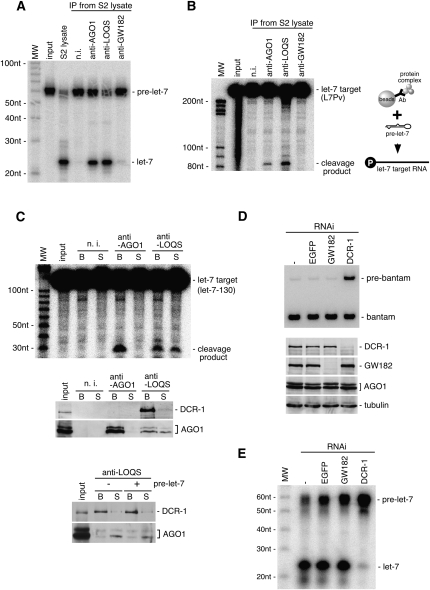
Characterization of two AGO1-containing complexes immunoprecipitated from S2 cells. (A) GW182 is not detected in the AGO1-DCR-1 complex immunoprecipitated from S2 cells using anti-LOQS. It should be noted that 10-fold more S2 lysate was used for n.i. (non-immune IgG; negative control) and anti-LOQS immunoprecipitations compared to those for anti-AGO1 and anti-GW182 immunoprecipitations to equalize the amounts of AGO1 in the immunoprecipitates. (Lowest panel, *) Bands corresponding to antibodies (heavy chains). (B) RNAs were extracted from the immunoprecipitated complexes (shown in A) and subjected to Northern blotting with DNA oligos for bantam and the opposite strand, bantam*. Anti-AGO1 and anti-GW182 immunoprecipitates almost exclusively contain single-stranded mature bantam. On the other hand, anti-LOQS immunoprecipitates show very weak signals for bantam precursor and bantam/bantam* duplexes. (C) Immunoprecipitated complexes (shown in A) were subjected to in vitro target RNA cleavage assays using a target RNA (bantam-130) showing full complementarity to bantam. The AGO1-GW182 complex, but not the AGO1-DCR-1 complex, exhibits bantam-dependent Slicer activity.
To compare miRNAs present in the AGO1 complexes, RNAs isolated from the complexes were probed with DNA oligos for bantam, an miRNA expressed in S2 cells, and its antisense strand, bantam* (Fig. 2B). The AGO1-GW182 complex contained abundant mature bantam (Fig. 2B; see anti-AGO1 and anti-GW182 lanes). In contrast, bantam* and pre-bantam were barely detected. The AGO1-DCR-1-LOQS complex (anti-LOQS lane) contained both bantam and bantam* at very low levels. Faint signals for pre-bantam also appeared. Both bantam and bantam* signals are ∼2.5-fold stronger compared with pre-bantam signal on each gel. These results suggest that miRNAs contained in the complex are mostly in either duplex or precursor forms and that AGO1 associated with DCR-1 might be mostly devoid of mature miRNA in vivo.
In vitro target RNA cleavage assays were performed using a target RNA harboring a sequence completely complementary to bantam miRNA (bantam-130). The AGO1-GW182 complex, which contains mature bantam, was able to cleave the target, whereas the AGO1-DCR-1-LOQS complex failed (Fig. 2C). These results confirm that AGO1 associated with DCR-1-LOQS is unloaded with mature, active miRNA.
