Abstract
The DExD/H-box Prp5 protein (Prp5p) is an essential, RNA-dependent ATPase required for pre-spliceosome formation during nuclear pre-mRNA splicing. In order to understand how this protein functions, we used in vitro, biochemical assays to examine its association with the spliceosome from Saccharomyces cerevisiae. GST-Prp5p in splicing assays pulls down radiolabeled pre-mRNA as well as splicing intermediates and lariat product, but reduced amounts of spliced mRNA. It cosediments with active spliceosomes isolated by glycerol gradient centrifugation. In ATP-depleted extracts, GST-Prp5p associates with pre-mRNA even in the absence of spliceosomal snRNAs. Maximal selection in either the presence or absence of ATP requires a pre-mRNA with a functional intron. Prp5p is present in the commitment complex and functions in subsequent pre-spliceosome formation. Reduced Prp5p levels decrease levels of commitment, pre-spliceosomal and spliceosomal complexes. Thus Prp5p is most likely an integral component of the spliceosome, being among the first splicing factors associating with pre-mRNA and remaining until spliceosome disassembly. The results suggest a model in which Prp5p recruits the U2 snRNP to pre-mRNA in the commitment complex and then hydrolyzes ATP to promote stable association of U2 in the pre-spliceosome. They also suggest that Prp5p could have multiple ATP-independent and ATP-dependent functions at several stages of the splicing cycle.
Keywords: Prp5, DEAD-box protein, U1 snRNP, U2 RNA, spliceosome, pre-mRNA splicing
INTRODUCTION
DExD/H-box proteins are proving to be crucial to most RNA processing pathways due to their ATP-dependent remodeling activities (Linder 2006). In the yeast Saccharomyces cerevisiae, eight DExD/H-box proteins including Prp2, Prp5, Prp16, Prp22, and Sub2 are essential for various stages in the nuclear pre-mRNA splicing cycle: assembling the spliceosome, forming the spliceosome's active catalytic site, releasing the spliced mRNA, and finally, disassembling the spliceosome (Staley and Guthrie 1998). Spliceosome assembly is thought to occur via a stepwise binding of the snRNPs and other factors on the pre-mRNA (Brow 2002). Although it is currently debated whether the spliceosome is pre-formed or assembled on pre-mRNA (Nilsen 2002; Stevens et al. 2002; Tardiff and Rosbash 2006), intermediates in assembly assays identify several points at which the DExD/H spliceosomal helicases act (Staley and Guthrie 1998).
One of the first DExD/H-box proteins required in the assembly pathway is the essential Prp5 protein (Prp5p). Prp5p and ATP hydrolysis are necessary for stable association of U2 snRNP with pre-mRNA and pre-spliceosome formation in S. cerevisiae, Schizosaccharomyces pombe, and human (Liao et al. 1992; Ruby et al. 1993; Xu et al. 2004). Heat inactivation of temperature-sensitive (Ts) prp5-1 mutant yeast whole-cell extract (WCE) in vitro specifically inhibits pre-spliceosome formation, but still allows binding of the U1 snRNP and commitment complex (CC) formation (Ruby et al. 1993). The Ts prp5-1 mutation lies between the conserved Q and I motifs of the helicase core that is responsible for Prp5p's ATP-dependent function, and therefore is predicted to reduce or abolish ATPase activity at the nonpermissive temperature (Abu Dayyeh et al. 2002). Other prp5 mutations that reduce or eliminate ATPase activity prevent pre-spliceosome formation as well (Perriman et al. 2003). Thus Prp5p's ATPase activity is normally required for pre-spliceosome formation.
Several lines of evidence suggest that Prp5p uses ATP hydrolysis to remodel U2 snRNP for binding to pre-mRNA. Prp5p can physically associate with U2 snRNP in S. cerevisiae, S. pombe, and humans (Abu Dayyeh et al. 2002; Will et al. 2002; Xu et al. 2004). U2 RNA stimulates Prp5p's ATPase activity more than other spliceosomal RNAs or synthetic homopolymers (O'Day et al. 1996). Heat inactivation of the Ts mutant prp5-1 protein reduces base-pairing of U2 with an oligonucleotide encoding the UACUAACA box of the pre-mRNA's branchpoint region (BR) (Wiest et al. 1996; Abu Dayyeh et al. 2002). Genetic studies in yeast also indicate that Prp5p interacts either functionally or physically with the 5′ end region of U2 RNA (Ruby et al. 1993; Wells and Ares 1994; Igel et al. 1998). This region of U2 RNA includes the nucleotides that base pair with the pre-mRNA's BR and that form two mutually exclusive stem–loops IIA and IIC, of which only IIA is active for pre-spliceosome formation (Zavanelli and Ares 1991). U2 RNA mutations that stabilize the inactive IIC are synthetically lethal with prp5 mutations predicted to reduce or eliminate ATP hydrolysis (Ruby et al. 1993; Wells and Ares 1994), whereas U2 mutations that stabilize the active IIA form can bypass some of these prp5 mutations (Perriman and Ares 2007). Thus Prp5 may convert IIC to IIA, thereby activating U2 for binding to pre-mRNA.
Yet there are still many unanswered questions as to when and how Prp5p acts during the splicing cycle. The order of association of Prp5p with U2 relative to other spliceosomal components has not been established. The order of U2 snRNP binding and base-pairing with pre-mRNA in the pre-spliceosome is also not known. Prp5p has been shown to have RNA-dependent ATPase activity, but unlike the other spliceosomal DExD/H-box proteins, it has yet to be shown to have RNA unwinding or other ATP-dependent activities (O'Day et al. 1996). The U2 switch between IIA and IIC also somehow involves the Cus2 protein. The cus2-1 mutation reduces IIC accumulation caused by some U2 RNA mutations. A cus2 deletion allows some ATPase-deficient prp5 mutant proteins to promote pre-spliceosome formation (Yan et al. 1998; Perriman and Ares 2007). Additionally, there is another DExD/H-box protein, Sub2, whose ATPase activity is required for pre-spliceosome formation (Fleckner et al. 1997; Kistler and Guthrie 2001). Sub2 may remodel or remove Bbp bound to the pre-mRNA's UACUAAC box in the transition from CC to pre-spliceosome, thereby aiding pre-mRNA to base-pair with U2 (Kistler and Guthrie 2001). The order in which Sub2 and Prp5 act is unknown.
Some recent in vivo results raise additional questions regarding how and when Prp5 functions in the spliceosome. Prp5p's ATP hydrolytic activity has been suggested to monitor the fidelity of BR pairing (Xu and Query 2007), possibly by unwinding abnormal U2-pre-mRNA duplexes. Additionally, Prp5p and U2-IIC are important for fidelity of 5′ splice site cleavage during the first transesterification reaction (Perriman and Ares 2007). Finally, U2 RNA switches between IIA and IIC at least twice within the spliceosome (Hilliker et al. 2007; Perriman and Ares 2007).
An important step toward understanding Prp5p's functions in spliceosome formation and splicing fidelity is to determine when Prp5p is present in the spliceosome. In S. pombe, SpPrp5p has been shown to be present in the pre-spliceosome as well as in a U1-U2 di-snRNP, but limitations of the S. pombe in vitro splicing assay prevent detecting Prp5p in steps both before and after pre-spliceosome formation (Xu et al. 2004). Here we examine interactions between Prp5p and the spliceosome of the yeast S. cerevisiae using a combination of in vitro splicing assays and affinity selections. The results show that ScPrp5p is in the commitment complex and thus is among the earliest spliceosomal factors to associate with pre-mRNA. It probably remains in the spliceosome through both transesterification reactions. Our data support a model in which Prp5p recruits U2 snRNP to the pre-mRNA in the commitment complex and then hydrolyzes ATP to promote stable association of U2 in the pre-spliceosome. Finally, the results also suggest that Prp5p could have multiple ATP-dependent functions in the spliceosome.
RESULTS
GST-Prp5p pulls down pre-mRNA, lariat intermediate, and lariat product from an in vitro splicing reaction
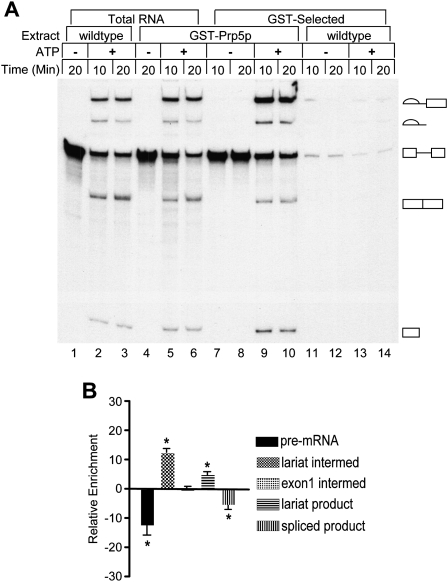
Previously we created a gene encoding a GST-Prp5 fusion protein and found it to be expressed and able to functionally replace the essential, wild-type Prp5p in vivo and in vitro (Abu Dayyeh et al. 2002). GST-Prp5p could pull down U2 snRNP from whole-cell extracts (WCE) without exogenously added pre-mRNA. To determine whether Prp5p associates with the spliceosome, we asked if GST-Prp5p could pull down added pre-mRNA substrate and its spliced products from an in vitro splicing reaction. Radiolabeled actin pre-mRNA was added to splicing reactions with or without ATP and with WCE that contained either GST-Prp5p or wild-type (untagged) Prp5p as a control. The reactions were incubated for 10 and 20 min at 23°C and immediately thereafter subjected to GST-affinity selection for 30 min at 0°C. RNAs from GST-selected samples as well as from the paired, unselected (total) reactions were extracted and fractionated by denaturing polyacrylamide gel electrophoresis (Fig. 1). In ATP-depleted reactions, GST-Prp5p pulls down unspliced pre-mRNA, the only species present in these reactions. In ATP-supplemented reactions, wild-type pre-mRNA is spliced, and GST-Prp5p pulls down the splicing intermediates and products as well as pre-mRNA. In general, selected RNA is enriched fivefold to 30-fold above background control levels from reactions with wild-type, untagged Prp5p (Fig. 1) or with GST tag alone (see, also, Fig. 4 below; data not shown), and selected RNAs comprise usually 5%–50% of total input radiolabeled transcript. Variation in selected RNA levels is due to differences among the RNA species as described below as well as to variability among extract preparations. The result that pre-mRNA is pulled down in the absence of ATP suggests that Prp5p associates with pre-mRNA before pre-spliceosome formation. Selection of splicing intermediates and products suggests that Prp5p associates with the spliceosome during the two splicing reactions.
FIGURE 1.
GST-Prp5p pulls down pre-mRNA, splicing intermediates, and lariat product from splicing reactions. (A) Total and selected RNAs from splicing reactions. Active whole-cell splicing extracts (WCEs) with wild-type Prp5p (lanes 1–3,11–14) or GST-Prp5p (lanes 4–10), were incubated at 23°C in splicing buffer with either glucose to deplete ATP or water. Radiolabeled actin pre-mRNA was then added with either no ATP (lanes 1,4,7,8,11,12) or ATP (lanes 2,3,5,6,9,10,13,14). Aliquots of splicing reactions were removed at the times indicated and incubated with glutathione-sepharose beads for 30 min at 4°C (lanes 7–14) or processed to isolate total RNA (lanes 1–6). One-tenth of total RNA and all of selected RNA samples were fractionated by denaturing gel electrophoresis and visualized by autoradiography. Lariat intermediate, lariat product, pre-mRNA, mature mRNA, and free exon 1 are indicated from the top to the bottom of the gel. (B) Relative RNA enrichment. Relative enrichment was calculated by subtracting the percentages of pre-mRNA, splicing intermediates, and products in unselected (total) isolated RNA in each 10 min and 20 min sample from the values for paired GST-selected RNAs. Data for the 10 min and 20 min samples were then combined. The means (±SD) for five independent experiments are plotted with asterisks indicating that those means differ significantly from 0; all p ≤ 0.02.
