Abstract
The U5 snRNA loop 1 aligns the 5′ and 3′ exons for ligation during the second step of pre-mRNA splicing. U5 is intimately associated with Prp8, which mediates pre-mRNA repositioning within the catalytic core of the spliceosome and interacts directly with U5 loop 1. The genome-wide effect of three U5 loop 1 mutants has been assessed by microarray analysis. These mutants exhibited impaired and improved splicing of subsets of pre-mRNAs compared to wild-type U5. Analysis of pre-mRNAs that accumulate revealed a change in base prevalence at specific positions near the splice sites. Analysis of processed pre-mRNAs exhibiting mRNA accumulation revealed a bias in base prevalence at one position within the 5′ exon. While U5 loop 1 can interact with some of these positions the base bias is not directly related to sequence changes in loop 1. All positions that display a bias in base prevalence are at or next to positions known to interact with Prp8. Analysis of Prp8 in the presence of the three U5 loop 1 mutants revealed that the most severe mutant displayed reduced Prp8 stability. Depletion of U5 snRNA in vivo also resulted in reduced Prp8 stability. Our data suggest that certain mutations in U5 loop 1 perturb the stability of Prp8 and may affect interactions of Prp8 with a subset of pre-mRNAs influencing their splicing. Therefore, the integrity of U5 is important for the stability of Prp8 during splicing and provides one possible explanation for why U5 loop 1 and Prp8 are so highly conserved.
Keywords: pre-mRNA splicing, U5 snRNA, Prp8, yeast, microarray
INTRODUCTION
Pre-messenger RNA (pre-mRNA) splicing, the removal of introns from pre-mRNA to form mRNA, involves two catalytic steps. In the first step the 2′OH group of the intron branch site reacts with the phosphodiester bond at the 5′ splice site releasing the 5′ exon and producing an intron lariat-3′ exon intermediate. The 3′OH of the released 5′ exon then reacts with the phosphodiester bond at the 3′ splice site, thus producing mRNA. These two transesterification reactions are mediated by the spliceosome. The spliceosome is a large protein–RNA complex comprising numerous proteins and five small nuclear RNAs (snRNAs); U1, U2, U4, U5, and U6. These snRNAs interact with each other, the pre-mRNA and the protein components of the spliceosome to catalyze the splicing process (Nilsen 1998).
Each snRNA is associated with a set of proteins in RNA–protein complexes termed small nuclear ribonucleoprotein particles (snRNPs). During splicing the snRNAs have distinct functions. Initially U1 snRNA contacts the pre-mRNA by base pairing to the 5′ splice site. The U2 snRNA base pairs to the branch site sequence and the spliceosome is completed by the binding of U4, U5, and U6 as a preformed tri-snRNP complex in which U4 and U6 snRNAs are extensively base paired through complementary sequences (Moore et al. 1993). Several RNA rearrangements then occur in the spliceosome. Briefly, these include removal of the U1 snRNA from the 5′ splice site and the disruption of base pairing between the U4 and U6 snRNAs. This allows U6 to base pair with conserved intron sequences at the 5′ splice site and with U2, to form part of the active site of the spliceosome. The U5 snRNA interacts with the 5′ exon near the 5′ splice site. After the first catalytic step of splicing the U5 snRNA continues to interact with the 5′ exon intermediate and forms a new interaction with the 3′ exon. These interactions, probably stabilized by Prp8, tether the 5′ exon intermediate and align the exons prior to the second catalytic step of splicing (Newman and Norman 1992; Sontheimer and Steitz 1993; Newman et al. 1995; Teigelkamp et al. 1995; O'Keefe et al. 1996; O'Keefe and Newman 1998).
The U5 snRNA contains a stem–loop that has a 9-nucleotide (nt) conserved sequence (GCCUUUYAY) (Patterson and Guthrie 1987; Guthrie and Patterson 1988) that forms part of a loop structure within stem–loop 1 in the yeast Saccharomyces cerevisiae (Frank et al. 1994). It is this conserved loop 1 sequence that interacts with the 5′ exon and 3′ exon adjacent to the 5′ and 3′ splice sites (Newman and Norman 1992; Wyatt et al. 1992; Sontheimer and Steitz 1993; Newman et al. 1995; O'Keefe et al. 1996; O'Keefe and Newman 1998; Alvi et al. 2001; McConnell and Steitz 2001). U5 loop 1 is not essential for the first catalytic step of splicing in vitro (O'Keefe et al. 1996); however, it is essential for the second step of splicing as it is believed that these U5 loop 1–exon interactions allow U5 to align the exons prior to the second catalytic step (Newman and Norman 1991, 1992; Newman et al. 1995; O'Keefe and Newman 1998). Mutagenesis of loop 1 indicated that altering the size of loop 1 resulted in a second step block in splicing and that changes to the sequence of loop 1 also influenced splicing (O'Keefe and Newman 1998; O'Keefe 2002).
The loop 1 region of U5 has also been implicated in interaction with Prp8 (for review, see Grainger and Beggs 2005). Prp8 is a large protein involved in both catalytic steps of splicing. It has no obvious homology with other proteins, yet it is highly conserved and is positioned within the catalytic core of the spliceosome. Prp8 is a component of the U5 snRNP and U4/U5/U6 tri-snRNP, in addition to crosslinking to a large proportion of U5 within loop 1 and both internal loops (IL) (Dix et al. 1998). Although Prp8 was only observed to crosslink to U5 loop 1 at position U97 it is likely that it makes direct contact with the entire stem–loop 1 structure. Prp8 is also known to crosslink to U6 (Vidal et al. 1999) and at, or near, all three (5′ splice site, branch site, and 3′ splice site) splicing-dependent features in the pre-mRNA (Wyatt et al. 1992; Teigelkamp et al. 1995; Umen and Guthrie 1995; Chiara et al. 1996; Reyes et al. 1996; Chiara et al. 1997; Reyes et al. 1999; Siatecka et al. 1999; Maroney et al. 2000; Ismaili et al. 2001; McPheeters and Muhlenkamp 2003). Mapping has revealed that interactions between Prp8 and the snRNA/pre-mRNA sequences involve the C-terminal and central regions of Prp8 (Reyes et al. 1999; Turner et al. 2006). It is believed that Prp8 acts to stabilize the RNA interactions at the catalytic core of the spliceosome or that the spliceosome is not a true ribozyme and requires Prp8 in the catalytic steps of splicing (Collins and Guthrie 2000; Abelson 2008).
A number of mutants in U5 loop 1 are temperature-sensitive and analysis of six intron-containing genes revealed that each mutant displayed differential splicing of these genes (O'Keefe 2002). Here we describe how three U5 loop 1 mutants influence pre-mRNA splicing in yeast by analysis of all intron-containing genes in S. cerevisiae utilizing a splicing microarray. Sequence analysis of pre-mRNAs exhibiting pre-mRNA accumulation in the mutant strains revealed a base bias at certain positions near the 5′ and 3′ exon–intron boundaries compared with other spliced pre-mRNAs. Pre-mRNAs that exhibited mRNA accumulation in the mutant strains displayed a base bias at only one position within the 5′ exon. Only two of these positions are known to interact with U5 loop 1, but the base bias does not relate to sequence changes made in U5 loop 1. Therefore, it appears that the mutations made in U5 loop 1 are not directly influencing the splicing of pre-mRNA and may influence another factor, such as Prp8. In line with this hypothesis we describe a reduced stability of Prp8 in one U5 mutant grown at the restrictive temperature, while the levels of the mutant U5 loop 1 snRNAs and the U5 snRNP protein Snu114 remained stable. In addition, in vivo depletion of the U5 snRNA also caused reduced Prp8 stability. Therefore, the integrity of U5 loop 1 and U5 as a whole is required for the stability of Prp8. We propose that the reduced stability of Prp8 influences the splicing efficiency of certain pre-mRNAs that have common bases near the important pre-mRNA sequences required for splicing.