The AGO1-DCR-1 complex shows activities to process pre-miRNA and load miRNA onto AGO1
AGO1 associated with DCR-1-LOQS is not loaded with mature miRNA (Fig. 2B,C). To examine if the miRNA loading step induces the dissociation of AGO1 from DCR-1-LOQS, pre-miRNA loading assays were performed. Both the AGO1 complexes with DCR-1-LOQS and GW182 were immunopurified from S2 cells (as in Fig. 2) and 32P-labeled pre-let-7 was added to the complexes and incubated. The AGO complex that contains DCR-1-LOQS was able to show processive activity of pre-let-7 to mature let-7, as expected (Fig. 3A; anti-AGO1, anti-LOQS). In contrast, the AGO1-GW182 complex failed to show the activity, confirming the earlier observation that the AGO1-GW182 complex is devoid of DCR-1 (Fig. 1D). Immunoprecipitates with anti-LOQS and anti-AGO1, after being exposed to pre-let-7 processing in vitro, were able to cleave the let-7 target (Fig. 3B). Thus, let-7 loading onto AGO1 that was originally contained in the complex was confirmed. When RNA cleavage assays were performed using the supernatant and the bead fractions separated after the miRNA loading, both fractions of anti-LOQS showed the Slicer activity (Fig. 3C). The same fractions were subjected to Western blotting using anti-AGO1, which showed the displacement of AGO1 from DCR-1. Under the same conditions, DCR-1 remained mostly on the beads (Fig. 3C). However, we noticed that under conditions in which pre-let-7 was not added, AGO1 was also detected in the supernatant (Fig. 3C), suggesting that some population of AGO1 might be displaced from DCR-1 independently of miRNA loading.
FIGURE 3.
The AGO1-DCR-1 complex exhibits pre-miRNA processing activity, but the AGO1-GW182 does not. (A) Immunoprecipitated complexes (Fig. 2A) were subjected to in vitro pre-miRNA processing assays using pre-let-7 labeled with 32P. The AGO1-GW182 complex does not show the activity because the complex contains little or no DCR-1. (B) Immunoprecipitated complexes (Fig. 2A) were first subjected to in vitro pre-miRNA processing using pre-let-7 (cold), and then target RNA cleavage assays were performed. The AGO1-GW182 complex, which did not show pre-let-7 processing activity in A, does not cleave the target (let-7 target; L7Pv), whereas the AGO1-DCR-1 complex shows the activity, indicating that the AGO1 that was originally contained in the complex was loaded with mature let-7 processed by the immunoprecipitated complex. (C) pre-let-7 processing was performed using let-7-130 as a target. After the reaction, the supernatant and bead fractions were separated by centrifugation, and then target RNA cleavage assays were performed as in B. The fractions were also subjected to Western blotting using anti-DCR-1 and anti-AGO1. AGO1 is detected both in the supernatant (S) and bead (B) fractions of anti-LOQS immunoprecipitates, whereas DCR-1 is only detected in the bead fraction, indicating that AGO1 dissociates from DCR-1 upon miRNA loading. Such dissociation was confirmed by performing target RNA cleavage assays. The cleavage products are observed in both the supernatant and bead fractions of anti-LOQS immunoprecipitates. The supernatant fraction of anti-AGO1 immunoprecipitates shows no activity to cleave the target RNA, because AGO1 stayed on the beads during the assays. The bead fraction of anti-LOQS shows stronger cleavage activity than expected. This was because pre-miRNA processing continuously occurred even after the target RNA cleavage reaction was started. The lowest panel shows that even under conditions where pre-let-7 was not added, AGO1 was released to the supernatant, indicating that AGO1 displacement from DCR-1 occurs in an miRNA-loading-independent manner. (D) GW182 and DCR-1 were depleted from S2 cells by RNAi, and RNAs isolated from individual cells were subjected to Northern blotting using a DNA oligo for bantam. When DCR-1 was depleted, pre-bantam was aberrantly accumulated, whereas GW182 depletion did not cause such an effect. The lower panel (Western blot) shows the high efficiency of DCR-1 and GW182 depletion in each sample. (E) GW182 and DCR-1 are depleted from S2 cells as in D, and the resultant cell lysates were subjected to pre-miRNA processing assays. Only DCR-1 depletion causes a severe decrease in pre-miRNA processing activity.
The AGO1-DCR-1 complex showed strong miRNA processing activity, although it excludes GW182 (Fig. 3A). To address the question of whether GW182 is unnecessary for the process, GW182 was depleted from S2 cells by RNAi, and Northern blotting was performed. In contrast to DCR-1 depletion, where pre-bantam accumulated, GW182 depletion did not cause such an effect (Fig. 3D). Neither did GW182 depletion abolish the pre-miRNA processing activity (Fig. 3E). Thus, GW182 is unnecessary for miRNA processing.
