Abstract
Hydroxyl-selective electrophiles, including N-methylisatoic anhydride (NMIA) and 1-methyl-7-nitroisatoic anhydride (1M7), are broadly useful for RNA structure analysis because they react preferentially with the ribose 2′-OH group at conformationally unconstrained or flexible nucleotides. Each nucleotide in an RNA has the potential to form an adduct with these reagents to yield a comprehensive, nucleotide-resolution, view of RNA structure. However, it is possible that factors other than local structure modulate reactivity. To evaluate the influence of base identity on the intrinsic reactivity of each nucleotide, we analyze NMIA and 1M7 reactivity using four distinct RNAs, under both native and denaturing conditions. We show that guanosine and adenosine residues have identical intrinsic 2′-hydroxyl reactivities at pH 8.0 and are 1.4 and 1.7 times more reactive than uridine and cytidine, respectively. These subtle, but statistically significant, differences do not impact the ability of selective 2′-hydroxyl acylation analyzed by primer extension-based (SHAPE) methods to establish an RNA secondary structure or monitor RNA folding in solution because base-specific influences are much smaller than the reactivity differences between paired and unpaired nucleotides.
Keywords: RNA SHAPE chemistry, bootstrap ANOVA, 2′-hydroxyl, RNA structure prediction, 2′-ribose pKa
INTRODUCTION
Chemical and enzymatic probing of RNA are critical tools in structural biology and have contributed enormously to our understanding of RNA structure and dynamics and of complex formation with proteins and other ligands. Central to these methods are probes that react with RNA, usually to induce cleavage or modification at flexible, unpaired, or solvent-accessible regions. Ideally, probe reactivity should depend exclusively on RNA structure or solvent accessibility, take place in vivo or under physiologically relevant conditions, be independent of nucleotide identity, and not require significant RNA-to-RNA optimization.
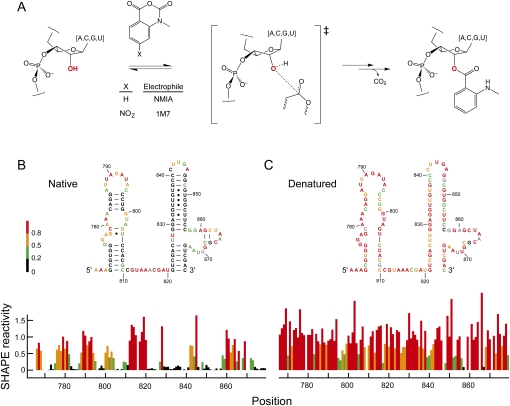
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), is well suited for analysis of local nucleotide structure and dynamics because it interrogates all four RNA nucleotides in a single, robust experiment (Merino et al. 2005). SHAPE uses hydroxyl-selective electrophiles such as N-methylisatoic anhydride (NMIA) and 1-methyl-7-nitroisatoic anhydride (1M7) (Merino et al. 2005; Mortimer and Weeks 2007) that react with the 2′-hydroxyl group at conformationally flexible or disordered nucleotides (Merino et al. 2005; Gherghe et al. 2008b) to form a 2′-O-ester product (Fig. 1A). Sites of modification are then identified by primer extension. SHAPE chemistry reports the positions of unpaired or otherwise conformationally unconstrained nucleotides under mild, structure-reinforcing conditions; shows good reactivity toward all four RNA nucleotides on the minute timescale; is suitable for in vivo RNA structure analysis; and does not require significant optimization because concurrent reagent hydrolysis makes a separate quench step unnecessary.
FIGURE 1.
Reaction of electrophiles with the 2′-hydroxyl position in RNA. (A) Scheme for SHAPE chemistry. (B,C) Secondary structures and normalized SHAPE reactivities analyzed under native and denaturing conditions. A representative region of 16S rRNA is shown.