RESULTS
Genome-wide analysis of pre-mRNA splicing with U5 snRNA loop 1 mutations
By randomization selection of the U5 snRNA loop 1 sequence, three mutations were previously isolated that displayed temperature-sensitive growth defects at 37°C and impaired splicing of certain pre-mRNAs (O'Keefe 2002). Compared to the wild-type loop 1, two of the mutants, U5-1 and U5-2, possess loop 1 structures of identical size, but not sequence, whereas the third, U5-3, has a single nucleotide deletion within loop 1 coupled with sequence alteration (Fig. 1). In the previous study the splicing of only six intron-containing genes (ACT1, ARP2, COF1, TUB1, SNR17A, and SNR17B) was assessed at 12 h following the switch to restrictive temperature (O'Keefe 2002). While this study indicated that there was an inhibition of splicing of only certain pre-mRNAs in the presence of the three U5 loop 1 mutants, the analysis was far from complete. We have now assessed the splicing of the complete set of 294 known or putative yeast introns to determine precisely how each U5 loop 1 mutant influences splicing during one cell division cycle. In examining splicing following one cell division cycle we hoped to identify the pre-mRNAs that were most influenced by mutations in U5 loop 1. Total RNA was isolated from strains containing either wild-type U5 snRNA or the mutants U5-1, U5-2, or U5-3 following growth for one cell division at restrictive temperature. As each strain has a different growth rate at the restrictive temperature the cell divison time for each strain varied. RNA was then used as a template for the synthesis of fluorescently labeled cDNA using an oligo dT primer. A genome-wide assessment of pre-mRNA splicing was performed by noncompetitive hybridization of the labeled cDNA to microarrays containing probes across the 5′ splice site, within the intron, across the exon boundaries of the mature mRNA, and within the 3′ exon of each intron-containing gene. Microarray data were standardized to account for different RNA levels, by dividing the background-subtracted signal from each probe by that of the corresponding 3′ exon probe, generating an intron index, 5′ splice site index and mature splice index. All indexes are expressed as the ratio of the U5 mutant index compared to the wild-type U5. The 5′ splice site index and intron index correlated well; therefore only analysis of the intron index is presented. The results of these and other pairwise comparisons of different sets of genes can be found at http://www.biology.ed.ac.uk/research/groups/jbeggs/microarray/U5/.
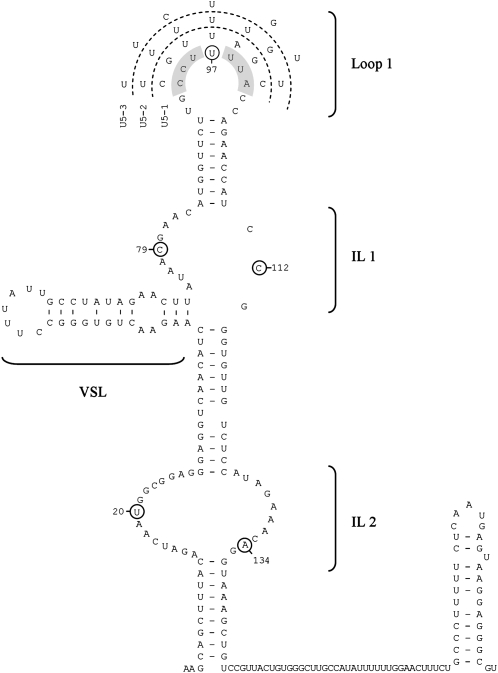
FIGURE 1.
Structure and mutations of U5 snRNA loop 1. The mutated nucleotides (highlighted) have been mapped onto the predicted secondary structure (Frank et al. 1994) of U5 snRNA and the alternative sequence of the three loop 1 mutants is described. The loop 1 structure, along with the internal loops (IL) and variable stem–loop (VSL) are indicated. Circled nucleotides indicate sites of crosslinking to Prp8 (Dix et al. 1998).
Confirmation of microarray analysis by in vitro U5 depletion/reconstitution and semiquantitative reverse transcription real-time PCR
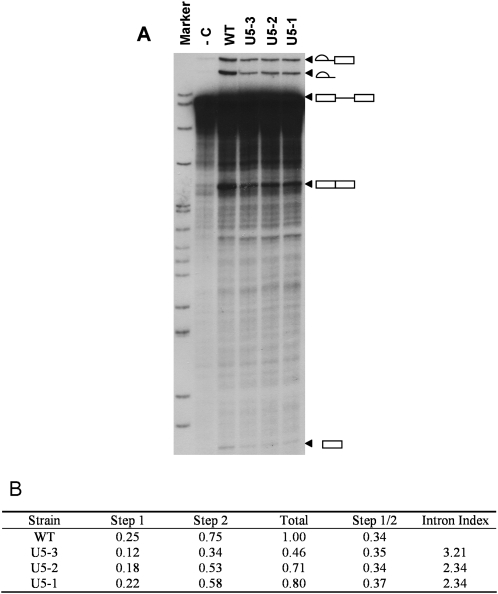
The intron index indicated that splicing of ACT1 pre-mRNA was defective in the three U5 loop 1 mutant strains compared to the wild-type. The ACT1 pre-mRNA is widely used to analyze pre-mRNA splicing in vitro, therefore we chose to analyze the splicing of ACT1 in the presence of the U5 loop 1 mutants in vitro. The U5 snRNA can be efficiently depleted from extracts produced from a yeast strain carrying a modified U5 snRNA and reconstituted with in vitro transcribed U5 snRNA (O'Keefe et al. 1996). The splicing efficiency of ACT1 pre-mRNA was assayed in U5 depleted whole cell extract reconstituted with wild-type U5, U5-1, U5-2, or U5-3 (Fig. 2A). The three U5 mutants do not exhibit a specific inhibition of the first or second catalytic step of splicing, but display a reduced ability to splice the ACT1 pre-mRNA. Quantitation of the splicing defects revealed that mutant U5-3 exhibited the greatest perturbation of splicing, followed by U5-2 and then U5-1 (Fig. 2B). This pattern corresponds with the intron index data for ACT1 in the microarray analysis, thus validating these microarray data (Fig. 2B).
FIGURE 2.
Reconstitution of ACT1 splicing activity in U5 depleted extracts. (A) U5 snRNA was depleted from whole cell yeast extract and reconstituted with either water (-C) or in vitro transcribed WT U5, or U5 mutants U5-1, U5-2, or U5-3. The splicing intermediates and products (indicated diagrammatically on the right) were resolved alongside a labeled pBR322 DNA-MspI digest marker. (B) Quantification of the splicing activity of the U5 mutants was obtained by phosphorimaging. Data were normalized for the amount of pre-mRNA per lane after subtraction of the background (-C depletion lane). Total splicing activity for WT U5 has been arbitrarily set to 1. The step 1 total is the combined values for the lariat-3′ exon intermediate and the released 5′ exon. The step 2 total is the combined values of the mature RNA and the released lariat. By dividing the step 1 total by that for step 2 it is possible to discern if there is a block in either step of splicing. Standard error of these data does not exceed 0.05. Also tabulated is the intron index data from the microarray for ACT1.