The above data imply that the two distinct AGO1 complexes found in Drosophila S2 cells, one with DCR-1-LOQS and the other with GW182, correspond to miRLC and miRISC, respectively.
Migration of miRLC and miRISC on native gels differs from that of siRLC and siRISC
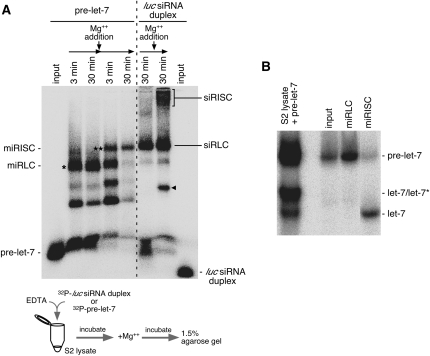
Gel shift assays have shown that siRLC and siRISC formed in RNAi migrated differently on native gel systems (Pham et al. 2004; Tomari et al. 2004b). We examined how miRLC and miRISC that are formed in the miRNA pathway migrate on the gel systems. 32P-labeled pre-let-7 was added into the S2 cell lysate and incubated for 30 min in the presence of EDTA, which should inhibit DCR-1 (Saito et al. 2005). During this time period, we observed that a particular complex, indicated by an asterisk in Figure 4A, accumulated. However, upon DCR-1 activation by the addition of Mg2+ (Saito et al. 2005), the RNP band gradually disappeared, but another complex, which shifted slightly up the gel, appeared (shown with a double asterisk in Fig. 4A). Examination of RNAs contained in both complexes on native acrylamide gels revealed that the lower (*) and upper (**) complexes hold predominantly pre-miRNA and single-stranded mature miRNA, respectively (Fig. 4B). Considering these data, we determined the lower and upper complexes as miRLC and miRISC, respectively.
FIGURE 4.
Visualization of miRLC and miRISC on native agarose gels. (A) pre-let-7 was labeled with 32P and incubated in S2 cell lysates in the presence of EDTA for 3 min and 30 min. After 30 min of incubation, Mg2+ was added to the reaction mixture to induce DCR-1 activity. Further incubation was carried out as indicated. The resultant samples at each time point were loaded onto an agarose gel, and complexes containing either pre-let-7 or mature let-7 were visualized after running the gel. Simultaneously, siRNA duplex labeled with 32P was incubated in the S2 lysates in the presence of EDTA or Mg2+. After 30 min of incubation, both the lysates were loaded on the same gel system. The input lanes on the far left and right sides show pre-miRNA and siRNA duplexes by themselves, respectively. The bands with an asterisk and two asterisks correspond to miRLC and miRISC, respectively. The bands corresponding to siRLC and siRISC are as indicated on the gel. The band shown with a black triangle is an unknown product, which did not appear in previous equivalent experiments (Miyoshi et al. 2005) but appeared in this study. (B) The bands indicated as miRLC and miRISC in A were isolated from the gel, and RNAs isolated were subjected to a native acrylamide gel. The miRLC contained pre-let-7, whereas miRISC contained mostly single-stranded let-7. (Input lane) Contains pre-let-7 as the starting material. Pre-let-7 incubated in S2 lysates was also subjected to the same gel (S2 lysate + pre-let-7), which shows the migration patterns of pre-let-7, let-7/let-7*, and mature let-7.
On the gel system, we noticed that siRLC (Tomari et al. 2004b) and siRISC migrated much slower than miRLC and miRISC, respectively (Fig. 4A), indicating that the components in siRLC and miRLC, as well as siRISC and miRISC, must be distinct from each other.