FIGURE 4.
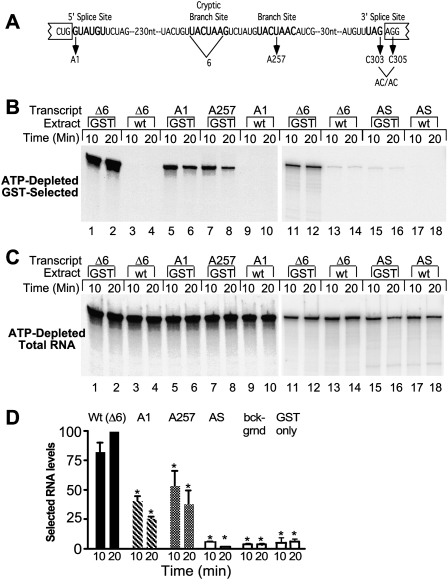
Efficient association of Prp5p with RNA requires a functional intron. (A) Diagram of actin wild-type and mutant pre-mRNAs. (From left to right) Bold lettering indicates the 5′ splice site, the cryptic branch site, the authentic branch site, and the 3′ splice site. Also depicted are mutant pre-mRNAs and the 6-nt cryptic branchpoint sequence deleted from the Δ6 transcript. The A1, A257, and AC/AC mutations were each made in the Δ6 background. (B) RNA in GST-Prp5p selected material. ATP-depleted GST-Prp5p (GST) or wild-type (WT) extracts in splicing buffer were incubated with equal amounts of functional actin (Δ6), 5′ splice site mutant (A1), branch site mutant (A257), and antisense (AS) RNAs for the indicated times. Splicing reactions were subjected to GST selection after which RNA was extracted, fractioned by denaturing PAGE, and visualized by autoradiography (12-h film exposure for lanes 1–10 and 96-h PhosphorImager screen exposure for lanes 11–18). (C) Total RNA from splicing reactions. RNA was extracted from samples removed from reactions before GST selection. RNA was visualized by autoradiography (2.5-h film exposure for lanes 1–12 and 67-h Phos-phorImager screen exposure for lanes 11–18). (D) Quantification of GST-selected RNA levels. Levels of selected wild-type and mutant A1 and A257 RNAs in three experiments as in A were measured and calculated as percentage of wild-type RNA pulled down at 20 min in the same experiment. The means (±SD) are plotted. Levels statistically significantly different from Δ6 levels are indicated by asterisks; all p ≤ 0.01. The levels of A1 and A257 are not statistically different from each other. Antisense RNA levels are the same as background levels (selections with an untagged Prp5 protein and selections with GST-tag only) measured in two, 10, and two experiments, respectively.
Although all RNA species pulled down from a splicing reaction are well above background levels, the percentage of each in selected samples differs from that in paired, unselected samples (relative enrichment) (Fig. 1B). Lariat intermediate and lariat product each comprises a higher percentage of selected versus unselected RNAs, and thus there is a relative enrichment of these two RNAs in selected samples. This enrichment supports the conclusion that Prp5p associates with active spliceosomes undergoing the first and second transesterification reactions. In contrast, there is a relative loss of pre-mRNA and spliced mRNA. Pre-mRNA loss probably occurs because some pre-mRNA added to reactions is incorporated into nonspliceosomal complexes lacking Prp5p. Spliced mRNA loss could occur if Prp5p no longer associates with this RNA when it is released from the spliceosome. Alternatively, the association between Prp5p and mRNA may change after the splicing reactions such that it is no longer stable to selection or that the GST tag is no longer accessible. We also considered that if only 50% of pre-mRNA added to a reaction is incorporated into spliceosomes, this would alter the percentages of the other RNA species in selected versus unselected samples, but this predicts a positive relative enrichment rather than the observed relative loss. Relative enrichment for the exon 1 intermediate is not statistically different from zero. This RNA may be more easily lost than lariat intermediate during selection.
Additional tests for the presence of Prp5p in an active spliceosome used AC/AC mutant RNA with a mutation in the 3′ splice site (Supplemental Fig. S1). This mutant RNA undergoes the first, but not the second transesterification reaction and accumulates arrested spliceosomes in vitro (Vijayraghavan et al. 1986; Cheng and Abelson 1987). GST-selection of AC/AC RNA shows substantial increases in the levels of exon 1 and lariat intermediate above background (Supplemental Fig. S1A). These results are consistent with Prp5 being present in the spliceosome through at least the first transesterification reaction.
To rule out the possibility that the N-terminal GST-tag might somehow enable Prp5p to associate with the spliceosome or splicing intermediates, we also tested Prp5p with a C-terminal TAP tag (Prp5-TAP). The TAP tag has previously been used to study other spliceosomal factors, and on its own, it does not bind RNA or snRNPs (Gavin et al. 2002; Ohi et al. 2002; Dziembowski et al. 2004). In our assays, WCE was made from yeast cells in which the wild-type PRP5 gene copy of Prp5p was replaced by plasmid shuffling with one encoding Prp5-TAP. With AC/AC pre-mRNA as splicing substrate, Prp5-TAP pulls down pre-mRNA in the absence of ATP as well as pre-mRNA, lariat intermediate, and exon 1 in the presence of ATP (Supplemental Fig. S1C; data not shown). Selected RNA levels are twofold to 3.5-fold those of controls from WCE with untagged Prp5p and, thus, usually lower than GST-Prp5p-selected levels. Several factors probably contribute to different amounts of RNA being selected by the two tagged Prp5 proteins: expression levels of GST-Prp5 from the galactose-inducible GAL10 promoter versus Prp5-TAP from the PRP5 promoter and plasmid vector copy number as well as tag position and selection efficiency, and higher nonspecific binding of extract factors to IgG-Sepharose than to glutathione-Sepharose.
If Prp5p is present in the spliceosome, then it will pull down spliceosomal snRNPs along with pre-mRNA and splicing intermediates. Northern analyses show that either GST- or TAP-tagged Prp5p can pull down spliceosomal snRNPs along with splicing intermediates from total splicing reactions with AC/AC pre-mRNA (Supplemental Fig. S1; data not shown). However, both GST-Prp5p and Prp5-TAP pull down more of some spliceosomal RNPs than others from total reactions even in the absence of added pre-mRNA. As previously reported (Abu Dayyeh et al. 2002), GST-Prp5p pulls down detectably larger amounts of U2 snRNP compared to other snRNPs. Prp5-TAP pulls down both U1 and U2 snRNPs at levels fourfold to sevenfold background levels (Supplemental Fig. S1). Both GST-Prp5p and Prp5-TAP pull down lower amounts of the other three snRNPs (U4, U5, and U6); levels vary from 0.5-fold to threefold background levels. Thus it is probable that Prp5p associates with snRNP complexes other than or in addition to functional spliceosomes in splicing reactions. It is also possible that either tag or both tags could destabilize one or more snRNPs in the spliceosome, thus promoting their loss during selection. We therefore sought another approach.
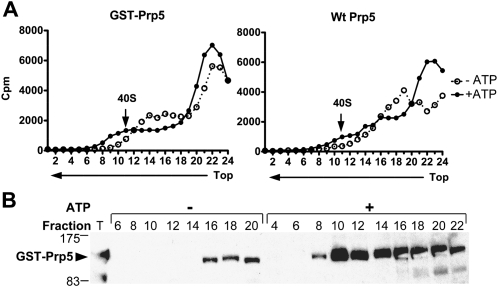
Previous work showed that spliceosomes formed on radiolabeled pre-mRNA in vitro sediment as 40S particles and contain either pre-mRNA or splicing intermediates (Brody and Abelson 1985; Lin et al. 1987). We assayed for GST-Prp5p in active spliceosomes isolated by glycerol gradient sedimentation (Fig. 2). Radiolabeled AC/AC RNA was used again as splicing substrate in order to arrest spliceosomes before the second transesterification reaction. Splicing reactions with either GST-Prp5p or untagged Prp5p WCEs, and under ATP-depleted or ATP-supplemented conditions were incubated for 15 min and then sedimented through 11%–23% glycerol gradients in low ionic conditions. Fractions were collected and assayed for radioactive counts as well as GST-Prp5p and total radiolabeled RNAs in unselected fractions. Radioactive counts across the gradients show small 40S peaks centered on fraction 11 in ATP-supplemented reactions but not in ATP-depleted reactions (Fig. 2A). Both radiolabeled pre-mRNA and splicing intermediates are present in unselected fractions 9–15, whereas less pre-mRNA and no intermediates are found in corresponding fractions from ATP-depleted reactions (data not shown). Similarly, GST-Prp5 is found in fractions 8–14 but little or no GST-Prp5p is detectable in corresponding fractions from the ATP-depleted reaction (Fig. 2B). Thus, GST-Prp5p sedimentation recapitulates that of the spliceosomal Prp11 protein, which peaks in fractions 10 and 11 under the same ionic and sedimentation conditions (Chang et al. 1988). Additional analyses showed that GST-Prp5p in the 40S fractions pulls down radiolabeled pre-mRNA, splicing lariat, and exon 1 as well as the five spliceosomal snRNAs (data not shown). Finally, sedimentation of parallel splicing reactions with ATP, but without added pre-mRNA, shows little GST-Prp5p in the 40S fractions (data not shown). The combined results from gradient sedimentation analyses show that Prp5p cosediments with active spliceosomes formed on added pre-mRNA.
FIGURE 2.
Cosedimentation of GST-Prp5p with 40S spliceosomes in glycerol gradients. Radiolabeled AC/AC mutant pre-mRNA was incubated for 15 min in splicing reactions depleted of or supplemented with ATP, and with WCEs with either the N-terminal GST-tag, or untagged Prp5p. Reactions were then fractionated by sedimentation in 11%–23% glycerol gradients in low ionic conditions (50 mM KCl, 2 mM MgCl2, and 20 mM HEPES at pH 7.6). Gradient fractions were collected and analyzed. (A) Radioactive counts in each fraction. Cherenkov emissions in aliquots of gradient fractions from reactions with GST-Prp5p (left panel) or with untagged Prp5p (right panel) were counted. An arrow indicates the 40S spliceosomal peak (fraction 11). (Left panel, fractions 13–17 with GST-Prp5p) A shoulder in the radioactivity profile from reactions depleted of ATP occasionally occurred, but it did not depend on the presence of a tag. (B) Western blot of 40S fractions probed with anti-Prp5p antibody. Proteins in even-numbered fractions with GST-Prp5p with or without ATP (left panel in A) were concentrated by precipitation and analyzed by a Western blot probed with anti-Prp5p antibody. An aliquot of WCE with GST-Prp5p (T) corresponds to 20% of the reaction loaded onto the gradient. The bulk of GST-Prp5p in the gradient from the ATP-depleted reaction resides in fractions 22–24 (not shown). Molecular weight markers are indicated on the left. (Arrow) Indicates full-length GST-Prp5p. A fragment of GST-Prp5p (at 83 kDa) was probably generated by proteolytic cleavage during WCE preparation.
In summary, we conclude that Prp5p associates with an active spliceosome through the first and second transesterification reactions. GST-Prp5p and active spliceosomes cosediment in glycerol gradients. Both GST-Prp5p and Prp5-TAP pull down pre-mRNA and splicing intermediates from splicing reactions. However, Prp5p may not associate with the mature, spliced mRNA released from the spliceosome as there is a reduced amount of mRNA in selected versus unselected samples. Prp5p may also associate with pre-mRNA before pre-spliceosome formation as GST-Prp5p and Prp5-TAP pull down pre-mRNA in the absence of ATP. Additionally, Prp5-TAP can pull-down U1 as well as U2 snRNP in the absence of added pre-mRNA, suggesting that it might contact U1 snRNP like its orthologs from fission yeast and humans (Xu et al. 2004).