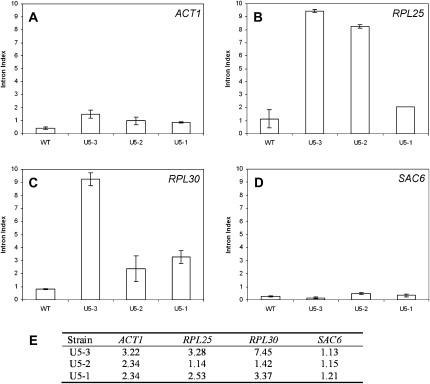
Semiquantitative reverse transcription real-time-PCR (RT-PCR) was also performed to confirm the microarray data with a selected number of pre-mRNAs. Three pre-mRNAs (ACT1, RPL25, and RPL30) that displayed significant intron accumulation in the microarray data were selected for analysis by RT-PCR, and one that displayed no intron accumulation (SAC6) was used as a control. The RT-PCR data support the microarray data (Fig. 3). RT-PCR data for ACT1 exhibit the same pattern as observed with both the microarray data and the depletion/reconstitution reactions, with mutant U5-3 displaying the greatest intron accumulation, followed by U5-2 and then U5-1 (Fig. 3 cf. A and E). The microarray data and the RT-PCR data for RPL30 corroborate well, with U5-3 mutant displaying the greatest intron accumulation, followed by U5-1 and then U5-2 (Fig. 3 cf. C and E). The microarray data and the RT-PCR data for RPL25 (Fig. 3 cf. B and E), however, are not entirely consistent. The intron accumulation of RPL25 for both the U5-3 and U5-1 mutants, when assessed by RT-PCR, follow the same pattern as measured by microarray, however, mutant U5-2 displays higher intron accumulation when measured by RT-PCR. The pre-mRNA SAC6 exhibits no intron accumulation in either the microarray data or the RT- PCR data (Fig. 3 cf. D and E). Although these RT-PCR data only assess the intron accumulation of certain pre-mRNAs in the mutant strains they support the microarray data well. In addition, as many pre-mRNAs that display intron accumulation are involved in ribosome biogenesis, we also performed polysome analysis. All three U5 loop 1 mutant strains displayed defects in ribosome assembly consistent with a defect in ribosomal protein production (data not shown).
FIGURE 3.
Intron accumulation measured by semiquantitative real-time PCR analysis of (A) ACT1/YFL039C, (B) RPL25/YOL127W, (C) RPL30/YGL030W, and (D) SAC6/YDR129C. Relative quantities of the introns were assessed by performing semiquantitative RT-PCR on the same RNA samples as used in the microarray experiment. As with the microarray experiment, intron accumulation is expressed relative to the total amount of each transcript in each strain, assessed by designing primers to the 3′ exon. These experiments were performed on biological triplicate samples and the error is shown as standard error from the mean. (E) Intron index data from the microarray for the indicated pre-mRNAs are tabulated.
Accumulated pre-mRNAs resulting from U5 loop 1 mutations display a change in base prevalence at pre-mRNA positions near the 5′ and 3′ splice sites
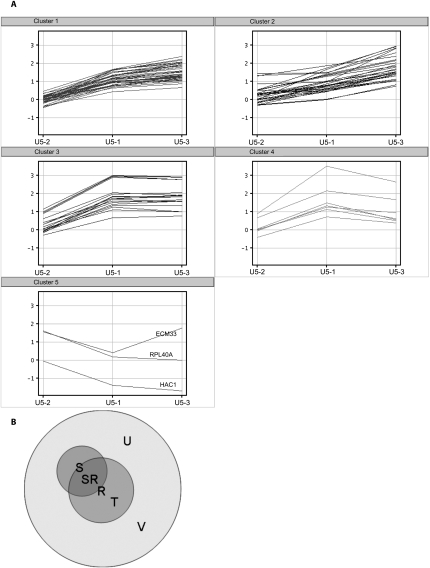
The U5 loop 1 interacts with the end of the 5′ exon before and after the first step of splicing (Newman and Norman 1991, 1992; Wyatt et al. 1992; Sontheimer and Steitz 1993; Newman et al. 1995; O'Keefe and Newman 1998) and in certain experimental situations U5 loop 1 complementarity with the 5′ exon and 3′ exon is important for splicing (Newman and Norman 1991, 1992; Bacikova and Horowitz 2005; Crotti et al. 2007). Therefore, one prediction is that pre-mRNAs that have been less or more efficiently spliced may have common sequences in the 5′ and 3′ exons that relate to the mutations made in U5 loop 1. To address this prediction the sequences at the 5′ and 3′ exon–intron junctions, as well as branch site of the accumulating pre-mRNAs, were analyzed. Microarray data were first grouped into five separate clusters dependent upon intron accumulation in each mutant, using the Pearson centered similarity measure in Genespring 9 (K-means clustering) (Fig. 4). Pre-mRNAs present in each cluster are displayed in Table 1. Statistical filters applied in the clustering analysis removed features that were not twofold more abundant in at least one pairwise comparison of the mutants. By clustering the data in this way it is possible to analyze the sequences around the splice sites and branch site of a cluster of pre-mRNAs that accumulate in a similar manner. To increase the number of pre-mRNAs that are grouped for the statistical analysis, K-means clusters 1 and 2 (dominated by U5-3 data) and clusters 3 and 4 (dominated by U5-1 data) were combined. Due to the low number of pre-mRNAs in cluster 5, statistical analysis of base frequency in this cluster was not possible.
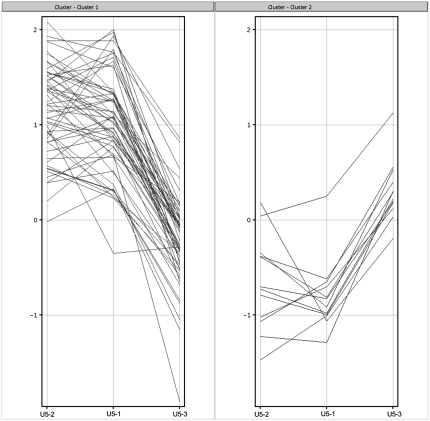
FIGURE 4.
K-means clustering of introns that are twofold more abundant in at least one pairwise comparison of the mutants. (A) This displays the intron accumulation profile of each pre-mRNA (log base 2 of the ratio mutant/wild-type) for each of the three mutants. These clusters were identified in Genespring (Version 9, similarity measure: Pearson centered, over 50 iterations). Although five clusters were identified (other numbers of seed points were tried but were no more informative; data not shown) there are many similarities, showing that generally the mutations affect similar features of the introns. Clusters 1 and 2 are similar in that U5-3 predominates, whilst clusters 3 and 4 are similar in that U5-1 predominates. As cluster 5 contains only two introns (HAC1 contains a different kind of intron, and is present only as a control) no further statistical analysis could be performed on this cluster. (B) Euler diagram showing the relationships of the introns analyzed for each of the clusters. U is the set of all introns on the microarray. R is the set of all ribosomal protein introns. S is all introns selected from the data and present in a K-means cluster. SR is the intersection set of R and S, i.e., ribosomal protein introns selected from the K-means clustering (S∩R). T is the ribosomal protein introns not selected by K-means clustering (R\S). V is the introns not selected by K-means clustering and not ribosomal protein introns [(S∪R)c]. This is a general representation of the sets used; in practice these are different sets for each of the K-means clusters, intron cluster 1 and 2, intron cluster 3 and 4 and mature. So clusters 1 and 2 set S is different from clusters 3 and 4 set S.