The requirement for various domains of AGO1 in miRNA, DCR-1, and GW182 binding
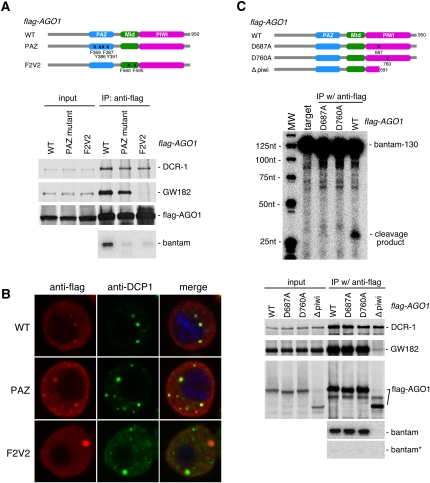
Argonautes are characterized by two domains, PAZ and PIWI (Farazi et al. 2008; Hutvagner and Simard 2008). The PAZ domain has the ability to bind to single-stranded RNA, whereas the PIWI domain endows Slicer activity to Argonautes by folding a three-dimensional structure similar to that of RNase H (Song et al. 2003, 2004; Ma et al. 2005). Another functional domain in Argonautes, the Mid domain, is located between the PAZ and PIWI domains, and is known to function as a 5′-phosphate binding module (Ma et al. 2005; Parker et al. 2005; Rivas et al. 2005). When two conserved phenylalanines in the Mid domain of AGO1, F560 and F595, were mutated to valines (F2V2), its binding to miRNAs and GW182 was abolished, suggesting that the Mid domain is important for AGO1 effector activity (Eulalio et al. 2008). We also produced the F2V2 mutant and found that it could associate with DCR-1 just as well as wild-type (WT) AGO1 (Fig. 5A); however, the mutant was not loaded with mature miRNA (Fig. 5A; Supplemental Fig. 2A), nor could it associate with GW182 (Fig. 5A), as shown previously. The F2V2 mutant does not accumulate in P-bodies but instead formed larger aggregations in the cytoplasm (Fig. 5B; Supplemental Fig. 2B), which correlates well with the fact that the mutant lacks the GW182-binding activity. In contrast, the PAZ mutant, in which four amino acids (F369, Y386, F387, and Y391) that are supposedly necessary for association with small RNAs (Song et al. 2003) were altered to alanines, associated with DCR-1 and GW182 to a similar extent as the WT, although the PAZ mutant was not loaded with mature miRNA (Fig. 5A; Supplemental Fig. 2A). These data suggest that miRNA loading is not a prerequisite for association with GW182 and P-body localization.
FIGURE 5.
The requirement for various domains of AGO1 in binding with miRNA, DCR-1, and GW182. (A) Two mutants of AGO1, the PAZ and F2V2 mutants, were produced by mutagenesis and employed for binding assays with miRNA, DCR-1 and GW182. Proteins associated with Flag-AGO1-WT and Flag-AGO1-PAZ and Flag-AGO1-F2V2 mutants were blotted with anti-DCR-1, anti-GW182, and anti-Flag antibodies. All interacted with DCR-1, but the F2V2 mutant failed to associate with GW182, unlike the wild type (WT) and the PAZ mutant. Northern blotting revealed that neither of the mutants associates with mature bantam. (B) The F2V2 mutant (red), which lacks binding capacity to GW182, fails to accumulate in P-bodies, whereas the PAZ mutant (red) co-localizes with DCP1 (green), a marker for P-bodies. (C) The D-D-H motif in the PIWI domain was mutated to A-D-H or D-A-H. Both mutants were expressed in S2 cells in a Flag-tagged form. Both mutants interacted with DCR-1 and GW182 and also mature single-stranded miRNA. In vitro target RNA cleavage assays show that both mutants lack Slicer activity, indicating that Slicer activity of AGO1 is dispensable for miRNA loading. The Δpiwi mutant interacts with DCR-1 but not with GW182, indicating that the domains required for binding with DCR-1 and GW182 are different.