Although the overall correlation between local nucleotide flexibility and SHAPE is now well established (Merino et al. 2005; Mortimer and Weeks 2007; Gherghe et al. 2008b), there are at least three current observations that suggest factors other than RNA structure might influence the reactivity of NMIA or 1M7. First, cytidine residues in flexible regions sometimes have lower SHAPE reactivities than other nucleotides with similar apparent local structures (Badorrek and Weeks 2006; Badorrek et al. 2006; Jones et al. 2008). Second, the pKa of the ribose 2′-hydroxyl group varies by as much as 0.5 units as a function of the nucleobase in RNA mono- and dinucleotides (Jarvinen et al. 1991; Velikyan et al. 2001; Acharya et al. 2003). Third, the rate of base-catalyzed in-line cleavage, which is also sensitive to the 2′-hydroxyl pKa, varies by up to fourfold as a function of nucleotide sequence (Li and Breaker 1999).
The most general model that explains the strong relationship between SHAPE reactivity and local nucleotide flexibility is that 2′-hydroxyl acylation occurs preferentially at rare conformations that are sampled more frequently at flexible or disordered sites (Merino et al. 2005). Any factor that modulates the nucleophilicity of the 2′-hydroxyl group, including potential electronic crosstalk between the RNA base and 2′-hydroxyl, will in turn modulate the yield of 2′-O-ester adducts and measured SHAPE reactivity.
In this work, we develop a systematic approach to explore the extent to which nucleotide identity modulates NMIA and 1M7 reactivity with the 2′-hydroxyl in diverse RNAs. Using a bootstrap analysis of variance (ANOVA), we show that all four RNA nucleotides react similarly with both NMIA and 1M7 but that there are small, statistically significant, differences such that purines are slightly more reactive than pyrimidines.
RESULTS
Strategy
To assess the influence of base identity on SHAPE reactivity, we analyzed four structurally diverse RNAs. For each RNA, structure was interrogated both under conditions that stabilize native secondary and tertiary folding and also under strongly denaturing conditions. The four RNAs included a transcript corresponding to the 5′ end of an HIV-1 genome, the specificity domain of Bacillus subtilis RNase P, and portions of the Escherichia coli 16S and 23S rRNAs. The HIV-1 transcript includes the first 976 nucleotides (nt) from the 5′ end of the genome and contains both a highly structured regulatory region as well as less structured RNA coding regions (Wilkinson et al. 2008). The RNase P specificity domain (154 nt) from the thermophilic prokaryote B. subtilis forms a compact, well-defined, and highly constrained structure (Krasilnikov et al. 2003). Finally, we analyzed ∼400-nt internal segments from authentic 16S and 23S rRNAs, isolated from E. coli (Deigan et al. 2009). These diverse RNAs represent a cross section of many typical RNA motifs.
Using these four RNAs, we obtained structural constraints using both the NMIA and 1M7 SHAPE reagents for a total analysis of 5128 nt. This data set is sufficiently large to establish rigorously the intrinsic SHAPE reactivities of each RNA nucleotide.
Statistical analysis of intrinsic reactivity in denatured RNA
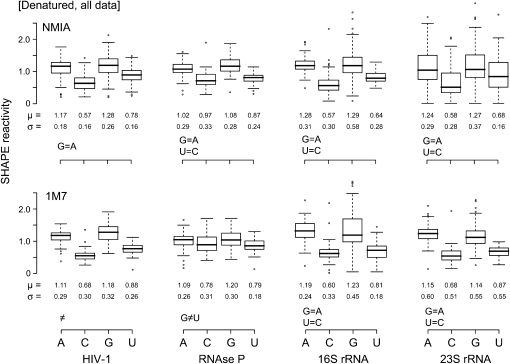
To measure the intrinsic reactivity of each of the four nucleotides, we performed SHAPE experiments on all four RNAs using both 1M7 and NMIA under denaturing conditions (20 mM Hepes [pH 8.0] at 90°C). As expected, nucleotides are consistently more reactive than is observed for the natively folded RNAs (for example, Fig. 1, cf. B and C). However, some nucleotides (Fig. 1C, green and black bars) remained unreactive under denaturing conditions. The existence of these unreactive positions may indicate that base pairs or other structural constraints still form in RNA under our denaturing conditions. In order to make clear conclusions regarding intrinsic nucleotide reactivities not confounded by residual RNA structure, we analyzed two subsets of our 2411 denaturing condition measurements. The first group consists of the entire data set, which assumes that the RNA is completely denatured and that low reactivities do reflect unconstrained positions (Fig. 2). The second group excluded nucleotides that form internal Watson–Crick base pairs in the accepted secondary structure for these RNAs. Our assumption was that nucleotides in internal pairs form the strongest interactions and are therefore the most likely to remain paired under denaturing conditions.