TABLE 1.
Clusters of genes that display intron accumulation
As the K-means clusters contain a large number of ribosomal protein transcripts that share sequence similarities with each other, as well as with ribosomal protein transcripts that are not in the clusters, the effect of such shared sequences was controlled by dividing the pre-mRNAs into sets and subsets, depending on whether or not they encode ribosomal proteins (Fig. 4B). Thus, set S represents all the pre-mRNAs in a cluster, set R represents all yeast ribosomal protein pre-mRNAs and subset SR, the overlap of these two, contains ribosomal protein pre-mRNAs in the cluster. For all pre-mRNAs in clusters 1 + 2 or 3 + 4, the bases found around the 5′ splice site, branch site, and 3′ splice site were tabulated and a χ2 test was used to compare the observed base composition with the expected base composition (sequences existing in these regions in the set of nonclustered, nonribosomal protein pre-mRNAs; set V). The probability (P-value) was then calculated that there is no significant variation of the base identity distribution at a specific position in the clustered pre-mRNAs compared to the base distribution at the same position in the set of nonclustered yeast pre-mRNAs (the null hypothesis; i.e., the K-means clustering of the transcripts was random). A low P-value here would indicate a significant difference between the K-means clustered pre-mRNAs (set S) and other nonribosomal protein transcripts (set V). The same analysis was performed to compare K-means clustered ribosomal protein pre-mRNAs (set SR) with nonclustered ribosomal protein pre-mRNAs (set T) as an indication of where a low P-value for bases at a specific position may be a characteristic of ribosomal protein transcripts, which are generally highly expressed and efficiently spliced.
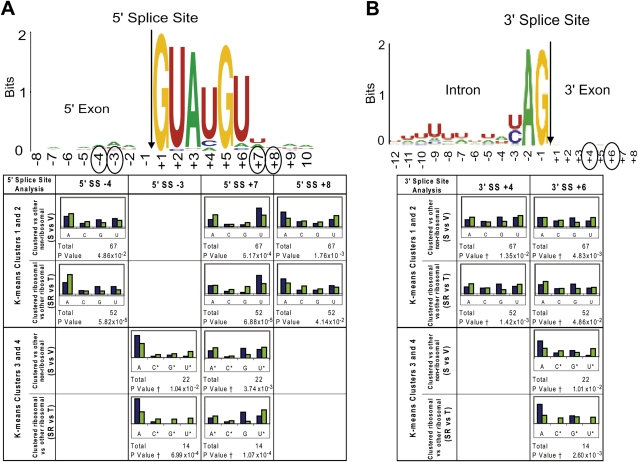
Figure 5 presents the results for positions with a P-value of less than 0.05 (where the null hypothesis can be rejected). In both groups of introns analyzed (clusters 1 + 2 and clusters 3 + 4), the frequency of the different bases at intron position seven (+7) downstream from the 5′ splice site (5′SS +7) and at exon position six (+6) downstream from the 3′ splice site (3′SS +6) varied significantly from the frequencies for nonclustered, nonribosomal protein pre-mRNAs. Notably, the base frequencies at these positions also differed between the clusters, there being a higher occurrence of uridine at 5′SS +7 in the pre-mRNAs in clusters 1 + 2 than in clusters 3 + 4, and a bias toward adenosine at 3′SS +6 in clusters 3 + 4, but not in clusters 1 + 2. At other positions around the 5′ and 3′ splice sites (5′SS −4, 5′SS −3, 5′SS +8, and 3′SS +4) a significant difference was observed only for clusters 1 + 2 or clusters 3 + 4, but not for both. For example, at exon position three (−3) upstream of the 5′ splice site (5′SS −3), only pre-mRNAs in clusters 3 + 4 display a bias toward adenosine. In each case, although the base frequencies in the set of all clustered pre-mRNAs (set S) are clearly influenced by the inclusion of a large proportion of ribosomal protein pre-mRNAs, a comparison of these clustered ribosomal protein pre-mRNAs (set SR) with other (nonclustered) ribosomal protein pre-mRNAs (set T) shows that the base distributions in the clustered pre-mRNAs do not simply reflect sequence features that are typical of all ribosomal protein transcripts. By these criteria, no position within 10 bases of the branch site was found to vary significantly from the expected and also differ from ribosomal protein introns. Interestingly, with the exception of 5′SS −3 and 5′SS −4, positions in the pre-mRNA that displayed variation of the base identity distribution are not known to interact directly with the U5 snRNA loop 1. Thus, either these positions represent novel sites in the pre-mRNA that interact with the U5 snRNA loop 1 or mutations in U5 loop 1 disrupt interactions of another factor at these positions, a likely candidate being Prp8.
FIGURE 5.
Base prevalence found in accumulated pre-mRNA resulting from mutations in U5 loop 1. For reference, the base prevalence found near the 5′ splice site (A) and the 3′ splice site (B) for all intron-containing yeast pre-mRNAs is displayed at the top as sequence logos (Crooks et al. 2004). The occurrence of each base (A, C, G, or U) at each position in the RNA around the 5′ splice site and 3′ splice site was tabulated. A χ2 test was used to compare observed base composition (set S pre-mRNAs; blue bars) with the expected base composition (set V pre-mRNAs; green bars) in that region and P-values were calculated (see Materials and Methods). “*” Indicates bases that were binned and “†” indicates where Yates correction for continuity was applied due to the small number of bases. For comparison, the same analysis was performed on the selected (set RS) versus other (nonselected; set T) ribosomal protein pre-mRNAs, to determine whether any differences seen may be due to features common to all ribosomal protein transcripts. Results are presented for positions where both P-values were less than 0.05.