The D-D-H triad necessary for exhibiting Slicer activity (Rivas et al. 2005) was subsequently mutated to either A-D-H (D687A) or D-A-H (D760A) in AGO1 to examine if such mutations could affect interactions with DCR-1 and GW182, as well as miRNA loading. That both the mutations abolished Slicer activity was confirmed by target RNA cleavage assays (Fig. 5C). It was revealed that both mutants were loaded with mature bantam (but not with bantam*) and interacted with DCR-1 and GW182 as did the WT (Fig. 5C). We further examined miRNA loading onto the AGO1 mutants, which revealed that even miRNA with a fully paired seed sequence (“seed” sequence corresponds to the second to seventh nucleotides of mature miRNAs from the 5′-ends) (Bushati and Cohen 2007), such as miR-277, was loaded onto the Slicer-deficient AGO1 mutants as efficiently as onto WT AGO1 (Supplemental Fig. 3A). These results indicate that Slicer activity is not necessary, in most cases, for efficient miRNA loading in vivo. The Δpiwi mutant, where the PIWI domain of AGO1 was deleted, did not interact with GW182 (Fig. 5C), as reported previously (Behm-Ansmant et al. 2006). This mutant interacted with DCR-1 but was not loaded with miRNA (Fig. 5C). It seems that the PAZ and Mid domains, which are thought to be necessary for small RNA association (Farazi et al. 2008), are not sufficient to receive mature miRNA upon processing by DCR-1. All these data indicate that DCR-1 and GW182 require different domains of AGO1 for their association and that active miRISC formation requires not only both the functional PAZ and MID domains but also the existence of the Piwi domain even without the slicing activity.
DISCUSSION
In this study, we determined that there are two distinct types of AGO1-containing RNP complexes in Drosophila S2 cells. One such complex, immunoprecipitated with anti-LOQS, contained DCR-1, but no GW182 (Fig. 2A). This complex contained little or no endogenous, mature miRNA and showed no target RNA cleavage activity in an endogenous miRNA-dependent manner (Fig. 2B,C). Pre-miRNA and miRNA/miRNA* duplexes were detected in the complex, but at negligible levels (Fig. 2B), suggesting that the majority of AGO1 in the complex are “empty” and thus ready to be loaded with mature miRNA. In fact, mature miRNA processed by the complex from the precursor was able to be loaded onto AGO1 that was originally contained in the complex (Fig. 3B). Earlier studies showed that the Ago2-Dicer-TRBP complex isolated from mammalian cells is able to process pre-miRNAs to mature miRNAs and the resultant miRNAs are loaded onto Ago2, the homolog of Drosophila AGO1, residing in the complex (Gregory et al. 2005; Maniataki and Mourelatos 2005). The Ago2-Dicer-TRBP complex was referred to as miRLC (Maniataki and Mourelatos 2005). Thus, we consider the AGO1-DCR-1-LOQS complex as miRLC in Drosophila.
In the other AGO1-containing complex, GW182 was detected, but DCR-1 and LOQS were absent (Fig. 2A). Mature bantam existed abundantly in the complex, but it excluded pre-bantam and bantam* (Fig. 2B). The complex showed strong activity in cleaving bantam target (Fig. 2C), although no pre-miRNA processing activity was observed (Fig. 3A). This correlated well with the fact that the AGO1-GW182 complex lacks DCR-1. Thus, we consider this particular complex as miRISC in Drosophila. It should be noted that AGO1 and GW182 might only be the core components of miRISC, and other unknown proteins might also be involved. The determination of such components is currently under investigation in our laboratory.