FIGURE 2.
Box plot analysis of SHAPE reactivities for the entire denatured RNA data set. Upper and lower panels illustrate experiments performed with NMIA and 1M7. Equalities at the bottom left of each group emphasize nucleotides showing statistically equivalent reactivities. Boxes outline the middle 50% of each data set; medians are indicated with bold lines. Whiskers above and below each box give the largest or smallest nonoutlier values; outliers are indicated by open circles and are >1.5 times the interquartile range (box).
We report our reactivity data using box plots (Tukey 1977), a convenient way to visualize statistics for large data sets (Fig. 2). The box spans the central half (from 25%–75%) of the data, and the median is shown with a heavy line. The vertical distance between the upper and lower box edges is termed the interquartile range (IQR). Whiskers in the box plot illustrate 1.5 times the IQR. Values outside this range are commonly taken to be outliers (Tukey 1977) and are shown explicitly as circles. We also report the mean (μ) and standard deviation (σ) for each data set.
Visual inspection of box plots showing nucleotide reactivities under denaturing conditions suggests that reactivities follow a clear trend, G ≈ A > U > C, for both reagents and all four RNAs (Fig. 2). We evaluated the statistical significance of these differences using bootstrap ANOVA (Higgins 2003) and multiple comparison procedures (Westfall and Young 1993). The bootstrap ANOVA showed statistically significant (P < 0.05) differences in nucleotide reactivity for all data sets.
The results of the multiple comparison procedures (Fig. 2, summarized at the lower left of each analysis) emphasize that the two purine residues, guanosine and adenosine, had statistically equivalent reactivities and that the pyrimidines also reacted similarly. A few data sets, notably RNase P with 1M7, showed a departure from this trend because all nucleotides showed equal reactivity.
The most critical result is how robust the overall reactivity trends are. The overall reactivity trend and mean reactivities are nearly identical for both the NMIA and 1M7 electrophiles, independent of whether internal pairs are removed (Fig. 2; data not shown). These results emphasize that we are measuring intrinsic nucleotide reactivities that are not influenced by residual structure under denaturing conditions.
Analysis of native state RNA
To determine whether nucleotide identity influences SHAPE reactivity in fully folded RNAs, we also analyzed nucleotide reactivities for the four RNAs equilibrated under conditions that enforce native structure (in the presence of Mg2+ at 37°C) prior to reaction with NMIA or 1M7.
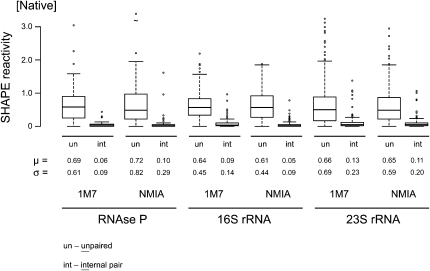
For the folded native RNAs, reactivities vary dramatically because nucleotides experience many different local nucleotide environments. We therefore separated nucleotide reactivities for each RNA into four groups: (1) unpaired; (2) paired, but adjacent to unpaired or noncanonically paired nucleotides (termed external paired); (3) paired, and adjacent to other canonically paired nucleotides (internal paired); and (4) noncanonically paired. As expected, unpaired nucleotides exhibited the highest mean (∼0.66 SHAPE units) and standard deviation (∼0.60) in reactivities, followed by the external pairs. Internal base pairs showed the lowest mean reactivity (∼0.09) and reactivity variability (σ ∼0.17) (Fig. 3). Noncanonically paired nucleotides had idiosyncratic profiles, characterized by variability in measurement means and standard deviations for each RNA.