Accumulated mRNAs resulting from U5 loop 1 mutations display a change in base prevalence in the 5′ exon that is not consistent with base pairing with U5 loop 1
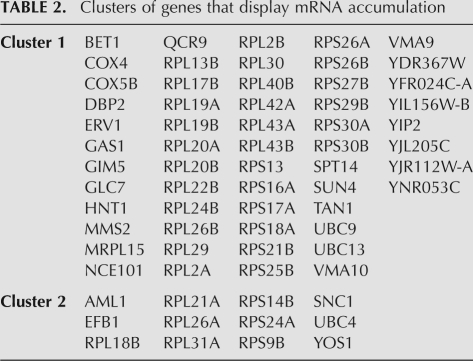
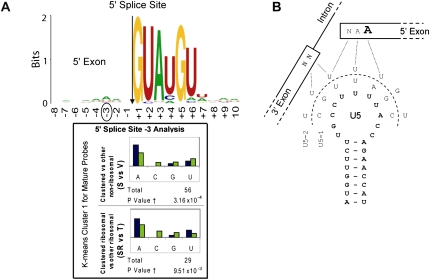
To investigate base frequency in pre-mRNAs that displayed accumulation of mRNA in the U5 mutant strains, a similar analysis was performed using the mature splice index. The data were sorted into two separate clusters (Fig. 6) using the same parameters that were applied to the intron index clustering analysis. Pre-mRNAs present in each cluster are listed in Table 2. Statistical analysis was performed on the sequences of the pre-mRNAs in cluster 1; however, this was not possible for cluster 2 due to its small size. Only one position was significantly different from the consensus sequences; 5′SS −3 displayed an increase in the incidence of adenosine (Fig. 7A) that is significantly greater than the bias for adenosine observed for other intron-containing transcripts in yeast (Long et al. 1997; Lopez and Séraphin 1999). The third base upstream of the 5′ splice site of the pre-mRNA has been shown to interact with positions C95 and U96 of U5 loop 1 (McGrail et al. 2006) and this position in the 5′ exon intermediate with position U98 of U5 loop 1 (Newman and Norman 1991; 1992; Crotti et al. 2007). However, neither of the two U5 mutants that display mRNAs in this group (U5-1 and U5-2) is completely consistent with mutations in these bases that would increase base-pairing interactions (Fig. 7B). Therefore, accumulation of specific mRNAs with these U5 mutants may not be a direct effect of changing the U5 loop 1 sequence, but may be due to disruption of another factor that interacts with the mRNA, possibly Prp8. As shown in Figure 5, there is also a bias toward adenosine at the 5′SS −3 position in the pre-mRNAs in clusters 3 + 4 that show intron accumulation. As these pre-mRNAs accumulate mainly in response to the U5-3 and U5-1 alleles, the pre-mRNA accumulation may result from decreased base-pairing interactions between adenosine at 5′SS −3 with the mutant U5 loops. Alternatively, as some of the same transcripts occur in pre-mRNA clusters 3 + 4 and in mRNA cluster 1, it is possible that these pre-mRNAs are slightly stabilized, giving rise to increased pre-mRNAs and mRNAs, however, it is not clear how U5 loop mutations could have such an affect.
FIGURE 6.
K-means clustering of spliced mRNAs that are two times more abundant in at least one pairwise comparison of the mutants. Analysis of the mature probe data was performed in the same way as for the intron probe data. Splitting the data into two clusters, Cluster 1 (left) and Cluster 2 (right), produced the best compromise between discriminating clusters and number of probes found in the clusters. Unlike the results with the intron probes, U5-1 and U5-2 behaved similarly in both clusters; in the first cluster U5-1 and U5-2 show accumulation of mature mRNA (implying that splicing of these transcripts is improved).
TABLE 2.
Clusters of genes that display mRNA accumulation
FIGURE 7.
Base prevalence found in accumulated mRNAs resulting from mutations in U5 loop 1. (A) Base prevalence found near the 5′ splice site for all intron containing pre-mRNAs is displayed at the top for reference. The sequences of the pre-mRNAs identified as giving rise to more spliced mRNA in U5-1 and U5-2 (cluster 1) were examined in the same way as described in Figure 5. Only one position had significant change from the consensus sequences; the third nucleotide from the end of 5′ exon (5′SS −3) shows a highly significant increase in the proportion of adenosine at that position and decrease in the proportion of cytidine. (B) U5 loop 1 interactions with the exons after the first step of splicing and the structure of the U5 loop 1 is shown (bold) along with the U5-1 and U5-2 sequences. Known interactions between U5 and the 5′ and 3′ exons are indicated (hashed lines). The third nucleotide upstream of the 5′ splice site in the 5′ exon that displayed a significant increase in the incidence of adenosine at that position in pre-mRNAs exhibiting an increase in mature RNA in U5-1 and U5-2 compared to WT U5 is emboldened.
Reduced stability of Prp8 with a U5 loop 1 mutation may explain increased nucleotide frequency in accumulated pre-mRNA and mRNA
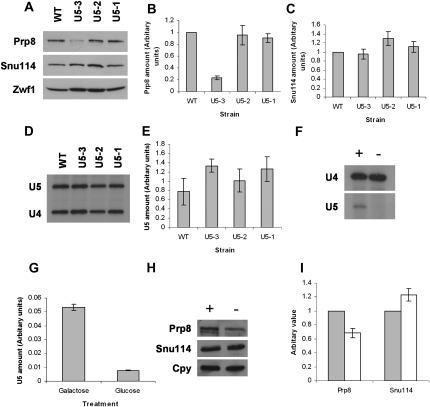
Our microarray results indicate that certain positions in pre-mRNAs that display either pre-mRNA or mRNA accumulation deviate in base prevalence compared to all other intron containing pre-mRNAs. Most of these positions are not known to interact directly with the U5 snRNA loop 1. One model to explain these results is that mutation in U5 loop 1 does not directly cause an alteration in splicing, but may perturb another factor that interacts directly with the pre-mRNA during splicing. An obvious candidate is the spliceosomal protein Prp8, which interacts extensively with U5 snRNA (Fig. 1; Dix et al. 1998). In addition, Prp8 is known to interact with bases at or next to each of the positions that we have found to deviate in pre-mRNAs and mRNAs that accumulate in the U5 mutants (Teigelkamp et al. 1995; Chiara et al. 1997; McPheeters and Muhlenkamp 2003). Interestingly, quantitation of Western blots of Prp8 protein revealed that mutant strain U5-3 appeared to possess less Prp8 than the wild-type strain, although, this was not seen with the U5-1 and U5-2 strains (Fig. 8). U5 snRNA was at comparable levels in these mutant strains (Fig. 8D,E), confirming that this effect on Prp8 stability was caused by the mutations in the U5 loop 1 and was not a result of a general reduction in U5 levels. The levels of the U5 snRNP protein Snu114 also remained constant in the U5-3 and U5-1 extracts (Fig. 8A,C). In the U5-2 extract, significantly higher concentrations of Snu114 were observed (Fig. 8C).
FIGURE 8.
Prp8 levels are reduced in U5 mutant strains and in response to a reduction of U5 in vivo. (A) Equal amounts of splicing extracts from the wild-type and U5 loop 1 mutant strains were resolved by electrophoresis and relative quantities of Prp8, Snu114, and Zwf1 were analyzed by immunoblotting. Immunoblotting was performed in triplicate and a representative blot is shown. Zwf1 (glucose-6-phosphate dehydrogenase) was used to confirm equal loading. (B,C) Quantification of Prp8 and Snu114 levels. Data were normalized against the loading control and are expressed as a difference from the WT value. Error is shown as standard error from the mean. (D) The U5 snRNA was quantified by primer extension to confirm that the observed reduction in Prp8 was not due to a general reduction in U5 snRNA. U4 snRNA was used to confirm equal loading. (E) Quantification of the primer extension of U5 snRNA. U5 amounts are expressed relative to the U4 snRNA loading control and error is shown as standard deviation from the mean. (F) U5 and U4 levels were confirmed by primer extension of RNA isolated from cells grown in galactose (indicated by “+”) or glucose (indicated by “−”) to confirm U5 was depleted. (G) Quantification of primer extension of U5 isolated from cells grown in galactose or glucose. U5 is expressed relative to U4, which was used to confirm equal loading. (H) U5 is required for Prp8 stability in vivo. U5 was depleted in vivo by switching from growth on galactose (indicated by “+”) to growth on glucose (indicated by “−”) and Prp8 levels were analyzed by immunoblotting as above. Anti-carboxypeptidase Y (Cpy/PRC1) was used to confirm equal loading (Young et al. 2001). Prp8 levels were reduced in samples where U5 is depleted. (I) Quantification of Prp8 and Snu114 levels following growth of cells in either galactose (gray bars) or glucose (white bars). Immunoblotting was performed in triplicate and data were normalized against the loading control and are expressed as a difference from the galactose value. Error is shown as standard error from the mean. All quantification was performed using Intelligent Quantifier (Bio Image Systems).