siRLC and siRISC were visualized in the native gel systems by incubating 32P-labeled siRNA duplex with Drosophila cell lysates (Pham et al. 2004; Tomari et al. 2004b). In this study, we visualized miRLC and miRISC on native agarose gels by incubating 32P-labeled pre-miRNA with S2 lysates. It was found that both miRLC and miRISC migrated differently from siRLC and siRISC, respectively (Fig. 4A). The migration distances between miRLC and siRLC were obvious, but not large. However, the situation observed for siRISC and miRISC clearly differs. siRISC is known to be associated with ribosomes (Pham et al. 2004); thus, it is massive and thus hardly runs on the native gels, even under conditions in which the obvious target RNAs were absent (Fig. 4A). However, miRISC migrated much faster, suggesting that miRISC does not seem to associate with polysomes, even under the same conditions. Although the Argonautes involved in miRISC and siRISC, AGO1 and AGO2, are similar in size and sequence, their behavior in terms of protein–protein interaction should be very different from one another. It was previously reported that miR-2 induces dense complexes, heavier than the 80S complex, termed pseudo-polysomes, in Drosophila embryo lysates (Thermann and Hentze 2007). Pseudo-polysomes were formed in the environment to which mRNA targets were exogenously added, hence the large apparent size. miRISC observed in our study was rather small, which may be due to the unavailability or accessibility of the targets for let-7 in S2 lysates utilized in the assays.
Upon miRNA loading, AGO1 was detected in a fraction separate from the DCR-1-containing fraction (Fig. 3C), indicating that the loading of mature miRNA frees AGO1 from the miRLC, as has been reported in mammalian systems (Maniataki et al. 2005). However, the amounts of DCR-1 associated with WT AGO1 and the PAZ mutant were approximately equal (Fig. 5A), indicating that DCR-1 can be displaced from AGO1 independently of miRNA loading. It is possible that the substitution of amino acids may have caused considerable conformational changes in AGO1, and thus DCR-1 was then easily dissociated; however, it also suggests that miRNA loading onto AGO1 might only partially contribute to such displacement and other systems might also operate in vivo.
The PAZ mutants of mammalian Ago2 (Ago2-PAZ9 and Ago2-PAZ10) did not localize to P-bodies (Liu et al. 2005). In contrast, the PAZ mutant of Drosophila AGO1 was associated with GW182 and was localized to P-bodies as in the case of the WT (Fig. 5A), meaning that, at least in Drosophila, GW182 does not discriminate between AGO1 with and without miRNA. Thus, it is speculated that even WT AGO1 could be localized to the P-bodies if it encounters GW182 prior to miRNA loading. However, P-bodies are not the place for the dead-ending of the AGO1 function. It is generally thought that once target mRNAs are degraded in P-bodies, AGO1 is released from P-bodies to the cytosol for subsequent silencing cycles. This step might displace GW182 from AGO1, although solid evidence for this scenario is still missing. Such displacement of GW182, upon release from P-bodies, might happen to AGO1 that was somehow accidentally localized to P-bodies without being associated with miRNA, which provides another chance for AGO1 to encounter DCR-1 and become activated.
The PIWI mutants of AGO1, D687A and D706A, showed no Slicer activity, but mature miRNAs were loaded onto the mutants as efficiently as onto WT AGO1 (Fig. 5C; Supplemental Fig. 3A). This means that the miRNA unwinding step occurs, in most cases, in a Slicer-independent manner. It was previously shown that in the Drosophila RNAi system, the Slicer activity of AGO2 largely functions in the siRNA unwinding process (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005). The miRNA/miRNA* duplex tends to contain a bulge in the middle of the molecule, where the slicing of the passenger strand would occur; therefore, unwinding should not be mediated by Slicer. In fact, Matranga et al. (2005) have previously shown that strand rejection of miRNA* is not accompanied by miRNA* strand cleavage by hAgo. In contrast, in mammals, Slicer activity of Ago2 (the homolog of Drosophila AGO1) was shown to be involved in let-7 maturation (Diederichs and Haber 2007). It seems that the requirements of Argonaute-Slicer activity in miRNA maturation in mammals and flies are different from each other. ATP depletion from S2 cell lysates caused less-efficient miRNA maturation, although such treatment did not affect the siRNA unwinding activity (MC Siomi, unpubl.). Thus, we can speculate that there are yet-to-be identified factor(s) that require ATP hydrolysis in the miRNA/miRNA* unwinding process, as opposed to the mammalian systems, where ATP is unnecessary for miRNA maturation (Gregory et al. 2005).