FIGURE 3.
Differential reactivity of unpaired (un) and internally (int) paired nucleotides toward NMIA and 1M7. Paired nucleotides react within a tighter range and have a smaller mean reactivity than do unpaired nucleotides.
Multiple comparison procedures confirmed that unpaired and internal pairs have very different reactivities (Fig. 3), consistent with the basic model that SHAPE measures nucleotide flexibility. The statistical analysis also confirmed that SHAPE measures small differences in nucleotide environment because most data sets also exhibited statistically significant differences between unpaired and externally paired nucleotides.
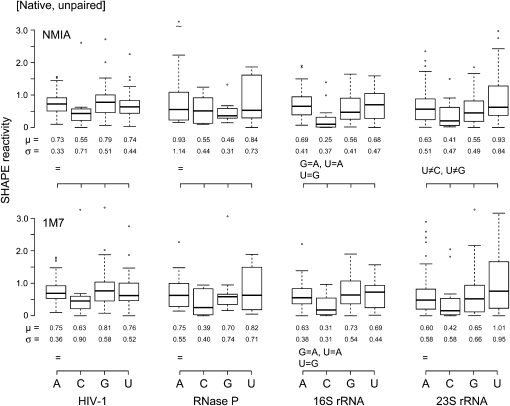
We also assessed intrinsic nucleotide reactivities for unpaired nucleotides (group 1) in the context of the folded RNAs (Fig. 4). These data sets were relatively small because fully unpaired nucleotides comprised only 32%–48% of each RNA. Statistical differences between nucleotide reactivities are less pronounced than those for the denatured RNAs, and it was not possible to detect base-specific trends at the P < 0.05 level. However, qualitative inspection of the reactivity data sets for unpaired nucleotides under native conditions reveals the same overall trend as observed under denaturing conditions. Adenosine and guanosine nucleotides are generally, but not always, more reactive than cytidine and uridine; cytidine residues were consistently the least reactive in all data sets (Fig. 4).
FIGURE 4.
Box plots of nucleotides that are single-stranded in natively folded RNAs. Reactivities toward NMIA and 1M7 are shown in the upper and lower panels, respectively. Statistical equalities are indicated at the lower left of each plot.
DISCUSSION
SHAPE chemistry is much more sensitive to RNA structure than to nucleotide identity
SHAPE chemistry reports local nucleotide flexibility and disorder via reaction at the ribose 2′-hydroxyl position, consistent with a mechanism in which flexible nucleotides are better able to sample relatively rare, but highly reactive, conformations (Fig. 1A). We establish, based on an analysis of over 5000 nucleotide reactivity measurements, that the nucleobase has only a weak influence on SHAPE reactivity (Fig. 5, summarized in the box), and that this influence is small relative to the contribution of local RNA structure. Two lines of evidence support these conclusions.