As mutation in U5 snRNA loop 1 influenced the stability of Prp8 it was of interest to ascertain whether the stability of Prp8 changed when the U5 snRNA was depleted. U5 snRNA was depleted from cells in vivo by placing the U5 gene under control of the GAL1 inducible promoter, and shifting the cells from galactose to glucose to turn off expression of the U5 snRNA gene. After 2 h in glucose the levels of U5 snRNA were depleted, whereas levels of U4 snRNA remained relatively constant (Fig. 8F,G). Depletion of U5 resulted in a decrease in Prp8 stability, whereas the level of Snu114 remained constant (Fig. 8H,I). Thus, Prp8 stability is dependent on the integrity of the U5 snRNA and may explain the observed base prevalence in accumulated pre-mRNAs and mRNAs resulting from the U5-3 mutation.
DISCUSSION
We present here a detailed investigation into the effects on pre-mRNA splicing of three temperature-sensitive mutations in the U5 snRNA loop 1. In this first microarray analysis of snRNA mutants, all three U5 alleles that were tested exhibited an alteration in splicing profile of subsets of pre-mRNAs. The microarray results have been validated by RT-PCR, polyribosome profiling and in vitro U5 depletion and reconstitution. Intriguingly, pre-mRNAs, whose splicing was influenced by the U5 mutants, display a bias for certain bases near the 5′ and 3′ splice sites.
Base pairing between the U5 snRNA and exon sequences can influence splicing when there are splice site mutations or mutation in the spliceosome protein Prp18 (Newman and Norman 1991, 1992; Bacikova and Horowitz 2005; Crotti et al. 2007). Additionally, in yeast there is a base bias found in the 5′ exon next to the 5′ splice site that corresponds to the sequence of U5 loop 1 (Long et al. 1997; Lopez and Séraphin 1999). Based on these observations one might expect that pre-mRNAs that are not spliced well in the presence of U5 loop 1 mutations would display base variation in exon sequences at the splice sites that would prevent base pairing with U5 loop 1. On the other hand, one might expect that pre-mRNAs that are spliced better in the presence of U5 loop 1 mutations would display base variation in exon sequences at the splice sites that would favor base pairing with U5 loop 1. However, our analysis of predominant bases near the 5′ splice site, 3′ splice site, and branch site of pre-mRNAs that display pre-mRNA accumulation with U5 mutants when compared to other pre-mRNAs, indicates that positions not known to interact with U5 loop 1, 5′SS +7, and 3′SS +6, display variation in both combined clusters. While positions known to interact with U5 loop 1, 5′SS −4 and 5′SS −3, do display variation in accumulated pre-mRNAs, the variation only occurs in one of the combined clusters and is not entirely consistent with disruption of base-pairing interactions. Only the 5′SS −3 position displayed variation in the base prevalence in pre-mRNAs that exhibit an increase in spliced mRNA in the mutant strains. This position has been shown previously to interact with U5 loop 1 (Alvi et al. 2001; McGrail et al. 2006). However, the base-pairing model of U5 loop 1 interactions with the exon sequences cannot account for the bias for adenosine at this position as well. Neither U5 loop 1 mutant that displays an increase in these specific mRNAs has a compensatory loop 1 mutation that would favor an adenosine at this position in the exon. In hindsight, these results are not surprising as base-pairing interactions of U5 loop 1 with exon sequences have only been found in situations where normal splicing is perturbed. Therefore, the mutations within U5 loop 1 may have a more indirect interaction with these bases within the pre-mRNA and mRNA, possibly through Prp8. Prp8 interacts extensively with both U5 and the pre-mRNA (for review, see Grainger and Beggs 2005). Photo-cross-linkable groups or single labels have been placed at several positions within the 5′ exon, intron, and 3′ exon to identify proteins that interact with these positions. Prp8 specifically crosslinks at position −3 in the 5′ exon upstream of the 5′ splice site (Ismaili et al. 2001), positions +7 and +8 downstream from the 5′ splice site within the intron (Ismaili et al. 2001) and position +6 downstream from the 3′ splice site within the 3′ exon (Chiara et al. 1997) in mammalian spliceosomes. In yeast spliceosomes, Prp8 crosslinks to the 5′ exon up to at least position −8 from the 5′ splice site and within the 3′ exon up to position +13 downstream from the 3′ splice site (Teigelkamp et al. 1995). These positions correlate with the positions that have an altered prevalence of bases in pre-mRNAs that exhibit intron or mature RNA accumulation in this study. Thus, Prp8 interactions at these positions may be influenced by U5 loop 1 mutations.
Previously, it was reported that Prp8 crosslinks to U97 of U5 loop 1; however, U5 loop 1 is not essential for U5-Prp8 interactions as depletion or substitution of loop 1 failed to prevent co-immunoprecipitation of U5 with Prp8 (Dix et al. 1998). Although deleting the entire loop 1 does not affect the co-immunoprecipitation of Prp8-U5, by mutating the sequence and size of loop 1 different forces may be exerted perturbing the Prp8-U5 interactions and thus the stability of Prp8.
It is conceivable that if Prp8 is destabilized then the number of catalytically active spliceosomes would be reduced. However, a reduction in active spliceosomes does not account for the differential splicing defects observed within the U5 loop 1 mutants. We propose that by mutating U5 loop 1 the stability of Prp8 within the spliceosome is reduced and therefore its interactions with certain pre-mRNAs would be perturbed, resulting in the observed differential splicing profiles. This is supported by our finding that the U5-3 mutant, which most severely influences splicing, has reduced stability of Prp8. While U5 mutants U5-1 and U5-2 do not display any significant reduction in Prp8 stability these mutants also do not display a strong influence on splicing. Therefore, we propose that Prp8 stability may also be reduced in these mutants, but that Western blotting may not be sensitive enough to detect the slight changes.
U5 snRNA, Prp8, and Snu114 interact within the spliceosome (Brenner and Guthrie 2006). Binding of U5 snRNA by Snu114 is dependent upon Prp8 (Dix et al. 1998) and an RNA-free complex containing both Prp8 and Snu114 has been reported in mammals (Achsel et al. 1998). Also, mutations in Snu114 cause Prp8 instability and it was proposed that Prp8 stability is dependent upon interaction with Snu114 (Brenner and Guthrie 2006). Here we describe the destabilization of Prp8 by mutation or depletion of the U5 loop 1 without any adverse influence upon levels of Snu114. We suggest that the integrity of both U5 and Snu114 are required for Prp8 stability. The apparent dependence on the U5 loop 1 for Prp8 stability may also explain why the loop 1 structure and Prp8 are so highly conserved. We also postulate that, since Prp8 is destabilized in the most severely affected mutant strain, there would be less interaction between Snu114 and U5, however these U5 loop 1 mutants do not affect the stability of Snu114.