MATERIALS AND METHODS
Plasmid constructs
The Flag-AGO1 (WT) expression plasmid was generated by inserting the coding region of AGO1 from pGEX-AGO1-PB (Miyoshi et al. 2005) into the pRH3xflag vector (Saito et al. 2005). The expression plasmids for AGO1 mutants, Flag-AGO1-PAZ (F369A, Y386A, F387A, Y391A), Flag-AGO1-F2V2 (F560V, F595V), Flag-AGO1-D687A, and Flag-AGO1-D760A were produced by site-directed mutagenesis using the QuikChange Kit (Stratagene). The expression plasmid for Flag-AGO1-Δpiwi (containing AGO1 peptide from the first methionine to P691; D687 is altered to alanine) was obtained as a by-product during the construction of the Flag-AGO1-D687A plasmid.
Immunoprecipitation, silver staining, and Western blotting
Monoclonal antibodies against GW182 were produced against full-length GW182 (CG31992) tagged with GST. Anti-AGO1, anti-Aub, and anti-Piwi antibodies were described previously (Miyoshi et al. 2005; Saito et al. 2006; Nishida et al. 2007). Monoclonal antibodies against LOQS were produced against full-length LOQS (CG6866) tagged with GST. Monoclonal antibodies against DCR-1 were produced against the N terminus of DCR-1 (750 amino acids). Monoclonal antibodies against DCR-2 were produced against the C terminus of DCR-2 (680 amino acids) tagged with GST. Immunoprecipitation from S2 cell lysates or ovary lysates were performed as described previously (Miyoshi et al. 2005). After immunoprecipitation, the beads were washed extensively with a buffer containing 30 mM HEPES (pH 7.4), 150 mM potassium acetate, 5 mM magnesium acetate, 5 mM DTT, 0.1% NP-40, 2 μg/mL pepstatin, 2 μg/mL leupeptin, and 0.5% aprotinin. The immunoprecipitated proteins were eluted with SDS sample buffer without DTT. After elution, DTT was added to the eluates to 100 mM. The eluates were then boiled and loaded onto SDS-acrylamide gels. After electrophoresis, protein bands were visualized using Silver Quest (Invitrogen). Western blotting was performed as described previously (Miyoshi et al. 2005).
Target RNA cleavage assay
The reactions were performed as previously described (Miyoshi et al. 2008). To make a let-7 target RNA (L7Pv in Fig. 3B), pBS-let-7-L7Pv was digested with PvuII, and in vitro transcription was performed using MEGAscript T3 (Ambion). To make a shorter let-7 target RNA (let-7-130 in Fig. 3C), a PCR fragment was amplified with T7 and T3 primers from pBS-let-7-130 and in vitro transcription was performed using MEGAscript T7 (Ambion). The pBS-let-7-130 and pBS-let-7-L7Pv plasmids were constructed as follows. pBS-let-7-130, a KpnI-EcoRI fragment containing a sequence completely complementary to mature let-7 was produced by annealing a set of DNA oligonucleotides (5′-CACTATACAACCTACTACCTCAG-3′ and 5′-AATTCTGAGGTAGTAGGTTGTATAGTGGTAC-3′) and inserting the resultant DNA fragment into pBluescript digested with KpnI and EcoRI. pBS-let-7-L7Pv; a KpnI-EcoRI fragment was produced by annealing two DNA oligonucleotides (5′-CTGAGGTAGTAGGTTGTATAGTG-3′ and 5′-AATTCACTATACAACCTACTACCTCAGGTAC-3′) and inserted into pBluescript digested with KpnI and EcoRI. To make miR-bantam target RNA (bantam-130), a PCR fragment was amplified with T7 and T3 primers from pBS-miR-ban-target (Miyoshi et al. 2005), and in vitro transcription was performed using MEGAscript T7. All target RNAs were 32P-labeled at the 5′-G cap by guanylyltransferase (Ambion) as previously described (Miyoshi et al. 2005).
Northern blot analysis
Northern blot analysis was performed as described previously (Miyoshi et al. 2005). The probe used for detecting miR-bantam* was 5′-GTCAAACCAAATCGAAAACC-3′.