An interaction between Prp5p and pre-mRNA is temperature-independent
To further examine the interaction between Prp5p and the spliceosome during the course of a splicing reaction, we sampled an in vitro splicing reaction at several times. GST-Prp5p efficiently pulls down pre-mRNA and some lariat intermediate as early as 1 min after initiating a splicing reaction at 23°C (Supplemental Fig. S2A). Surprisingly, pre-mRNA was even pulled down in the “zero-time control” in this experiment. This control was collected by first adding ATP-supplemented GST-Prp5p extract/splicing mixture directly to glutathione-Sepharose beads at 0°C, then adding radiolabeled actin transcript and holding the reaction at 0°C thereafter. Previous findings show that the yeast U1 snRNP does not form a complex with pre-mRNA at 0°C that is stable to pull-down conditions (Ruby and Abelson 1988) and that ATP hydrolysis is severely hindered at 0°C (O'Day et al. 1996). However, GST-Prp5p is able to pull down pre-mRNA as efficiently from reactions incubated at 0°C as at 23°C (Supplemental Fig. S2A), suggesting that the association is independent of temperature and ATP hydrolysis.
To examine the effects of temperature further, we varied the order of addition of WCE and pre-mRNA at 0°C and 23°C in tandem with affinity selection under increasing dilutions (Supplemental Fig. S2B). In one case, an in vitro splicing reaction was initiated by adding radiolabeled transcript to an ATP-depleted GST-Prp5p extract/splicing buffer mixture. This reaction was allowed to incubate for 10 min at 23°C. Aliquots of the reaction were then diluted with onefold, 25-fold, 50-fold, or 100-fold volume of buffer when added to a constant amount of glutathione-Sepharose beads. In the second case, a splicing reaction with added radiolabeled transcript was incubated for 10 min at 0°C and selected in an identical fashion. In the third case, the order of addition was examined by first adding ice-cold GST-Prp5p extract in splicing buffer to the various amounts of buffer with glutathione-Sepharose beads (at 0°C) and then adding radiolabeled transcript. In all three cases, selection of pre-mRNA is not drastically affected by either a 25-fold or a 50-fold dilution. Selection of pre-mRNA was hindered upon a 100-fold dilution, but it was still significantly above background. In a fourth case, a mixture of GST-Prp5p WCE and splicing buffer was incubated for 10 min at 23°C and then added to ice-cold glutathione-Sepharose beads before adding radiolabeled transcript. This case exhibited a nearly identical pre-mRNA selection profile when compared to the three previous selection scenarios (data not shown). Therefore, pull-down of pre-mRNA by GST-Prp5p is independent of temperature and order of transcript addition. However, we do not know if the association at 0°C under dilute conditions occurs by the same mechanism as in a splicing reaction at 23°C.
Heat-inactivated Ts mutant prp5-1 protein in splicing extracts still pulls down pre-mRNA
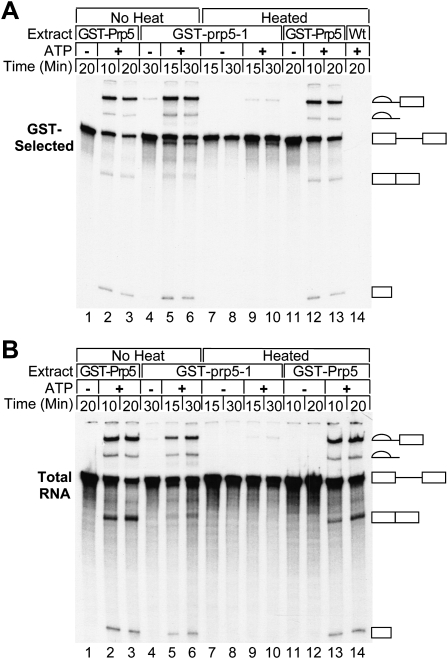
The ability of GST-Prp5p to pull down pre-mRNA from ATP-depleted reactions suggests that Prp5p associates with pre-mRNA before U2 snRNP binding and pre-spliceosome formation. To test this, we used heat-inactivated extracts with the Ts prp5-1 mutant protein defective for pre-spliceosome formation in vitro (Ruby et al. 1993). Plasmid shuffling was used to replace the wild-type PRP5 gene with a construct encoding the mutant prp5-1 protein as a GST fusion. Yeast cells expressing GST-prp5-1p as well as their respective WCEs exhibit the same temperature-sensitive profile as those with untagged, mutant prp5-1p (data not shown). Active splicing extracts were made from cells expressing GST-prp5-1p grown at the permissive temperature. In vitro heating of the GST-prp5-1 WCEs abolishes splicing of radiolabeled actin pre-mRNA in the presence of added ATP (Fig. 3B). Untreated GST-prp5-1p pulls down splicing intermediates, pre-mRNA, and lariat product similarly to wild-type GST-Prp5p (Fig. 3A). Heat-inactivated GST-prp5-1p pulls down pre-mRNA in the presence or absence of added ATP as efficiently as untreated GST-prp5-1p in the absence of ATP. These results indicate that the interaction between Prp5p and pre-mRNA occurs before pre-spliceosome formation and that Prp5p is present in the CC along with the U1 snRNP, as heat-inactivated prp5-1 WCE still efficiently forms CC (Ruby et al. 1993). Furthermore, the interaction is independent of a functional helicase domain. The prp5-1 mutation is predicted to result in a G293D substitution in a conserved region between helicase Q and I motifs and to result in compromised ATPase activity (Abu Dayyeh et al. 2002). Therefore, these results further support the conclusion that the association of pre-mRNA with Prp5p is independent of ATP.
FIGURE 3.
Heat-inactivated Ts GST-prp5-1 protein pulls down pre-mRNA from splicing reactions. (A) RNA in GST-selected material. Active WCE with GST-prp5-1p, GST-Prp5p, or wild-type Prp5p was either heated for 10 min at 37°C or incubated on ice (no heat) before adding splicing buffer with either glucose or water. After incubation for an additional 15 min at 30°C to deplete ATP, radiolabeled pre-mRNA and either no ATP (to the glucose treated reactions) or ATP (to water-treated reactions) was then added. Samples were removed at the times indicated and subjected to GST selection. RNA was extracted, fractionated by denaturing PAGE, and visualized by autoradiography (23 h exposure). (B) Total RNA from splicing reactions. RNA was extracted from samples removed from the reactions in A before GST selection. An amount from a sample equivalent to that in A was fractionated by denaturing PAGE and visualized by autoradiography (6 h exposure).
GST-Prp5p's interaction with pre-mRNA is sensitive to point mutations in the 5′ splice site and BR
To determine if the interaction of Prp5p with pre-mRNA in splicing extract requires that RNA have a functional intron, we performed GST-Prp5p pull-downs of radiolabeled actin (Act) mutant pre-mRNAs. The 5′ splice site mutation A1 and branch site mutation A257 in Act pre-mRNA (Fig. 4A) were previously shown to inhibit splicing in vivo (Vijayraghavan et al. 1986) and in vitro (Cheng and Abelson 1987) and to reduce or prevent CC formation (Ruby and Abelson 1988). We found that pull-downs of the A1 and A257 mutant RNAs are significantly reduced to 50% or less of the wild-type (Δ6) levels in the presence and absence of ATP (Fig. 4B–D; data not shown). GST-Prp5p selects more A257 mutant than A1 mutant in conditions with or without ATP; however, this difference is not statistically significant. Equivalent mutations in the Rp51a pre-mRNA gave similar results (data not shown). Combined with the results that the association of Prp5p with pre-mRNA can occur at 0°C, we also surmise that recognition of the 5′ splice site and BR is maintained at 0°C during the GST-selection.
The absence of an intron has an even larger effect than the intronic point mutations. One of the RNAs tested is antisense to wild-type pre-mRNA and therefore is the same size as pre-mRNA but lacks a functional intron. The levels of selected antisense RNA (Fig. 4B) are <0.5% of the total RNA (Fig. 4C) in reactions and are not statistically different from background levels (Fig. 4D). A second RNA (ΔIVS), synthetically transcribed as the Act RNA without the intron, gives results similar to those of antisense RNA (data not shown).
We conclude that Prp5p's interaction with RNA in splicing extracts requires an intron and is partly dependent on the 5′ splice site and UACUAAC-box sequences.
snRNPs are not required for GST-Prp5 to associate with pre-mRNA
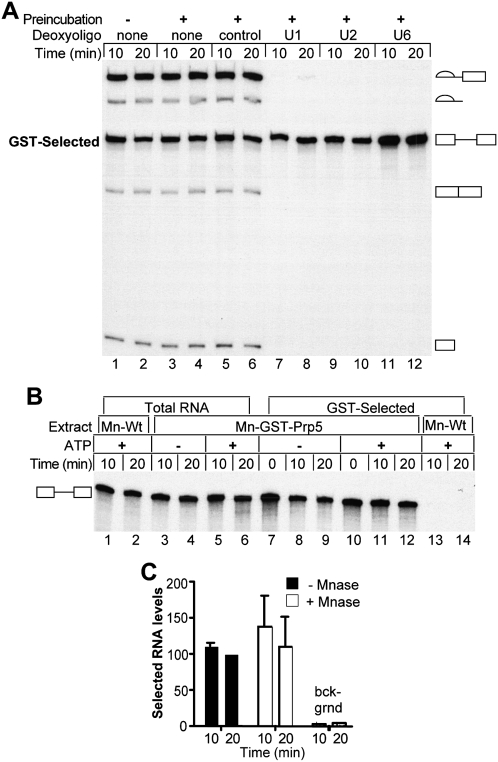
Previously Prp5p has been shown to associate with U2 snRNP in extracts from S. cerevisiae (Abu Dayyeh et al. 2002). Here we also observed that Prp5p pulls down U1 (Supplemental Fig. S1); so, it may physically associate with U1 like the Prp5 proteins from S. pombe or human (Xu et al. 2004). Therefore, Prp5p's association with pre-mRNA may be indirectly mediated via a snRNP. To rule out this possibility, we assayed the effects of RNase H cleavage of U1, U2, and U6 snRNAs on GST-Prp5p pull-downs of pre-mRNA from the treated extracts. Oligonucleotide-directed RNase H degradation of the 5′ end regions of U1, U2, or U6 RNA in WCEs has previously been shown to prevent functional association of each snRNP with the pre-mRNA, thereby inhibiting formation of functional CCs, pre-spliceosomes, and spliceosomes, respectively (Ruby and Abelson 1988; Fabrizio et al. 1989).
The minimal concentration of oligonucleotides necessary for degrading each snRNA was determined by titration using in vitro splicing and primer extension assays (data not shown). GST-Prp5p is still capable of efficiently pulling down pre-mRNA from splicing reactions with degraded U1, U2, or U6 (Fig. 5A). As controls, WCEs were incubated in parallel at either 0°C or 30°C without an oligonucleotide, or at 30°C with a control oligonucleotide. These control treatments have no effect on splicing or subsequent GST-selection. We conclude that the association between Prp5p and pre-mRNA is independent of U1, U2, and U6 RNA integrity. Furthermore, these data indicate that Prp5p is present in the commitment and pre-spliceosomal complexes as these complexes accumulate in reactions in which the U2 and U6 RNAs, respectively, are degraded.
FIGURE 5.