By performing the first microarray analysis of a set of mutations in an RNA component of the spliceosome we have uncovered a new role for the highly conserved U5 loop 1. These data suggest that when the U5 loop 1 is mutated Prp8 is destabilized, leading to a differential effect on the splicing of certain pre-mRNAs. This dependence of Prp8 stability on the U5 loop 1 is a novel role for this snRNA and we propose that this may be one reason why the U5 loop 1 sequence is so highly conserved. It will be of interest to determine whether any of the other snRNA components of the spliceosome display transcript-specific defects in splicing when mutated, similar to that seen here with the U5 snRNA and previously with mutations of the core protein components of the spliceosome (Clark et al. 2002; Burckin et al. 2005; Pleiss et al. 2007a,b).
MATERIALS AND METHODS
Strains and growth conditions
S. cerevisiae strain YROK2 (MAT a; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7∷kanMX6; pRS416-U5) (O'Keefe 2002) with the U5 encoding gene (SNR7) disrupted and complemented with pRS416-U5 was transformed with a TRP1-based plasmid containing either the wild-type U5 gene, U5 mutant U5-1, U5-2, or U5-3 (O'Keefe 2002). Growth on 5-FOA was used to evict the pRS416–U5 plasmid to produce strains YROK2-U5-WT (MAT a; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7∷kanMX6; pRS314-U5-WT), YROK2-U5-1 (MAT a; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7∷kanMX6; pRS314-U5-1), YROK2-U5-2 (MAT a; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7∷kanMX6; pRS314-U5-2) and YROK2-U5-3 (MAT a; ura3-52; trp1Δ63; leu2Δ1; his3Δ200; GAL2; snr7∷kanMX6; pRS314-U5-3). These strains were confirmed by PCR, sequencing and growth analysis at permissive and restrictive temperatures. For RNA isolation, strains were cultured in YPD at the permissive temperature (30°C) to an OD600 of 0.5 and were then switched to the restrictive temperature (37°C) and RNA was isolated from cultures at OD600 of 1. The duration spent growing at the restrictive temperature was on average 1.5 h for WT, 2.3 h for U5-1, 2.7 h for U5-2, and 2.7 h for U5-3. To deplete U5 in vivo a GAL1 inducible promoter was inserted in the promoter region of the SNR7 gene encoding U5. This was achieved by amplifying the GAL1 promoter, along with a selectable marker, from plasmid pFA-kanMX-PGAL1 (Longtine et al. 1998) using U5 Gal F: 5′-GAAAATTTTTGTAGAAACGGAGTGCTCAGTATAAAAAGCGCATAGTAAGACGAATTCGAGCTCGTTTAAAC-3′, and U5 Gal R: 5′-CAGTTCTTGATGTTGACCTCCCTCCGCCATTGATCTGTAAAGCTGCTTCATTTTGAGATCCGGGTTTT-3′. The PCR product created was transformed into BY4742 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) and the resulting strain U5-GAL was confirmed by PCR.
Sample preparation, microarray design, and analysis
RNA was isolated from triplicate cultures of strains grown as described above. Total RNA was isolated from 20 mL of culture using Trizol reagent (Invitrogen). A custom oligonucleotide microarray was designed and printed via the Nimblegen system, a single color, noncompetitive microarray system. Each array slide possessed four spots for each probe. Labeling, using Cy3, hybridizations and array scanning were performed by Nimblegen using their standard protocols. All relevant sample preparation, microarray design and microarray data can be viewed at http://www.biology.ed.ac.uk/research/groups/jbeggs/microarray/U5/ (for correspondence regarding microarray analysis, contact Jean D. Beggs; e-mail: jbeggs@ed.ac.uk). The background-subtracted signal channel-specific intensities obtained from the scanned arrays were analyzed by first taking the median of the four spots for each probe. The median values of each 5′ splice site, intron, and mature mRNA probes were divided by the median value of their respective exon probe to standardize for RNA level. These values for each U5 snRNA mutant were then divided by the equivalent values obtained from the wild type. Therefore, these data are expressed as a function of the wild type, irrespective of the amount of transcript in the two different cell types. These data were then used in clustering analysis using Genespring Version 9 (Agilent Technologies). In the clustering analysis a filter was applied such that only features that were two times more abundant in at least one pairwise comparison of the mutants would pass. K-means clustering was performed on the filtered data and five clusters were identified. To improve the reliability of subsequent statistical analysis, clusters 1 and 2 were combined, and clusters 3 and 4 were combined. As cluster 5 contains only two introns no further statistical analysis was preformed on it. In order to take into consideration the possibility of bias due to the large number of ribosomal protein transcripts in the clusters, pre-mRNAs were grouped into sets depending on whether or not they encoded ribosomal proteins (Fig. 4B). For clusters 1 + 2 and clusters 3 + 4, the occurrence of each of the four bases (A, C, G, or U) at each position in the RNA from 10 bases upstream to 15 bases downstream from the 5′ splice site, 10 bases upstream to 10 bases downstream from the branch site, and from 14 bases upstream to 10 bases downstream from the 3′ splice site was tabulated and a χ2 test was used to compare the observed base composition with the expected base composition in that region, based on the set of all nonclustered, nonribosomal protein pre-mRNAs (set V). Using the χ2 test for independence at three degrees of freedom, P-values were determined for all these sites. Where the small sample size precluded this simple analysis, the smallest numbers in the expected values were combined (“binned”) and a comparison was made between Yates' correction for continuity (Yates 1934) and Fisher's exact test to obtain more reliable P-values. The more conservative Yates results were used although this eliminated some marginal results. The same analysis was performed on the ribosomal protein pre-mRNAs in these clusters (subset SR), comparing these with other (nonclustered) ribosomal protein pre-mRNAs (set T), in order to determine whether any bias observed in the clustered pre-mRNAs was due to features shared by ribosomal protein pre-mRNAs, which are generally well spliced. Results are presented only for positions where both P-values were less than 0.05.
Semiquantitative reverse transcriptase real-time-PCR
Real-time PCR was performed using the QuantiTect SYBR Green RT-PCR kit (Qiagen) according to manufacturer's protocols and a PTC-200 Peltier Thermo cycler (MJ Research). Primers were used at a final concentration of 1 μM to amplify 40 ng of total RNA from the same samples as used in the microarray experiments. Primers were designed to amplify the intron or exon two of ACT1 intron forward primer (IFP); 5′-CGATTGCTTCATTCTTTTTG-3′, intron reverse primer (IRP); 5′-TTTTGACCCATACCGACCAT-3′, exon two forward primer (EFP); 5′-TTCCAATTTACGCTGGTTTC-3′, and exon two reverse primer (ERP); 5′-GTCCAAGGCGACGTAACATA-3′, of SAC6 IFP; 5′-CTCTTCRGCGATTAGAGA-3′, IRP; 5′-ATAAGTAGCATCACCATCTT-3′, EFP; 5′-TAAACATATCAATAGCGTCT-3′, ERP; 5′-CTTGTAYCGATAGTATCAGG-3′, of RPL25 IFP; 5′-CGAATTCATCTCCTATGAA-3′, IRP; 5′-CTTGTATGAGTCCAATCTGT-3′, EFP; 5′-ACTCATACAAGGTCATTGAG-3′, ERP; 5′-AATCTAACGTAAGCCTTCTT-3′ and of RPL30 IFP; 5′-CATTTCAACAGGCCCCAGTT-3′, IRP; 5′-CTGGAGTGTTAGCGGCAATG-3′, EFP; 5′-CATTGCCGCTAACACTCCA-3′, ERP; 5′GCTTCCAAAATAGAGACAACACC-3′. Real-time PCR data were analyzed using the Opticon Monitor 3 software. Primers were designed to amplify a section of both the intron and exon of each pre-mRNA, ensuring that amplicon sizes were identical in all cases. Data obtained from the RT-PCR were standardized for transcription by dividing the intron values by the exon values. A standard curve of the wild-type U5 RNA of 40 ng/reaction, 4 ng/reaction, and 0.4ng/reaction was used as a comparison.