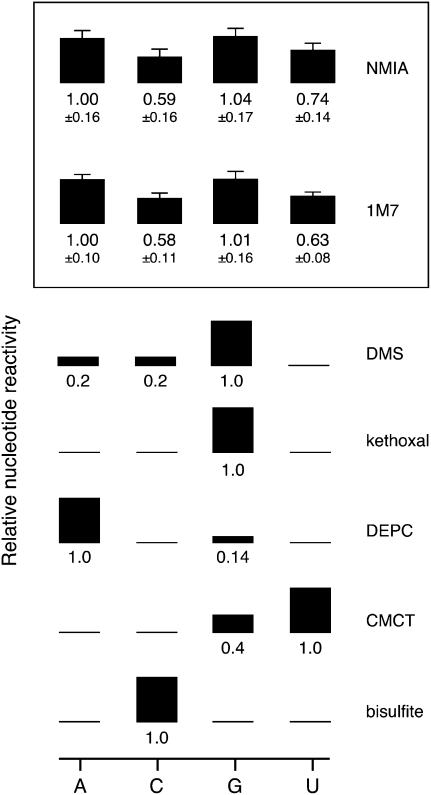
FIGURE 5.
Nucleotide-specific reactivities for NMIA and 1M7. Reactivities for NMIA and 1M7 are reported as the mean plus an error term (root-mean square of the coefficient of variation). These values are compared with other reagents that form stable covalent adducts with RNA. Numerical estimates for the relative nucleotide-specific reactivity of each reagent were obtained from: DMS (Lawley and Shah 1972), bisulfite and kethoxal (Ehresmann et al. 1987), DEPC (Peattie 1979), and CMCT (Gilham 1962).
First, stably paired nucleotides consistently react to a lower yield and exhibit a smaller range in reactivities than do unpaired nucleotides (Fig. 3). The average ratio of mean SHAPE reactivity for unpaired and internally paired nucleotides is 7.3. Thus, SHAPE chemistry has a strong positive predictive ability for differentiating paired and unpaired nucleotides.
Second, no consistent, statistically enforceable trend in reactivity as a function of nucleotide identity can be discerned based on unpaired nucleotides in natively structured RNAs. Thus, the weak structural interactions that occur in the single-stranded regions of typical folded RNAs have a larger effect on reactivity than any intrinsic difference imposed by nucleotide identity (Fig. 4). If nucleotide reactivity were more important, a consistent trend would be obvious in the single-stranded regions of native-state RNAs.
Intrinsic nucleotide reactivities are also independent of the choice of SHAPE electrophile. In general, we recommend the use of 1M7 for routine analysis of RNA structures at equilibrium. 1M7 is less sensitive to variations in ion concentrations (Mortimer and Weeks 2007) and to the possible contributions of slow conformational dynamics at specific RNA nucleotides (Gherghe et al. 2008a).
Accurate prediction of RNA structure based on experimental chemical modification information requires a pseudo-free-energy-change approach
SHAPE reactivities strongly discriminate between paired and unpaired nucleotides, regardless of nucleotide identity (Fig. 3). However, it is also evident that some unpaired nucleotides have reactivities of zero (Fig. 3, lower whiskers in the “un” box plot), while some internally paired nucleotides have moderate to high reactivities (Fig. 3, upper whiskers in the “int” box plot). There is no clear demarcation between paired and unpaired nucleotides. Thus, any RNA structure prediction algorithm that imposes a hard cutoff between paired and unpaired nucleotides is guaranteed to introduce inaccuracies in a structure calculation. Although we have performed our statistical analysis using SHAPE information, this result likely applies even more strongly to conventional chemical probes for RNA structure like base-selective chemical probes and nucleolytic enzymes (Lavery and Pullman 1984; Mathews et al. 2004; Mortimer et al. 2009).
SHAPE reactivities can be converted into pairing probabilities or pseudo-free-energy-change terms and used to constrain existing thermodynamic models for RNA folding to determine RNA secondary structures with accuracies that are often greater than 95% correct (Deigan et al. 2009). This pseudoenergy approach is highly tolerant of experimental and other errors because each reactivity measurement provides an energetic bias to the final structure calculation but does not impose an abrupt cutoff (Deigan et al. 2009; Wilkinson et al. 2008).