Association of Prp5p with pre-mRNA does not require intact U1, U2, or U6 RNA. (A) Pre-mRNA in GST-Prp5p selected material from reactions with RNase-H-degraded RNAs. Deoxyoligonucleotides complementary to the 5′ end regions of U1, U2, and U6 RNAs specifically target these RNAs for cleavage by endogenous RNase H activity. Active GST-Prp5p WCEs supplemented with 0.2 mM ATP were first incubated with added water or the following deoxyoligonucleotides: 1.0 μM control, 5.0 μM anti-U1, 1.0 μM anti-U2, and 1.0 μM of anti-U6 for 30 min at 30°C. The splicing reactions were initiated by adding radiolabeled pre-mRNA and fresh ATP and incubated at 23°C. Samples were removed at 10 and 20 min and subjected to GST-selection before RNA extraction and denaturing PAGE. GST-Prp5p WCE that did not undergo the first incubation at 30°C was used as an additional control (lanes 1,2). (B) GST-selected pre-mRNAs from micrococcal nuclease-treated WCE. WCEs of GST-Prp5p or wild-type Prp5p were first incubated with micrococcal nuclease (MNase) to destroy RNAs, after which reactions were stopped by adding EGTA. Splicing buffer with either glucose (−ATP) or water (+ATP) was then added to MNase-treated extracts (Mn-Wt or Mn-GST-Prp5p) and incubated for an additional 15 min at 30°C. After addition of fresh ATP (+ATP), radiolabeled pre-mRNA was added, and the reactions were incubated at 23°C. Samples were removed at the times indicated and either subjected to GST-selection (lanes 7–14) or processed to isolate total RNA (lanes 1–6). Zero time points were collected as described in Materials and Methods. Equal volumes of undiluted GST-selected material and 10-fold diluted total extracted RNA were fractionated by denaturing PAGE and visualized by autoradiography. (C) Quantification of RNAs selected from MNase-treated WCE. WCE with GST-Prp5p or untagged Prp5p (“bckgrnd”) was incubated with MNase (Mn-GST-Prp5p) or water and depleted of ATP. The extracts were then analyzed by pull-down assays as in B. Levels of selected RNAs in two experiments were measured and calculated as a percentage of RNA pulled down from reactions with untreated WCE at 20 min in the same experiment. The means (±SD) are plotted. There is no significant difference due to MNase treatment.
RNase H degradation of U1 RNA leaves truncated forms that can still interact with pre-mRNA and splicing factors even though they are not fully functional for splicing (Du and Rosbash 2001). To eliminate further the possibility that Prp5p associates with pre-mRNA through the U1 snRNP, we tested the effects on GST-Prp5p's ability to pull down pre-mRNA when all RNAs in the extract were degraded by micrococcal nuclease (MNase). MNase was added directly to GST-Prp5p or wild-type Prp5p WCE supplemented with calcium, an essential cofactor for the enzyme's activity, and allowed to react for 45 min at 30°C. EGTA was then added to chelate calcium, thereby inactivating MNase. MNase treatment of GST-Prp5p extract cleaved 99% of the spliceosomal RNAs (Supplemental Fig. S3) and inhibited pre-mRNA splicing (Fig. 5B). Strikingly, GST-Prp5p still efficiently pulls down pre-mRNA from splicing reactions with MNase-treated WCE (Fig. 5B,C). On average, 10% of the input pre-mRNA was selected from reactions with either MNase-treated or untreated WCE, and the levels of selected pre-mRNA averaged 38-fold that of the background control, nuclease-treated wild-type Prp5p extracts. Therefore, intact spliceosomal snRNPs are not required for Prp5p to associate with pre-mRNA. However, it is still possible that some snRNP proteins may be required as these are still present in the MNase-treated extracts.
Prp5p in the CC is functional for formation of an active pre-spliceosome
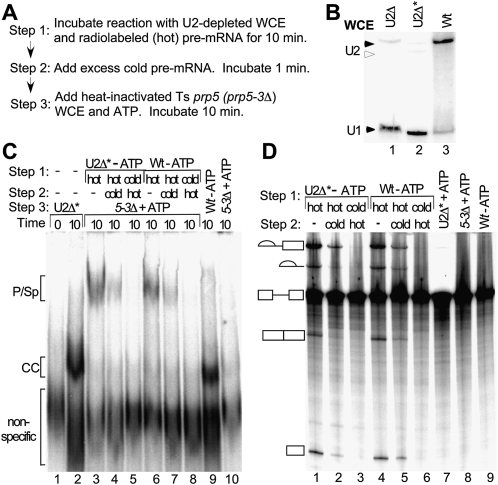
The association of Prp5p with pre-mRNA in the absence of ATP, intact U2 snRNA, or an active helicase core indicates that it is present in the CC. Does Prp5p bind first to the CC and then function in subsequent pre-spliceosome formation? We tested this via a variation of a “commitment-chase” experiment (Fig. 6A; Legrain et al. 1988; Seraphin and Rosbash 1989); wild-type Prp5p in the CC should subsequently promote formation of a functional pre-spliceosome and spliceosome in heat-inactivated Ts prp5 mutant extract that has U2 snRNP but an inactive Prp5 protein. In Step 1 of the experiment to test for functional Prp5p in the committed complex, radiolabeled (hot) pre-mRNA was incubated in reactions with U2-depleted WCE containing wild-type Prp5p for 10 min at 23°C, during which time CC with active Prp5p would form. To ensure that only CC forms, we used WCE from a yeast strain with U2 snRNA metabolically depleted as previously described (Seraphin and Rosbash 1989) and further biochemically depleted U2 by RNase H treatment (Fig. 6B). In Step 2, a 190-fold excess of nonradiolabeled (cold) pre-mRNA was then added, and the reaction was incubated for 1 min. Finally, in Step 3, ATP and heat-inactivated, Ts mutant prp5 WCE were added, and the reaction was incubated for another 10 min. After Step 3, part of the reaction was analyzed by native gel electrophoresis for pre-spliceosomes and spliceosomes. The other part of the reaction was processed to detect spliced radiolabeled RNAs by denaturing PAGE. If wild-type Prp5p is present and relatively stable in the CC and it functions in pre-spliceosome formation, then hot pre-mRNA formed on the CC in Step 1 will be preferentially chased into the pre-spliceosome and spliceosome and subsequently spliced in Step 3. In contrast, if Prp5p is not present in the CC with hot pre-mRNA, then the CC formed on excess cold pre-mRNA in Step 2 will out-compete the hot CC for wild-type Prp5 as well as U2 snRNP, and no hot spliceosomal complexes nor splicing of hot pre-mRNA will be seen.
FIGURE 6.
Prp5p in the commitment complex functions in pre-spliceosome formation. (A) Diagram of commitment-chase experiment. In Step 1, radiolabeled (hot), pre-mRNA was incubated for 10 min at 23°C in reactions with U2-depleted WCE to form commitment complexes. In Step 2, 190-fold excess unlabeled (cold) act pre-mRNA was added, and the reaction was incubated for 1 min. In Step 3, heat-inactivated Ts prp5-3 mutant WCE and ATP were added, and the reaction was incubated for 10 min. After Step 3, part of the reaction was analyzed by native gel electrophoresis, and the other part was analyzed for splicing activity. (B) Northern analysis of U2 RNA in WCEs used in the commitment experiment. RNA was extracted from equal amounts of wild-type WCE (Wt) and WCE from yeast cells metabolically depleted of U2 (U2Δ). U2Δ WCE was treated further by deoxyoligonucleotide-directed RNase H cleavage (U2Δ*) to inactivate any residual U2 before RNAs were extracted. RNAs were measured by Northern blot analysis with U1 and U2 RNA probes. (Solid arrowheads) Intact U2 and U1 RNAs. (Open arrowhead) U2 RNA degraded by RNase H cleavage. RNAse H treatment reduced levels of intact U2 to ≤ 2% that of wild type. (C) Spliceosomal complexes assayed by native gel electrophoresis. In Step 1, hot (lanes 3,4,6,7) or excess cold (lanes 5,8) pre-mRNA was first incubated in reactions with WCEs with wild-type Prp5p. Either U2-depleted (U2Δ*, lanes 3–5) or ATP-depleted wild type (Wt, lanes 6–8) WCE was used. In Step 2, water (lanes 3,6), excess cold pre-mRNA (lanes 4,7), or hot pre-mRNA (lanes 5,8) was added and the reactions were incubated for 1 min for Step 2. In Step 3, ATP and heat-inactivated prp5-3 WCE were added and the reactions were incubated for 10 min (lanes 3–8). Three control reactions were incubated with hot pre-mRNA for 10 min as follows: (lane 2) U2-depleted WCE (U2Δ*); (lane 9) ATP-depleted wild-type WCE; (lane 10) heat-inactivated prp5-3 WCE and ATP. Lane 1 is a “zero” time control in which U2Δ* WCE and splicing buffer are incubated with loading buffer and competitor RNA for 10 min on ice before hot pre-mRNA is added. In this native gel electrophoretic assay, the pre-spliceosomal and spliceosomal complexes comigrate, and CC1 and CC2 comigrate (Ruby 1997). Low levels of CC in the reaction with heat-inactivated prp5-3 WCE (lane 10) did not result from the prp5-3 mutation or interfere with the assay. (D) Splicing assay. In Step 1, hot (lanes 1,2,4,5) or excess cold (lanes 3,6) pre-mRNA was incubated in reactions with WCEs with wild-type Prp5p. Either U2-depleted (U2Δ*, lanes 1–3) or ATP-depleted wild type (Wt, lanes 4–6) WCE was used. In Step 2, water (lanes 1,4), excess cold pre-mRNA (lanes 2,5), or hot pre-mRNA (lanes 3,6) was added and the reactions were incubated for 1 min for Step 2. In Step 3, ATP and heat-inactivated prp5-3 WCE were added and the reactions were incubated for 10 min (lanes 1–6). Control reactions were incubated with hot pre-mRNA for 10 min as follows: (lane 7) U2-depleted WCE and ATP (U2Δ* + ATP); (lane 8) heat-inactivated prp5-3 WCE, and ATP; (lane 9) ATP-depleted wild-type WCE. Pre-mRNA and splicing intermediates and products are indicated as in Figure 1.
When hot pre-mRNA forms CCs with wild-type Prp5p in a committed reaction according to the scenario in Figure 6A, it is subsequently chased into pre-spliceosomes and spliceosomes when inactivated Ts prp5-3 mutant extract and ATP are added (Fig. 6C, lane 4). Note that in this native gel assay, CC1 and CC2 comigrate, and pre-spliceosomes comigrate with spliceosomes (denoted P/Sp) (Ruby 1997). The P/Sp level is at least threefold that in the control reaction in which excess cold pre-mRNA is added first in Step 1 and hot pre-mRNA is added second in Step 2 (Fig. 6C, lane 5); indeed, no detectable CC or other spliceosomal complexes are formed on hot pre-mRNA in this control reaction. The P/Sp level in the committed reaction is also 40% that in an unchallenged reaction in which no excess cold pre-mRNA is added in Step 2 (Fig. 6C, lane 3), so not all hot pre-mRNA in CCs can be chased into active spliceosomes, as we have previously observed (Ruby 1997). Similar levels of P/Sp are observed in reactions in which ATP-depleted wild-type extract is used instead of U2-depleted extract (Fig. 6C, lanes 6,7). These reactions with wild-type extract rule out the possibility that the reduced level of P/Sp in the committed reaction with U2-depleted extract (Fig. 6C, lane 4) is due to residual deoxyoligonucleotide cleaving U2 from the prp5-3 extract added in Step 3. Other controls show that both extracts in Step 1 (U2-depleted extract in the presence of ATP, and ATP-depleted wild-type extract), as well as the heat-inactivated Ts prp5-3 mutant extract, form only CC (Fig. 6C, lanes 2,9,10). We also note that heat-inactivated prp5-3 extract forms low levels of CC (Fig. 6C, lane 10), but additional experiments showed that this did not result from the prp5-3 mutation or interfere with the commitment assay (data not shown).