Depletion and reconstitution of U5 in vitro and U5 depletion in vivo
Whole cell yeast extract was made from the S. cerevisiae strain SC261.8 as described previously (Alvi et al. 2001). U5 depletion and reconstitution reactions were performed also as previously described (O'Keefe et al. 1996). Briefly, U5 was depleted from yeast extract at 34°C and reconstituted with wild-type or U5-1, U5-2, or U5-3 in vitro transcribed RNA. Splicing was performed at 23°C for 20 min. To deplete U5 in vivo the U5–GAL strain was grown at 30°C in YP-GAL to an OD600 of 1, harvested, resuspended in YPD, and cultured for a further 2 h and splicing extract made as previously described (Alvi et al. 2001).
Protein and snRNA quantification
Whole cell extracts were produced from strains cultured at 30°C to an OD600 of 1 after which they were cultured at 37°C to an OD600 of 2–3. Protein concentration of each extract was determined using a BCA Protein Assay (Pierce) and protein concentrations were standardized before being resolved on a 12% Tris-glycine Duramide gel (Cambrex). Immunoblotting was performed using Anti-Prp8 8.6 antibody (Grainger and Beggs 2005) at 1:5000, anti-Snu114 polyclonal antibody at 1:20,000 (raised against a peptide encompassing amino acids 1–15), anti-glucose-6-phosphate dehydrogenase (Zwf1) (Sigma) at 1:20,000, and anti-carboxypeptidase Y (Cpy) at 1:30,000 (Young et al. 2001). Primary antibodies were detected using protein A-HRP conjugate (BioRad) or anti-sheep-HRP antibody (BD Biosciences). Detection was performed using Supersignal West Dura substrate (Pierce). Quantification of band density was performed using Intelligent Quantifier (Bio Image Systems). U5 and U4 snRNA were quantified by primer extension analysis as previously described (O'Keefe 2002). U5 primer extension was performed using either U5RT (5′-AAAAATATGGCAGGCCTACAGTAACGG-3′), when analyzing WT, U5-1, U5-2, and U5-3 RNA samples, or U5RT 125 (5′-GGAGACAACACCCGGATGGTTCTGGTA-3′), when analyzing RNA samples isolated from the U5–GAL strains. U4RT All (5′-GGTATTCCAAAAATTCCCTACATAGTC-3′) was used to quantify amounts of U4.
ACKNOWLEDGMENTS
We would like to thank all members of the O'Keefe group for their invaluable discussion during this work, Alastair Kerr (U. Edinburgh) for advice on the statistical analyses, Andy Hayes for advice on real-time PCR, Leo Zeef for initial microarray data analysis, and Colin Stirling for the CPY antibody. This research was funded by Wellcome Trust Grants 067311 (to J.D.B.) and 071683 (to R.T.O.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1347409.
REFERENCES
- Abelson J. Is the spliceosome a ribonucleoprotein enzyme? Nat Struct Mol Biol. 2008;15:1235–1237. doi: 10.1038/nsmb1208-1235. [DOI] [PubMed] [Google Scholar]
- Achsel T, Ahrens K, Brahms H, Teigelkamp S, Lührmann R. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol Cell Biol. 1998;18:6756–6766. doi: 10.1128/mcb.18.11.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi RK, Lund M, O'Keefe RT. ATP-dependent interaction of yeast U5 snRNA loop 1 with the 5′ splice site. RNA. 2001;7:1013–1023. doi: 10.1017/s135583820101041x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacikova D, Horowitz DS. Genetic and functional interaction of evolutionarily conserved regions of the Prp18 protein and the U5 snRNA. Mol Cell Biol. 2005;25:2107–2116. doi: 10.1128/MCB.25.6.2107-2116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner TJ, Guthrie C. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA. 2006;12:862–871. doi: 10.1261/rna.2319806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Chiara MD, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Kramer RF, Reed R. Evidence that U5 snRNP recognizes the 3′ splice site for the catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Sugnet CW, Ares M. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Collins CA, Guthrie C. The question remains: Is the spliceosome a ribozyme? Nat Struct Biol. 2000;7:850–854. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti LB, Bacikova D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes & Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix I, Russell CS, O'Keefe RT, Newman AJ, Beggs JD. Protein–RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Roiha H, Guthrie C. Architecture of the U5 small nuclear RNA. Mol Cell Biol. 1994;14:2180–2190. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Ismaili N, Sha M, Gustafson EH, Konarska MM. The 100-kDa U5 snRNP protein (hPrp28p) contacts the 5′ splice site through its ATPase site. RNA. 2001;7:182–193. doi: 10.1017/s1355838201001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, de Souza SJ, Gilbert W. The yeast splice site revisited: New exon consensus from genomic analysis. Cell. 1997;91:739–740. doi: 10.1016/s0092-8674(00)80462-6. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopez PJ, Séraphin B. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: Implications for splice-site recognition. RNA. 1999;5:1135–1137. doi: 10.1017/s135583829999091x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Romfo CM, Nilsen TW. Functional recognition of the 5′ splice site by U4/U6•U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- McConnell TS, Steitz JA. Proximity of the invariant loop of U5 snRNA to the second intron residue during pre-mRNA splicing. EMBO J. 2001;20:3577–3586. doi: 10.1093/emboj/20.13.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail JC, Tatum EM, O'Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–3822. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Muhlenkamp P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol. 2003;23:4174–4186. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNA by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 303–357. [Google Scholar]
- Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW. RNA–RNA interactions in nuclear pre-mRNA splicing. In: Simons RW, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. pp. 279–307. [Google Scholar]
- O'Keefe RT. Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic Acids Res. 2002;30:5476–5484. doi: 10.1093/nar/gkf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Newman AJ. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Patterson B, Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987;49:613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007a;27:928–937. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007b;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Kois P, Konforti BB, Konarska MM. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA. 1996;2:213–225. [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Gustafson EH, Luo HR, Moore MJ, Konarska MM. The C-terminal region of hPrp8 interacts with the conserved GU dinucleotide at the 5′ splice site. RNA. 1999;5:167–179. doi: 10.1017/s1355838299981785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes & Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as the active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S, Newman AJ, Beggs JD. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggests a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner IA, Norman CM, Churcher MJ, Newman AJ. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA. 2006;12:375–386. doi: 10.1261/rna.2229706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- Vidal VPI, Verdone L, Mayes AE, Beggs JD. Characterization of U6 snRNA-protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt JR, Sontheimer EJ, Steitz JA. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- Yates F. Contingency tables involving small numbers and the χ2 test. J R Stat Soc. 1934;1:217–235. [Google Scholar]
- Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]