We investigated whether correcting the observed SHAPE reactivities by the difference in intrinsic reactivities for the four nucleotides changes or improves secondary structure prediction for the HIV-1, RNase P, or ribosomal RNAs when energy bonuses are introduced as a base-pair stacking term (Xia et al. 1998; Deigan et al. 2009). The predicted structures are essentially identical and exhibit the same overall topologies as those reported previously (Krasilnikov et al. 2003; Mortimer and Weeks 2007; Wilkinson et al. 2008; Deigan et al. 2009). Small differences were observed at the ends of some helices and at multihelix junctions. We infer that nucleotide-to-nucleotide variation in reactivity is not a significant source of error in SHAPE-directed RNA structure prediction.
Comparison of NMIA and 1M7 reactivities to other reagents used to map RNA structure
We developed general parameters for the intrinsic nucleotide reactivities for NMIA and 1M7 (Fig. 5). These relative reactivities were calculated from experiments performed under denaturing conditions (Fig. 2) and therefore represent the largest possible difference in reactivity between the four RNA nucleotides. The key result is that NMIA and 1M7 react broadly with all four nucleotides. The intrinsic reactivity order is A ≈ G > U > C. However, the maximal bias between purines and pyrimidines is less than twofold. This even reactivity stands in stark contrast to traditional structure-selective reagents that also react to form stable covalent adducts with RNA (Fig. 5). The local nucleotide environment can also be probed in degradative reactions using the lead(II) ion (Lindell et al. 2002) and base catalyzed in-line probing (Soukup and Breaker 1999). Both of these approaches share with SHAPE the feature that they react broadly with the four RNA nucleotides. Structure-specific cleavage with lead(II) is influenced by ion affinity toward specific RNA structures (David et al. 2001), and in-line probing reactivities vary by up to fourfold as a function of base identity (Li and Breaker 1999).
The physical basis for the differences in SHAPE reactivities between purine and pyrimidine residues is not completely clear. The pKa of the 2′-hydroxyl in dinucleotides is higher for the pyrimidines (at ∼12.8) than for adenosine (∼12.5) or guanosine (∼12.7) (Velikyan et al. 2001). Since 2′-O-adduct formation involves loss of this proton, the trend in pKa values offer a partial explanation for differing intrinsic reactivities. In some RNAs, a subset of cytidine residues, drawn as single-stranded, have low SHAPE reactivities (Badorrek and Weeks 2006; Badorrek et al. 2006; Jones et al. 2008). Given that the intrinsic reactivity of this nucleotide is, at most, only twofold different from the other nucleotides, we postulate that these cytidines may participate in a locally constraining interaction that remains to be fully characterized.
In sum, the differential effects of base identity on 2′-hydroxyl reactivity, while statistically significant, are small compared to the larger influence of local RNA structure on 2′-hydroxyl reactivity. That the least structured nucleotides show the largest reactivities strongly supports the initial model (Merino et al. 2005; Wilkinson et al. 2005) that SHAPE reactivity is primarily governed by local nucleotide flexibility.