Analysis of radiolabeled RNAs extracted from the other aliquot of the reactions reveals that some of the hot pre-mRNA in the committed reaction is spliced (Fig. 6C, lane 2). The levels of splicing intermediates (lariat and exon 1) are at least sevenfold to 11-fold higher, respectively, in the committed reaction than in the following control reactions: (Fig. 6C, lane 7) ATP but no inactivated prp5-3 WCE was added; (Fig. 6C, lane 3) cold pre-mRNA was added first in Step 1; or (Fig. 6C, lane 8) hot pre-mRNA was incubated in reactions with only inactivated prp5-3 WCE. A very small amount of spliced lariat product and mRNA can also be seen in the committed reaction (Fig. 6C, lane 2). Additionally no splicing is observed in reactions when hot and excess cold pre-mRNA are added together to active prp5 WCE or when excess cold pre-mRNA is added to either active prp5 or wild-type WCE 1 min before hot pre-mRNA (data not shown). Both committed reactions (Fig. 6C, lanes 2,5) have lower amounts of spliced RNAs than control, unchallenged reactions in which no cold RNA is added (Fig. 6C, lanes 1,4). The differences in levels of splicing in the committed versus unchallenged reactions are consistent with the levels of P/Sp complexes observed by native gel electrophoresis (Fig. 6C). Additionally, similar levels of splicing in committed reactions were also obtained with heat-inactivated prp5-1 mutant WCE (data not shown). Finally, these levels are also comparable to those from previous chase experiments of the CC (Seraphin and Rosbash 1989). The results from our modified commitment-chase experiment including both native gel electrophoresis and splicing assays indicate that wild-type Prp5p in the commitment complex functions for subsequent formation of an active pre-spliceosome.
Prp5p enhances commitment and pre-spliceosomal complex levels
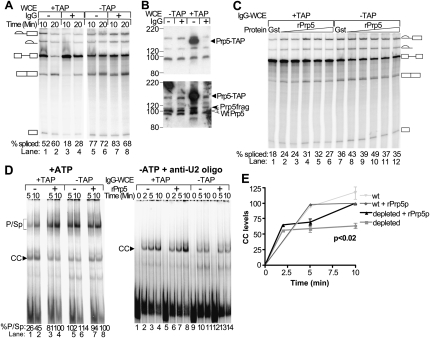
Because Prp5p is present in the commitment complex, perhaps it is required for U1 snRNP binding to pre-mRNA. To test this, we used WCE from the yeast strain with Prp5-TAP to deplete the protein by affinity selection with IgG-Sepharose. WCEs prepared from yeast strains with either the tagged or untagged form of Prp5p were incubated with either IgG-Sepharose or unmodified Sepharose and analyzed for splicing activity (Fig. 7A). IgG-treatment of Prp5-TAP WCE results in a 50%–65% reduction in splicing activity: the levels of spliced RNAs are 18%–28% versus 52%–60% in reactions with Prp5-TAP treated with IgG-Sepharose versus mock-treated with Sepharose only. Splicing reactions with similarly treated WCEs with untagged Prp5p show no difference due to the type of treatment. Splicing activity of the IgG-treated, TAP-tagged extracts is nearly doubled by adding recombinant Prp5p (rPrp5p), whereas added control GST protein has little or no effect (Fig. 7C). Adding rPrp5p to IgG- or mock-treated wild-type WCE as well as to mock-treated Prp5-TAP WCE does not stimulate splicing (Fig. 7C; data not shown). Splicing reactions with rPrp5p added at the highest concentration tested (2 μM) showed somewhat reduced activity for both IgG-treated WCEs. This decrease may be due to some inactive protein present in the preparation or to inhibition at high concentrations.
FIGURE 7.
Decreased Prp5p levels reduce levels of splicing and spliceosomal complexes. (A) Splicing assay of depleted and mock-treated WCEs. WCEs with either TAP-tagged or untagged Prp5p were treated with IgG-Sepharose or mock-treated with unmodified Sepharose. The WCEs were then assayed for splicing activity using radiolabeled wild-type pre-mRNA. The spliced intermediates and products derived from the wild-type act pre-mRNA are depicted as in Figure 1A. The mean percent total spliced RNA from two assays was calculated as described in Materials and Methods and indicated at the bottom of each lane; standard deviations were <10% of the means. (B) Western analysis of depleted and mock-treated WCEs. Samples of WCEs with either untagged or TAP-tagged Prp5 protein and either mock-treated or treated with IgG-Sepharose were fractionated by SDS-PAGE. Prp5 protein was detected by Western blotting and by probing first with anti-CBP antibody to detect the TAP-tag (top panel) and second with anti-Prp5p antibody to detect the Prp5 protein (bottom panel). (Open arrowhead) Wild-type Prp5p. (Solid arrowheads) Prp5-TAP or prp5p fragments derived from Prp5-TAP (Prp5frag). Positions of molecular weight markers are indicated on the left. The blot was not stripped between probings, so the Prp5-TAP signal is stronger than the signals for the wild-type Prp5p due to residual anti-CBP antibody. (C) Complementation of depleted and mock-treated extracts. Recombinant GST (1.6 μM protein) or Prp5p (from 126 nM to 2 μM protein in twofold increments) was added to splicing reactions containing radiolabeled pre-mRNA and IgG-treated WCE (IgG-WCE) with originally either TAP-tagged or untagged Prp5p and incubated for 10 min at 23°C. Total RNA was extracted, fractionated by denaturing PAGE, and visualized with a PhosphorImager. The spliced intermediates and products derived from the wild-type act pre-mRNA are depicted as in Figure 1. The mean percent total spliced RNA from two (lanes 1–6) and three (lanes 7–12) assays was calculated as described in Materials and Methods, and are indicated at the bottom of each lane; standard deviations were <14% (lanes 1–6) and 23% (lanes 7–12) of the means. (D) Native gel electrophoretic assays of spliceosomal complexes in depleted and mock-treated extracts. Radiolabeled wild-type act pre-mRNA was added to splicing reactions with IgG-treated WCEs (IgG-WCE) with either Prp5-TAP or untagged Prp5p and with or without added recombinant wild-type Prp5p. Samples were removed at the times indicated and fractionated by native gel electrophoresis. The complexes were visualized and measured with a PhosphorImager. The pre-spliceosomal and spliceosomal complexes migrate close to one another and appear as one band (p/sp). Only one CC band is formed in this native gel electrophoretic assay. (Left panel) ATP was added to the splicing reactions. (Bottom of each lane) The percentage of RNA in p/sp [p/sp divided by (CC + p/sp)] is listed. (Right panel) The WCEs were incubated with glucose to deplete ATP and a 2′-O-methyl oligonucleotide complementary to U2 RNA to block pre-spliceosome formation before radiolabeled pre-mRNA was added. (E) Quantification of CC levels. CC levels as measured in experiments in the right panel of D were calculated as the percentage of the maximal levels reached in that particular extract supplemented with rPrp5p. The means (±SD) were calculated from three and two experiments for IgG-treated WCEs with either TAP-tag or untagged Prp5p, respectively.
Western analysis shows that depleting TAP-tagged Prp5p from WCE with IgG-Sepharose removes >90% of the tagged Prp5p (Fig. 7B, upper panel; data not shown) as detected with antibody to the tag. However, probing with anti-Prp5 antibody reveals that small amounts of near-full-length Prp5 without the tag remain (Fig. 7B, lower panel). These untagged forms were probably produced by proteolytic cleavage during extract preparation. Northern analysis of RNAs extracted from the IgG-treated and mock-treated WCE shows no differences in the levels of the spliceosomal snRNAs due to the treatments (data not shown). The results of these splicing and complementation assays indicate that under the conditions used for depletion, Prp5p is the only factor that is being significantly reduced in the extract.
Prp5p reduction also results in lowering the levels of pre-spliceosomal and spliceosomal complexes (P/Sp) that can form in the presence of ATP as detected by native gel electrophoresis (Fig. 7D, left panel). The P/Sp complex levels increase with added rPrp5p. Adding rPrp5p to the IgG-treated, wild type, untagged extract (lanes 7,8) or to mock-treated Prp5-TAP WCE (data not shown) makes no difference in the levels of P/Sp complexes. The levels of CC, formed when U1 snRNP binds to the pre-mRNA, are higher in reactions with IgG-treated Prp5-TAP WCE in the absence versus presence of rPrp5p, but much of the CC may be chased into pre-spliceosomal and spliceosomal complexes in reactions with added rPrp5p.
In order to assess any effect on CC levels, we depleted the extracts of ATP and blocked U2 snRNP binding and pre-spliceosome formation by adding a 17-nt 2′-O-methyl oligonucleotide complementary to the branchpoint-recognition region of U2 RNA. This oligonucleotide has previously been shown to form a stable hybrid with U2 RNA and to block pre-spliceosome formation (Abu Dayyeh et al. 2002). In reactions with the blocked, Prp5-depleted extracts, CC levels initially increase similarly to those of parallel reactions with added rPrp5p (Fig. 7D, right panel; Fig. 7E). However, by 10 min, the amounts stop increasing, while those of reactions with rPrp5 protein increase further. By 10 min, CC levels in reactions with depleted extracts reach only 64% (±6% SD) of those in reactions with added rPrp5p. In contrast, adding rPrp5p to the reaction with the control, IgG-treated, wild-type, untagged extract has no effect. Northern blots of native gels probed for the spliceosomal snRNAs show no detectable differences in the level or mobility of either U1 or U2 snRNP due to depletion for two different extracts. The control extract used in Figure 7D has somewhat lower overall levels of U1 snRNP compared to the depleted extract (data not shown). This difference between extracts probably occurred during extract preparation before IgG treatment.
We conclude that Prp5p modestly stimulates CC formation, stabilizes the complex, or both. It promotes pre-spliceosome formation, consistent with the results of the above commitment-chase experiment (Fig. 6) and previous observations of heat-inactivated Ts prp5 mutant WCEs (Ruby et al. 1993). Whether it effects later complexes independent of its affects on the CC and pre-spliceosome could not be determined by this assay.
DISCUSSION
In this study, interactions between the DExD/H-box protein Prp5p and the spliceosome were assayed by measuring radiolabeled, synthetic pre-mRNAs that pull down with tagged Prp5p from in vitro splicing reactions. Several experimental results show that Prp5p associates with an active spliceosome: (1) Prp5p pulls down all RNA species present in an active spliceosome (pre-mRNA, and its splicing intermediates and products); (2) It cosediments with active 40S spliceosomes isolated by glycerol gradient centrifugation; (3) It does not pull down RNAs lacking an intron; (4) it pulls down pre-mRNAs with a mutation in the 5′ splice site or BR less efficiently than wild type pre-mRNA; and (5) reduced levels of Prp5p result in reduced levels of commitment, pre-spliceosome, and spliceosomal complexes. Additional data indicate that Prp5p associates with pre-mRNA during the early stages of spliceosome assembly: (1) Prp5p pulls down pre-mRNA in each of three conditions that cause CC accumulation and block pre-spliceosome formation (ATP-depletion, RNase H degradation of U2 snRNA, and heat-inactivation of the mutant Ts prp5-1 protein); (2) it functions within the commitment complex to form a functional pre-spliceosome; and (3) it associates with pre-mRNA in the absence of intact spliceosomal snRNAs. Finally, Prp5p may be present in the spliceosome after splicing, as it pulls down more lariat intron product than spliced mRNA.