RNAi
RNAi in S2 cells was performed as described previously (Saito et al. 2005). dsRNA utilized for GW182 depletion corresponded to nucleotides 1–698 of the GW182 coding sequence. The dsRNAs used for DCR-1 and EGFP were described previously (Saito et al. 2005).
Pre-miRNA processing assay
The pre-miRNA processing assays were performed as described previously (Saito et al. 2005). Pre-let7 (UGAGGUAGUAGGUUGUAUAGUAGUAAUUACACAUCAUACUAUACAAUGUGCUAGCUUUCU; Dharmacon) was labeled with 32P at the 5′-end and self-annealed.
Immunofluorescence
Immunofluorescence was performed by fixing S2 cells with 2% formaldehyde for 15 min. Cells were permeabilized using 0.1% Triton X-100. To visualize Flag-tagged AGO1 proteins, cells were stained with anti-Flag M2 antibody (Sigma). Alexa-546 anti-mouse IgG was used as a secondary antibody. After extensive washing, the cells were further stained with anti-DCP1 antibody coupled to fluorescein using a Fluorescein Labeling kit-NH2 (Dojindo). Monoclonal antibodies against DCP1 were produced against full-length DCP1 (CG11183) tagged with GST. Cells were also mounted with SlowFade Gold Antifade Reagents with DAPI (Invitrogen). All images were collected using a Zeiss LSM510 laser scanning microscope.
Native gel electrophoresis
Native gel electrophoresis was performed as described previously (Miyoshi et al. 2005). 32P-labeled pre-let-7 or luc siRNA duplex was first incubated in S2 lysates in the presence of 5 mM EDTA at 26°C, and then 30 mM magnesium acetate was added to the mixture. After incubation, samples were adjusted to 6% (w/v) glycerol and resolved by submarine native 1.5% agarose gel electrophoresis. The RNP bands were visualized by exposure to an imaging plate of a BAS 2500. To purify RNAs from the RNP complexes, corresponding bands were excised from the gels and RNAs were purified using SuprecEZ (Takara). Purified RNAs were solubilized in loading buffer (5 mM HEPES-KOH at pH 7.4, 5 mM EDTA, 5% glycerol, and 0.5% SDS) and resolved by 15% native acrylamide gel electrophoresis. As a control, the reaction mixture was incubated in the presence of magnesium acetate. After reaction, the sample was mixed with the same volume of the stop solution (5 mM HEPES-KOH at pH 7.4, 5 mM EDTA, 5% glycerol, 0.5% SDS, and 0.5 mg/mL proteinase K) and incubated for 10 min 37°C before loading.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank K. Saito, K.M. Nishida, T. Sunohara, and A. Ishizuka for technical assistance, and other members of the Siomi laboratory for discussions and comments on the manuscript. This work was supported by MEXT grants to H.S., the New Energy and Industrial Technology Development Organization (NEDO) grants to M.C.S., and JSPS Research Fellowships for Young Scientists to K.M. M.C.S. is supported by CREST from JST. H.S. is a member of the Genome Network Project (MEXT). M.C.S. is Associate Professor of Global COE for Human Metabolomics Systems Biology by MEXT.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1541209.
REFERENCES
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. MicroRNA function. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNA. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for Argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Séraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EK. The role of GW/P-bodies in RNA processing and silencing. J Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembry of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004a;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004b;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegal J, Yates JR, III, Parker R, Hannon GJ. A role for the P-body component Gw182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Paters L, Chan PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel Argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Uejima H, Okada TN, Siomi H, Siomi MC. In vitro RNA cleavage assay for Argonaute-family proteins. Methods Mol Biol. 2008;442:29–43. doi: 10.1007/978-1-59745-191-8_3. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine–piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Argaon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1 Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MD, Najand N, Chaker S, Pare JM, Haskins J, Hughes SC, Hobman TC, Locke J, Simmonds AJ. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA-binding motif in RNAi effector complexes. Nat Struct Mol Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L. Slicer and the Argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004a;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004b;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;23:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]