MATERIALS AND METHODS
SHAPE on HIV-1, RNase P, and ribosomal RNAs
The general procedures for SHAPE analysis of the four RNAs studied in this work—the HIV-1, RNase P, and 16S and 23S rRNAs—were described previously. The HIV-1 RNA is a transcript of the 5′-most 975 nucleotides from the NL4-3 strain (Wilkinson et al. 2008); the RNase P specificity domain RNA is imbedded in 5′ and 3′ structure cassette sequences (Merino et al. 2005; Mortimer and Weeks 2007); and 16S and 23S rRNA are authentic ribosomal RNA, purified from E. coli (Deigan et al. 2009). Nucleotides analyzed for each RNA were as follows: HIV-1, 7–402; RNase P, 86–239; 16S, 431-900; and 23S, 213–674 (precise ends varied slightly depending on the experiment). Accepted secondary structures for these RNAs were taken from the following sources (Cannone et al. 2002; Krasilnikov et al. 2003; Wilkinson et al. 2008). Experiments performed under denaturing conditions employed 20 mM Hepes (pH 8.0) for 4 min at 90°C. Native-state modification experiments were performed at 37°C in 50 mM Hepes (pH 8.0), 200 mM potassium acetate, and 5 mM MgCl2, except for the RNase P RNA, which were performed in 100 mM Hepes (pH 8.0), 100 mM NaCl, and 10 mM MgCl2. RNAs were generally allowed to equilibrate in buffer for 30 min prior to addition of reagent. RNAs were initially incubated in a buffer containing 10/9 of these concentrations, and reactions were initiated by addition of 1/10 volume of DMSO containing 1M7 or NMIA. No-reaction controls contained neat DMSO. The 10× NMIA stock concentration was 130 mM for all RNAs. The 1M7 stock (10×) concentration was 30 mM for 16S and 23S rRNAs, 100 mM for RNase P, and 50 mM for the HIV-1 RNA. Following modification, RNAs were recovered by ethanol precipitation and resuspended in 1/2× TE (5 mM Tris at pH 8.0, 0.5 mM EDTA). Each SHAPE reaction product ([+] and [–] reagent and one or two sequencing ladders) was analyzed using primers labeled with distinct fluorophores as described (Mortimer and Weeks 2007; Wilkinson et al. 2008; Deigan et al. 2009).
SHAPE data processing
Primer extension products were resolved on an ABI 3130 capillary electrophoresis DNA sequencer using custom fluorescence spectral calibration. Runs typically yielded more than 400 nt of structural information for the long RNAs. Raw electropherograms were analyzed using the signal processing framework in ShapeFinder (Vasa et al. 2008); areas were calculated for all peaks in the (+) and (–) reagent channels by Gaussian peak fitting. Absolute SHAPE reactivities were calculated by subtracting the (–) peak areas from the (+) peak areas. Positions exhibiting high background were discarded; reactivities that were slightly less than zero were reset to zero. SHAPE data sets were scaled such that a generic reactive nucleotide has an intensity of 1.0 and an unreactive nucleotide is 0. Reactivities for the native data sets were therefore normalized by dividing by the average of the 10% of the most reactive positions, after discarding points with reactivities greater than the third quartile plus 1.5 times the IQR. For denatured data, we assume that nearly all nucleotides are unconstrained: Peak areas were normalized by dividing each data point by the average reactivity of all peaks.
Statistical analysis of intrinsic nucleotide reactivities
A standard one-way ANOVA relies on assumptions of independence, normality, and homogeneous variances between groups (termed homoscedasticity) (Howell 2002). Quantile-Quantile (Q-Q) plots and the randomized Levene's test (Francis and Manly 2001) indicated that SHAPE data are not normally distributed and can be heteroscedastic. Therefore, a bootstrap ANOVA (Higgins 2003), which does not rely on assumptions of normality, was used to assess if the observed reactivities reflect intrinsic reactivity differences or chance. In the bootstrap ANOVA, reactivities, independent of nucleotide identity, were randomly sampled from the measured SHAPE reactivities and used to re-form the original group sizes. Resampling was performed 15,000 times and an F statistic calculated for each iteration. The proportion of F values that are greater than or equal to the F statistic for the original data is reported as a P-value; P-values less than 0.05 indicate that differences observed between groups in the original data are statistically significant. When the bootstrap ANOVA found statistically significant differences in reactivity as a function of nucleotide type, randomized multiple comparison procedures for homoscedastic and heteroscedastic nucleotide groups were performed to identify statistically equivalent or unequal groups (Westfall and Young 1993). For Figure 5, nucleotide reactivities are reported as the mean and the root mean square coefficient of variation. Statistical analyses were performed using R (R Development Core Team 2007).
Structure prediction
SHAPE-directed structure determination was performed using RNAstructure (Mathews et al. 2004) using SHAPE reactivity information as a pseudo-free energy change term (Deigan et al. 2009).