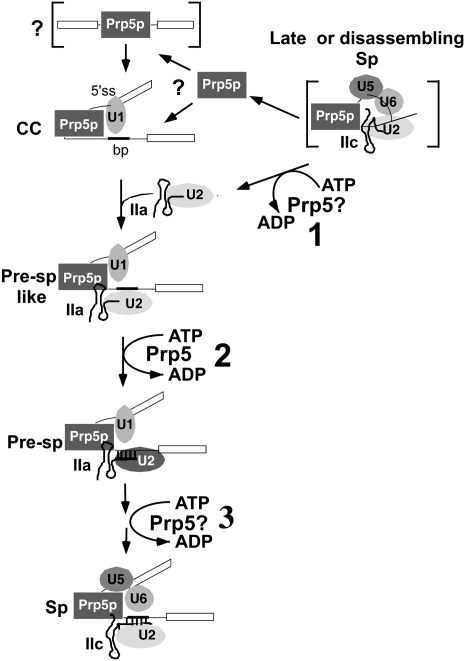
We conclude that Prp5p's association with pre-mRNA occurs before or during addition of U1 snRNP in spliceosome assembly and is sustained through the first and second transesterification reactions (Fig. 8). Although we cannot exclude the possibility that Prp5p is dissociating and reassociating at some stages in the splicing cycle after pre-spliceosome formation, the most parsimonious explanation is that Prp5p associates with pre-mRNA very early in spliceosome assembly and stays in the spliceosome until splicing is completed and the spliceosome disassembles. Therefore, Prp5p is an integral component of the spliceosome. It is like Brr2p, one other spliceosomal DExD/H -box protein that is an integral, spliceosomal component (Lauber et al. 1996). As discussed below, Prp5p also resembles several other spliceosomal DExD/H-box proteins by having multiple functions at various stages in the splicing cycle.
FIGURE 8.
Model of Prp5p in the splicing cycle. In this working model, the DEAD-box Prp5 protein in a late or disassembling spliceosome hydrolyzes ATP to convert U2 RNA from the IIC form to the IIA form (“1” in upper right corner). The bracketed complex indicates that the conversion of U2 could occur anytime after exon ligation through disassembly of the spliceosome. (Left side of diagram) U2 and Prp5 are recycled for a new round of splicing. The bracketed complex with pre-mRNA and Prp5p may precede CC, but the order of Prp5p and U1 snRNP binding to pre-mRNA is uncertain. In the commitment complex (CC), Prp5p may interact with U1 snRNP and other proteins to bridge the 5′ splice site and the UACUAACA box. It may also recruit U2 snRNP via protein–protein interactions in the absence of ATP to form a pre-spliceosome-like complex (Pre-Sp-like) in which U2 RNA is not yet paired with pre-mRNA. Prp5p then hydrolyzes ATP (“2” in the diagram) to form the stable pre-spliceosome (Pre-Sp) with U2 base-paired with pre-mRNA. Finally, Prp5p may have additional ATP-dependent functions after pre-spliceosome formation (“3” in the diagram) and switch U2 between the IIA and IIC forms. Only those splicing factors relevant to this study are shown, and they are arbitrarily arranged relative to one another. Prp5p may or may not bind pre-mRNA directly.
Prp5p may have several ATP-independent functions including recruiting U2 snRNP during pre-spliceosome assembly
Prp5p is a protein component of the CC, but it can also associate with U2 snRNP in the absence of ATP and independently of pre-mRNA (Abu Dayyeh et al. 2002; Xu et al. 2004). Therefore, Prp5p may recruit U2 snRNP to the pre-mRNA during pre-spliceosome formation. An important observation in support of this idea is that inactivated, Ts prp5-1p still efficiently pulls down pre-mRNA (Fig. 3) and forms the CC, but it is defective in pre-spliceosome formation (Ruby et al. 1993). Perriman, Ares, and colleagues (Perriman and Ares 2000; Perriman et al. 2003) found that in the absence of Cus2p, a functional pre-spliceosome can form with some ATPase-defective prp5 mutant proteins, but not with the heat-inactivated mutant prp5-1 protein. Therefore, they proposed that Prp5p has an ATP-independent function at this step in addition to an ATP-dependent one. Our data suggest that the ATP-independent function during pre-spliceosome formation is U2 snRNP recruitment. During recruitment, Prp5p in the CC would first interact with U2 snRNP in an ATP-independent manner to form an intermediate pre-spliceosome-like complex with U1 and U2 snRNPs. This complex would be less stable than the ATP-dependent pre-spliceosome in which U2 and pre-mRNA are base paired. Such an ATP-independent, pre-spliceosome-like complex has been observed in both budding yeast and human splicing assays under certain conditions (Michaud and Reed 1991; Jamison and Garcia-Blanco 1992; Liao et al. 1992; Michaud and Reed 1993; Hong et al. 1997; Perriman and Ares 2000; Stevens et al. 2002; Perriman et al. 2003; Xu et al. 2004).
Although both hPrp5p and ScPrp5p may recruit U2 snRNP to form an ATP-independent pre-spliceosome-like complex, SpPrp5p apparently differs (Xu et al. 2004). SpPrp5p's association with pre-mRNA depends on the integrity of both the U1 and U2 SnRNPs; RNase H degradation of either U1 or U2 inhibits binding of SpPrp5p to pre-mRNA. This suggests that SpPrp5p may bind during pre-spliceosome formation. However, S. pombe splicing extracts contain predominantly a U2/U5/U6 spliceosomal complex (Huang et al. 2002). ATP may be required to release the snRNPs or otherwise remodel the spliceosome in preparation for association with newly added pre-mRNA. In fact, although ATP is required for SpPrp5 to bind to pre-mRNA, ATPase-defective SpPrp5 proteins can associate with U1 and U2 snRNPs.
Our results also indicate interactions between Prp5p and U1 snRNP. The effects of the intronic A1 5′ splice site and A257 branch site mutations on pull-downs of radiolabeled pre-mRNA are similar for Prp5p and CC formation. In ATP-depleted splicing reactions with untreated extracts, GST-Prp5p pulls down 46% or less of wild-type levels for the A1 and A257 mutant RNAs (Fig. 4C). Levels of CC formation on the same mutant RNAs are 44% or less of wild-type levels as assayed by solid-phase splicing (Ruby and Abelson 1988). Furthermore, GST-pull-downs with the S. pombe and human Prp5 proteins showed that they can physically associate with U1 snRNP in the absence of pre-mRNA in vitro (Xu et al. 2004). Although previously we did not observe a similar association using GST-tagged Prp5p in S. cerevisiae (Abu Dayyeh et al. 2002), the C-terminal TAP-tagged Prp5p can pull down U1 snRNP in the absence of added pre-mRNA (Supplemental Fig. S1). Whether Prp5-TAP's interaction with U1 is direct or mediated via some other factor has yet to be tested. It is interesting to note that the U1-binding region in both the human and fission yeast Prp5 maps to the first 170 amino acids of the N-terminal region (Xu et al. 2004), which is apparently lacking in the shorter protein from S. cerevisiae when sequences are compared. Nonetheless, we found that 12 different algorithms (for review, see Oldfield et al. 2005) predict most of the N-terminal region (amino acids 1– 200) of ScPrp5p to be intrinsically disordered or unfolded, and therefore lacks defined secondary and tertiary structure. This predicted disorder is conserved for nearly the entire N-terminal region of Prp5 orthologs. Intrinsically disordered regions in other proteins bind RNA or interact with other proteins (Dunker et al. 2001; Tompa and Csermely 2004).
Whether Prp5 is directly recognizing the 5′ splice site and BR or interacting indirectly with the pre-mRNA via U1 snRNP or other factors also remains to be determined. Prp5p has at least three regions that could potentially bind RNA directly. The helicase domain in other DExD/H-box proteins can bind RNA (Caruthers and McKay 2002). Prp5p's N terminus could encode RNA-binding activity as just discussed. Its C-terminal regions flanking the helicase domain is 230 amino acids and thus large enough to encode RNA-binding activity as well (Dalbadie-McFarland and Abelson 1990). However, we previously found that Prp5p cannot directly pull down an oligonucleotide encoding the BR's UACUAAC box from splicing extracts (Abu Dayyeh et al. 2002). Alternatively, there are several other factors including U1 that could mediate the association. Bbp and Mud2p bind the pre-mRNA's UACUAAC box region in the CC and physically associate with the U1 snRNP Prp40 protein to form a bridge that brings the 5′ splice site and BR together (Abovich and Rosbash 1997; Berglund et al. 1997, 1998). Prp5p could be interacting with this bridge in the CC or even be a part of it. In S. pombe, Prp5p is thought to bridge the U1 and U2 snRNPs in the pre-spliceosome, but this interaction is dependent on an intact U2 snRNP (Xu et al. 2004). Our data suggests that in S. cerevisiae, Prp5p could be part of a bridge in the CC as well as in the pre-spliceosome.
Prp5p may have additional roles in the CC as we also observed that Prp5p stimulates CC levels. If Prp5p precedes U1 snRNP binding, then it may recruit U1 snRNP. Alternatively, if it binds concomitant with U1 snRNP, it may stabilize the CC. Unfortunately, because CC formation is rapid at 23°C and because Prp5p can associate with pre-mRNA even at 0°C, we could not use GST-pull-downs to resolve the order. That Prp5p can associate with pre-mRNA efficiently at 0°C does argue that Prp5p binding precedes U1 base-pairing with pre-mRNA's 5′ splice site and, therefore, CC formation. Neither base-pairing of U1 RNA or CC formation occurs at 0°C (Ruby and Abelson 1988; Du and Rosbash 2001, 2002). Before U1-intron pairing and CC formation, however, U1 snRNP does bind to the 5′ splice site via its proteins as detected by UV-cross-linking of some U1 snRNP proteins to pre-mRNA at 0°C (Du and Rosbash 2002). This protein-mediated U1-pre-mRNA complex formed at 0°C can be chased by increased temperature into the CC (Du et al. 2004). Determining the relationship of Prp5p with this earliest, protein-mediated U1 snRNP-pre-mRNA complex will further elucidate any role that Prp5p may have in the CC.
In summary, Prp5p has at least one ATP-independent activity in that it binds to the CC in the absence of ATP. It may also have additional ATP-independent activities or functions as just discussed. By having one or more ATP-independent activities or functions, Prp5p behaves like several other spliceosomal DExD/H-box proteins. For example, Prp22p has an ATP-independent function before or during the second transesterification reaction (Schwer and Gross 1998; Mayas et al. 2006), as well as ATP-dependent functions for monitoring exon ligation and for release of mature mRNA after the second transesterification reaction (Company et al. 1991; Wagner et al. 1998; Mayas et al. 2006).
Prp5p's ATP-dependent function for pre-spliceosome formation
Previous in vitro experiments showed that Prp5p's ATPase activity is required for pre-spliceosome formation (Perriman and Ares 2000; Perriman et al. 2003). Its ATPase activity has also long been implicated by genetic and biochemical data in catalyzing a conformational switch between two mutually exclusive forms of U2 RNA; stem–loop IIC and stem IIA (Ruby et al. 1993; Wells and Ares 1994; O'Day et al. 1996). These stem–loops begin just 8 nt downstream from the region that base-pairs with pre-mRNA (Zavanelli and Ares 1991; Ruby et al. 1993). IIA but not IIC is active for pre-spliceosome formation, and IIA is more efficient than IIC for base-pairing with pre-mRNA (Zavanelli and Ares 1991; Ruby et al. 1993). Prp5p may catalyze switching from IIC to IIA, thereby activating U2 snRNP for pre-spliceosome formation and for intron pairing. But because it was not known when IIC and Prp5p would be present together, it was not known when Prp5p could catalyze this change. Previously, it has been suggested that Prp5p switches U2 to the IIA form before U2 is incorporated into the pre-spliceosome (Perriman and Ares 2000; Perriman et al. 2003). Our results indicating that Prp5p is present in the spliceosome at both the beginning and end of a splicing cycle suggest two possibilities for when Prp5p executes ATP-dependent functions required for pre-spliceosome formation: at the end of the splicing cycle when the spliceosome is remodeled in preparation for a new round of splicing (Fig. 8, “1”); and within the pre-spliceosome (Fig. 8, “2”). As discussed below, these two possibilities are not mutually exclusive, and, in fact, both may be required.
Because GST-Prp5p pulls down lariat product, Prp5p is likely present in the spliceosome during and just after exon ligation. At this time, U2 is thought to be in the IIC form (Hilliker et al. 2007; Perriman and Ares 2007). Therefore, if Prp5p catalyzes a switch from IIC to IIA, it could do so within the spliceosome sometime after exon ligation and before a new cycle of splicing (Fig. 8, “1”). However, this switching event alone would not suffice for U2 snRNP addition during pre-spliceosome formation in a new cycle. Most U2 is in the IIA form in normal splicing extracts, but ATP hydrolysis by Prp5p is still required at the time of pre-spliceosome formation (Perriman and Ares 2000, 2007). Thus the second, possibility is that Prp5p executes an ATP-dependent function during pre-spliceosome formation, perhaps to allow U2 to base pair. Whether this function involves U2 RNA or some other target is arguable. Prp5's ATPase activity is stimulated more by U2 RNA than by other spliceosomal RNAs or polynucleotides (O'Day et al. 1996). Additionally, ATP and Prp5p stimulate base-pairing of an oligonucleotide to U2 in splicing extracts, suggesting that Prp5p is acting on U2 in an ATP-dependent manner to somehow promote pairing (Abu Dayyeh et al. 2002). Cross-linking studies also indicate that the human U2 snRNP has already undergone a structural change by the time it base-pairs with the intron in the pre-spliceosome (Dybkov et al. 2006). Alternatively, Cus2p is thought to somehow govern Prp5p's ATPase function during pre-spliceosome formation, and it has been proposed that Prp5 may alter Cus2p's association with U2, thereby activating U2 for pre-spliceosome formation (Perriman and Ares 2000; Perriman and Ares 2007). Prp5p could also be like some other DExD/H-box proteins that both remove a protein and catalyze RNA rearrangements (Jankowsky et al. 2000, 2001), but Prp5p has yet to be shown to have these activities (O'Day et al. 1996). Finally, Prp5p's ATPase activity could be required to form a protein–protein interaction as has been recently discovered for another spliceosomal DEAD-box protein, UAP56 (Shen et al. 2008).
Prp5p may have multiple ATP-dependent functions
Prp5p's presence in the spliceosome during most of the splicing cycle is consistent with recent in vivo data suggesting that Prp5 has additional ATP-dependent functions after pre-spliceosome formation (Fig. 8, “3”). Others (Xu and Query 2007) observed from in vivo, genetic experiments that some prp5 mutants with reduced ATPase activity improve splicing of pre-mRNAs with mutations in the UACUAAC box of actin pre-mRNA. They suggest that Prp5p normally catalyzes a conformational change that further stabilizes U2-intron pairing after pairing occurs. Prp5p's ATPase activity has also been shown to be required for accurate splicing during the first transesterification reaction because an ATPase-deficient prp5 mutant shows decreased splicing fidelity at this step (Perriman and Ares 2007). Furthermore, U2 RNA may normally switch between IIA and IIC after pre-spliceosome formation but before the first transesterification reaction of splicing (Perriman and Ares 2007). Yet two more switches may occur in the transition between the first and the second transesterification reactions and just before or after the second transesterification reaction (Hilliker et al. 2007; Perriman and Ares 2007). Although not essential for splicing, IIC and the switching between IIC and IIA are necessary for splicing accuracy and efficiency. Our finding that Prp5p is present in the spliceosome during both the first and second catalytic steps suggests that it could act in one or more of these switching events.
MATERIALS AND METHODS
Plasmids
The following plasmids were previously constructed or obtained from others: pSR284-3 and pSR194-4 (Abu Dayyeh et al. 2002); pSP6-ActΔ6, pSP6-Act-A1, pSP6-Act-A257, pΔIVS and pAct-AS (antisense) (Vijayraghavan et al. 1986); pT7-Act from A. Zaug; pFA64XTAP (Tasto et al. 2001) from K. Gould; pRS413 (Sikorski and Hieter 1990) from R. Sikorski; pGEX4T from GE Life Sciences; and pRSetA from Invitrogen.
Deoxyoligonucleotides (oligos) were chemically synthesized by Sigma-Genosys:
oSR266 (5′-GGTATAGGGTTTCTAGACCACATGATACATGCTC-3′);
oSR268 (5′-GAAGAGTACTAAATACGGCGCCTCTTAAGTTTTTAGAATTTAAC-3′);
oSR270 (5′-CAATAGTTTCCATATGTCAAATAAACCC-3′);
oSR325 (5′-ACTACGGCGCCAAGATGAAGCGACGATGGAA-3′);
oSR326 (5′-CGTCGCTTAAGATTCTTTGCTCACCGAAGGAT-3′);
oHK13 (5′-CGATAGTCTTAAGGTTAGTAGGGTCGACGG-3′);
oHK14 (5′-AATTCCGTCGACCCTACTAACCTTAAGACTAT-3′);
oHK15 (5′-CCCAACTAGTGCTTCTGAAAATCTATATTTTCAAGGTGGTCAT-3′);
oHK16 (5′-CGATGACCACCTTGAAAATATAGATTTTCAGAAGCACTAGTTGGGGTAC-3′);
oHK17 (5′-ATGGTCATCGATTCATGTCCCCTATACTAGG-3′); and
oHK18 (5′-TTTAACCTTAAGATTTTGGAGGATGGTCGCC-3′).
Constructions of plasmids used standard cloning techniques (Sambrook et al. 1989). Plasmid pTK4 encoding GST-prp5-1p was made by ligating the 2.5-kb AatII-SalI fragment of pSR194-4 with the 11.0-kb AatII-SalI fragment of pSR284-3. Plasmid pSR318 expressing the Prp5-TAP protein under control of the PRP5 promoter has the HincII-ClaI PRP5 fragment cloned into the SmaI and ClaI sites of vector pSR413 with the following modifications created by several steps using in vitro mutagenesis and subcloning: (1) a new NdeI site at PRP5 ORF translational start codon with oligo oSR267; (2) HIS3 NdeI site deleted and a new XbaI site created with oligo oSR266; (3) new NarI and AflII sites introduced just before PRP5 stop codon with oligo oSR268; (4) 4XTAP-encoding DNA fragment amplified by PCR with oligos oSR and oSR and inserted between the NarI and AflII sites. The NdeI-SalI PRP5-TAP DNA fragment from pSR318 was subcloned into pRSetA vector cut with NdeI and XhoI to yield plasmid pSR326 for expression in Escherichia coli. Plasmid pHK45 encoding a C-terminal tandem affinity selection tag (TCG, for tandem calmodulin binding peptide and GST separated by a TEV protease site) was constructed from pSR326 in three successive steps. Plasmid pSR326 was cut with ClaI and EcoRI to remove part of the TAP-tag, and hybridized oligos oHK13 and oHK14 were inserted to create pHK43. Plasmid pHK43 was cut with KpnI and ClaI and hybridized oligos oHK15 and oHK16 were inserted at those sites to create pHK44. Plasmid pHK44 was cut with AflII and ClaI, and a DNA fragment encoding GST was inserted. The DNA fragment encoding GST was amplified by PCR using plasmid pGEX4T as template and oligos oHK17 and oHK18. All plasmids resulting from the use of PCR amplified products, mutagenesis, or oligos were sequenced for confirmation.
Yeast strains
TSR489-7-3A [Mata prp5d1815:LEU2 his3d200 leu2-3,-112 ura3-52 (pCEN-URA3-PRP5)] and TSR1673 [MATa prp5d1815:LEU2 his3d200 leu2-3,-112 ura3-52 (pCEN-HIS3-GST-PRP5)] were previously described (Abu Dayyeh et al. 2002). Mutants prp5-1 (RJL172 and SRY5-1c) and prp5-3 (SRY5-3a) were as described (Ruby et al. 1993). Strain BSY88 with the galactose-inducible and glucose-repressible U2 RNA gene was as described (Seraphin and Rosbash 1989). Plasmid shuffling (Sikorski and Boeke 1991) was used to replace the wild-type copy of PRP5 in TSR489-7-3A with the following plasmids: pTK4 to generate TSR2112 expressing GST-prp5-1p; and pSR318 to generate two strains TSR2309-2 and TSR2310 expressing Prp5-TAP.
WCE preparation and treatments
Yeast whole cell splicing extracts (WCE) were prepared using the liquid nitrogen method (Umen and Guthrie 1995; Ruby 1999b). For treating WCEs with IgG-Sepharose (Amersham Biosciences) or unmodified Sepharose 4B-CL (Sigma-Aldrich Chemical Co.), the supernatant obtained after ultracentrifugation was brought to 1 M KCl and added to packed Sepharose equilibrated in AGK buffer [10 mM HEPES-K+ at pH 7.8, 1.5 mM MgCl2, 200 mM KCl, 10% (v/v) glycerol, and 0.5 mM dithiothreitol] supplemented with additional KCl to a final concentration of 1 M. The slurry was mixed gently on a Nutator for 1 h at 0°C, after which the Sepharose was removed by sedimentation in a Sorvall RT6000B centrifuge at 3000 rpm for 10 min at 4°C. The supernatant was dialyzed against Buffer D [20 mM HEPES-K+ at pH 7.5, 0.2 mM EDTA, 0.5 mM DTT, 50 mM KC1, 20% (v/v) glycerol]. Any precipitate was removed by sedimentation in a microfuge at 14,000 rpm for 10 min at 4°C before the final WCE was flash-frozen in liquid nitrogen. IgG- and mock-treated WCEs were made in parallel; twice from strains TSR2309-2 and DSR489-7-3A, and once from strain TSR2310.
Heat inactivation of WCEs from strains TSR2112, RJL172, SRY5-1c, and SRY5-3a was accomplished by bringing WCE to 1.5 mM MgCl2 and 0.2 mM ATP, incubating the mixture for 10–15 min at 38°C, and terminating on ice. In vitro splicing reaction conditions were restored upon adding splicing buffer and adjusted to bring the final MgCl2 concentration to 2 mM.
Targeted cleavage of U1, U2, and U6 snRNA was carried out with deoxyoligonucleotides (deoxyoligos) oSR19, oSR20 (Ruby and Abelson 1988), and d1 (Fabrizio et al. 1989), respectively. oSR4, a control deoxyoligo encoding the phage T7 promoter, was also utilized (Ruby and Abelson 1988). Each 30- or 40-μL cleavage reaction contained 2.5 mM DTT, 75 mM KPO4 (pH 7.4), 3.75% PEG8000 (v/v), 3.75 mM MgCl2, 0.2 mM ATP, 50% WCE (v/v), in 20 mM HEPES K+ (pH 7.8 at 0°C), 0.2 mM EDTA, 50 mM DTT, and 20 mM (v/v) glycerol, and either 5 μM oSR19, 1 μM oSR20, 1 μM d1, 1 μM oSR4, or water. The minimal concentrations of deoxyoligos necessary for degradation of the target U RNAs were determined by titration using in vitro splicing assays (data not shown). After a 15-min incubation at 30°C, D-glucose was added (to 0.2 mM) to the cleavage mixture and incubated for an additional 15 min at 30°C to deplete ATP. Following this second incubation, 10 or 20 μL of reaction mixture were removed and extracted for primer extension analysis. A splicing reaction and subsequent GST affinity selection was carried out upon adding ATP (to 2.0 mM) and fresh 32P-radiolabeled yeast actin pre-mRNA to the remaining cleavage mixture.
Micrococcal nuclease (MNase) digestions were performed and terminated with the addition of EGTA to a final concentration of 2 mM as described (Cheng and Abelson 1986). Water was substituted for MNase in mock-treated reactions. For the reactions in Figure 5, splicing buffer containing 400 μM D-glucose (to deplete ATP) or water was then added to the digest mixture and incubated for an additional 15 min at 30°C. Splicing reaction conditions were restored upon adding ATP (to 2.0 mM) or water (to retain ATP-depleted conditions) and radiolabeled pre-mRNA. RNAs from MNase-treated reactions were extracted using hot phenol and analyzed by Northern blots with snRNA-specific probes as described previously (Ruby 1999a) to ascertain that >99% of the spliceosomal snRNAs were degraded. For Figure 5C, 2 μL of each reaction was counted in a liquid scintillation counter to ensure that equal amounts of radiolabeled pre-mRNA were added to the reactions: the samples differed by 10% or less of the total.