ACKNOWLEDGMENTS
We thank many members of the Weeks and Giddings laboratories for insightful suggestions in preparing this manuscript. This work was supported by a grant from the U.S. National Institutes of Health (AI068462 to K.M.W. and M.C.G.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1536209.
REFERENCES
- Acharya S, Foldesi A, Chattopadhyaya J. The pKa of the internucleotidic 2′-hydroxyl group in diribonucleoside (3′ →5′) monophosphates. J Org Chem. 2003;68:1906–1910. doi: 10.1021/jo026545o. [DOI] [PubMed] [Google Scholar]
- Badorrek CS, Weeks KM. Architecture of a γ retroviral genomic RNA dimer. Biochemistry. 2006;45:12664–12672. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- Badorrek CS, Gherghe CM, Weeks KM. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization. Proc Natl Acad Sci. 2006;103:13640–13645. doi: 10.1073/pnas.0606156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The Comparative RNA Web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Lambert D, Gendron P, Major F. Leadzyme. Methods Enzymol. 2001;341:518–540. doi: 10.1016/s0076-6879(01)41174-8. [DOI] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis RICC, Manly BFJ. Bootstrap calibration to improve the reliability of tests to compare sample means and variances. Environmetrics. 2001;12:713–729. [Google Scholar]
- Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008a;130:8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008b;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilham PT. An addition reaction specific for uridine and guanosine nucleotides and its application to the modification of ribonuclease action. J Am Chem Soc. 1962;84:687–688. [Google Scholar]
- Higgins JJ. An introduction to modern nonparametric statistics. Brooks/Cole; Pacific Grove, CA: 2003. [Google Scholar]
- Howell DC. Statistical methods for psychology. Duxbury/Thomson Learning; Pacific Grove, CA: 2002. [Google Scholar]
- Jarvinen P, Oivanen M, Lonnberg H. Interconversion and phosphodiester hydrolysis of 2′,5′- and 3′,5′-dinucleoside monophosphates: Kinetics and mechanisms. J Org Chem. 1991;56:5396–5401. [Google Scholar]
- Jones CN, Wilkinson KA, Hung KT, Weeks KM, Spremulli LL. Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA. 2008;14:862–871. doi: 10.1261/rna.909208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- Lavery R, Pullman A. A new theorectical index of biochemical reactivity combining steric and electrostatic factors. Biophys Chem. 1984;19:171–181. doi: 10.1016/0301-4622(84)85017-6. [DOI] [PubMed] [Google Scholar]
- Lawley PD, Shah SA. Methylation of ribonucleic acids by the carcinogens dimethyl sulphate, N-methyl-N-nitrosourea and N-methyl-N′-nitro-N-nitrosoguanidine: Comparisons of chemical analyses at the nucleoside and base levels. Biochem J. 1972;128:117–132. doi: 10.1042/bj1280117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Breaker RR. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc. 1999;121:5364–5372. [Google Scholar]
- Lindell M, Romby P, Wagner GH. Lead(II) as a probe for investigating RNA structure in vivo. RNA. 2002;8:534–541. doi: 10.1017/s1355838201020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. A fast acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Johnson JS, Weeks KM. Quantitative analysis of RNA solvent accessibility by N-silylation of guanosine. Biochemistry. 2009;48:2109–2114. doi: 10.1021/bi801939g. [DOI] [PubMed] [Google Scholar]
- Peattie D. Direct chemical method for sequencing RNA. Proc Natl Acad Sci. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2007. [Google Scholar]
- Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. Exploratory data analysis. Addison-Wesley; Reading, MA: 1977. [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikyan I, Acharya S, Trifonova A, Foldesi A, Chattopadhyaya J. The pKa's of 2′-hydroxyl groups in nucleosides and nucleotides. J Am Chem Soc. 2001;123:2983–2984. doi: 10.1021/ja0036312. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based multiple testing: examples and methods for P-value adjustment. Wiley; New York: 1993. [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]