Recombinant Prp5 and GST proteins
An overnight culture of E. coli strain BL21 cells containing plasmid pLysS and either the pHK45 or pGEX4T plasmid was grown at 30°C in 2XYT medium (Miller 1972) supplemented with ampicillin (100 μg/mL), chloramphenicol (200 μg/mL), and thiamine (200 μg/mL). The culture was diluted 20-fold in fresh, supplemented medium and incubated for another 2 h. Isopropyl-β-D-thiogalactopyranoside (IPTG; US Biologicals) was added to 2 mM, and the culture was incubated for 16–20 h at 18°C. The cells were harvested by centrifugation at 8360g for 15 min at 4°C, washed once in ice-cold lysis buffer [50 mM Tris-HCl at pH 7.6, 0.75 M NaCl, 10% (v/v) glycerol, 20 mM β-mercaptoethanol, and 1 mM phenylmethyl-sulfonyl fluoride] and resuspended in 2.5 mL of lysis buffer per gram of wet cell weight. Cell lysis was initiated on ice by adding 1 mL of 0.5 mg/mL lysozyme per gram of wet cell weight and Triton X-100 to a final concentration of 0.1%. Cells were sonicated with four 30-sec bursts in a Branson sonicator. Cell debris was removed by centrifugation at 25,430g for 15 min at 4°C followed by a second centrifugation at 125,730g for 1 h at 4°C. The supernatant was either flash-frozen in liquid nitrogen or immediately used for affinity selection with glutathione-Sepharose that had been prepared by washing three times each with a 10-fold volume of ice-cold 1 M NET (1 M NaCl, 50 mM Tris-HCl at pH 7.6, 0.05% Nonidet-P-40). The slurry was gently mixed for 30 min at 0°C after which the Sepharose was collected by sedimentation in a Sorvall RT6000B centrifuge at 3000 rpm for 10 min at 4°C. The Sepharose was washed three times with 1 M NET and twice with TK9 buffer (150 mM NaCl, 50 mM Tris at pH 9.0). GST-Prp5 was eluted in TK9 buffer supplemented with 1 mM dithiothreitol and 20 mM glutathione. The eluate was cleared of any extraneous beads by filtration through a disposable filter column (200 μm; Fisher Scientific), concentrated in a Millipore Amicon Ultra filter (10,000 kDa molecular weight cut-off) as per the supplier's recommendations, and dialyzed in Buffer D for 2 h. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C. Protein concentration was determined by the Bradford colorimetric assay.
In vitro transcription, splicing, commitment, and native gel electrophoretic assays
Plasmid DNA templates for wild-type, Δ6, and mutant Act RNAs were cut with HindIII to give transcribed pre-mRNAs with exon 1, intron, and exon 2 of 96, 303, and 197 nucleotides, respectively. Plasmid DNA template for antisense RNA was cut with EcoRI. Plasmid DNA templates were also cut with HpaII for native gel assays in Figure 7. Plasmid DNA template was cut with AccI for the AC/AC RNA used for the glycerol gradient sedimentation (Fig. 2) and for Prp5-TAP selection (Supplemental Fig. S1C). In vitro synthesis of 32P-radiolabeled wild-type and mutant yeast actin pre-mRNAs was as described (Ruby 1999b). A lower specific activity AC/AC RNA used in Prp5-TAP selection (Supplemental Fig. S1C) was synthesized (Cheng and Abelson 1987).
In vitro splicing assays were as described (Ruby 1999b). To deplete splicing extracts of ATP, WCE in splicing buffer was incubated with D-glucose (depletion) or water (mock-incubation) for 15 to 30 min at 30°C. A typical ATP-depletion reaction contained 2.5 mM DTT, 75 mM KPO4 (pH 7.4), 3.75% PEG8000 (v/v), 3.75 mM MgCl2, 200–400 μM mM D-glucose, and 50% WCE (v/v) in 20 mM HEPES K+ (pH 7.8 at 0°C), 0.2 mM EDTA, 50 mM DTT, and 20 mM (v/v) glycerol. Subsequent in vitro splicing reactions received added ATP (to 2.0 mM) or water (to retain ATP-depleted conditions) and fresh 32P-radiolabeled pre-mRNA.
Commitment-chase assays used WCE from BSY88 grown in galactose to mid-log phase, after which glucose was added to repress synthesis of U2 RNA during subsequent incubation as described (Seraphin and Rosbash 1989). U2-depleted, BSY88 WCE was incubated with 0.3 mM MgCl2, 0.2 mM ATP, and 200 nM oSR20 for 30 min at 30°C to degrade residual U2 RNA such that 2% or less of wild-type levels of intact U2 RNA remains. Wild-type extract was depleted of ATP by incubation in splicing buffer with 200 μM D-glucose for 30 min at 30°C. Reaction conditions were as described in Figure 6. Radiolabeled and cold pre-mRNAs were added as 1-μL aliquots, with the final reaction volumes in Step 3 being 20 μL. Seven microliter aliquots were analyzed by native gel electrophoresis, and RNAs in the remainder were processed for analysis of splicing by denaturing PAGE.
Native gel electrophoretic assays were as previously described (Ruby 1997, 1999a) except that the competitor RNA was total RNA extracted from yeast WCE rather than from mouse intestine. No differences in levels or types of complexes were observed when these two competitor RNAs were compared in several assays. Splicing reactions for native gels in Figure 6 used normal Act RNA with an exon 2 of 197 nt and those in Figure 7, C and D, used Act RNA with an exon 2 of 54 nt. Splicing extracts for the reactions in the right panel of Figure 7D were depleted of ATP and blocked by adding glucose and the 2′-O-methyl oligonucleotide mU2-0 to final concentrations of 200 μM each and incubating for 15 min at 23°C. Recombinant Prp5p was added where indicated, followed by radiolabeled pre-mRNA to initiate the reaction. The zero time sample was collected as previously described (Ruby 1999a). To ensure that equal amounts of radiolabeled pre-mRNA were added to the reactions, 2 μL of each reaction were removed at the end of the incubation for counting in a liquid scintillation counter: the samples differed by 10% or less of the total.
Tag-affinity selection
Ten microliter samples of splicing reactions were added to an equal volume of packed glutathione-Sepharose 4B beads or IgG-Sepharose (both obtained from Amersham Pharmacia Biotech, Inc.). The beads were prepared by three consecutive washes with ice-cold NET-150 (50 mM Tris-HCl at pH 7.6, 150 mM NaCl, and 0.05% Nonidet P-40). Samples were gently mixed on a Nutator for 30 min (GST-selection) or 1 h (TAP-selection) at 0°C, after which the beads were washed four times each with a 100-fold volume of NET-150. A solution of 50 mM sodium acetate, 10 mM EDTA, 1% SDS, and 10 μg of E. coli tRNA was added to each sample before extraction of RNAs with hot phenol (Ruby 1999a, b). To compare levels of RNA species in GST-selected material to total RNA in splicing reactions, equal amounts of extracted material for GST-selections and splicing reactions were loaded onto denaturing gels and quantified by identical exposure times to PhosphorImager screens. The exposure times used for film autoradiography for figures are stated in the figure legends.
For zero time reaction points and order of addition studies, 9 μL of splicing reaction without added radiolabeled actin pre-mRNA transcript were added to a 10 μL volume of packed glutathione-Sepharose 4B beads on ice. One microliter of radiolabeled transcript was added to this mixture before incubation for 30 min at 0°C.
The effect of diluting the splicing reaction on GST-Prp5p/pre-mRNA selection was assayed by adding 10 μL of splicing reaction mixture to 10 μL packed volume of glutathione-Sepharose beads suspended in various volumes of NET-150: 25×, 50×, and 100× dilutions contained 240, 490, and 990 μL of NET-150, respectively.
Radiolabeled RNA measurement and statistical analyses
Polyacrylamide gels were fixed in a 15% (v/v) methanol, 5% acetic acid solution, and dried before exposure to Molecular Dynamics PhosphorScreens (Molecular Dynamics). PhosphorScreens were scanned using a Molecular Dynamics Storm 840 PhosphorImager or SI PhosphorImager and quantified using Image-Quant software (Macintosh version 1.11 or PC version 5.2). Percent spliced RNA was calculated by the equation 100 ×[(LI+ LP + M + E1)/(LI + LP + P M + E1)], where LI, LP, P, M, and E1 represent lariat intermediate, lariat product, pre-mRNA, mRNA product, and exon 1 intermediate, respectively. Data were analyzed in Minitab (Windows version 13) by the Anderson-Darling test for normal distribution and by both the Bartlett and Levene tests for variance homogeneity. Differences in the means were analyzed using the General Linear Model ANOVA procedure. The means in Figure 1B were compared to zero by one-sample T-tests.
Glycerol gradient sedimentation
Sixty microliter reactions containing 540,000 cpm of AC/AC pre-mRNA were incubated for 15 min at 23°C. A 10-μL aliquot was removed and processed to analyze total radiolabeled RNAs in the splicing reaction by denaturing PAGE. The remaining 50 μL were mixed with 50 μL of gradient buffer without glycerol [50 mM KCl, 2 mM MgCl2, 20 mM HEPES-K+ (potassium-N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) at pH 7.5]. The samples were layered onto a 4.6 mL, linear 11%–23% glycerol gradient, and sedimented by ultracentrifugation in a SW55 rotor (Beckman Instruments) at 50,000 rpm for 90 min as previously described (Chang et al. 1988). Twenty-four fractions of 200 μL each were collected from which 20 μL aliquots were counted by Cherenkov emissions in a liquid scintillation counter. Fractions were then flash-frozen in liquid nitrogen. Proteins in even-numbered fractions (4–24) were precipitated by adding 10 μg of bovine serum albumin (New England Biolabs) and trichloroacetic acid to a final concentration of 5% and by freezing overnight at −20°C. Protein precipitates were neutralized with 2 μL of 2 M Tris base and dissolved in 15 μL of load buffer [62.5 mM Tris at pH 6.8, 2% (w/v) sodium dodecyl sulfate (SDS), 8 M urea, and 0.015% (w/v) each of bromophenol blue and xylene cyanol]. Odd-numbered fractions (7–15) were subjected to GST-selection as described above except that the supernatants were recovered from the beads and extracted to analyze the unselected (total) RNA.
Western analyses
WCE samples or gradient fractions along with protein molecular weight markers (New England Biolabs) were fractionated on a 10% polyacrylamide gel according to Laemli (Sambrook et al. 1989) but supplemented with 8 M urea. Proteins in the gel were transferred to Immobilon-P membrane (Millipore) in Towbin buffer without SDS. Blots were probed with either anti-CBP (calmodulin binding peptide) antibody (Fig. 7; Ghaemmaghami et al. 2003) or rabbit anti-Prp5 antibody #9 (Fig. 2) diluted 1:2000. They were then probed with either goat anti-mouse antibody (Fig. 7) or goat anti-rabbit (Fig. 2) secondary antibody conjugated with horseradish peroxidase (Bio-Rad Laboratories) and diluted 1:10,000. The proteins were detected with ECL-Plus detection reagent (Amersham Biosciences) or Femto detection reagent (Pierce) and autoradiography. The blot in Figure 7 was subsequently probed with rabbit anti-Prp5 antibody #9 and goat anti-rabbit-HRP antibodies.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank D. Ibarra, K. Gruchalla, and J. Stam for technical assistance; J. Weissman for anti-CBP antibody; G. Dalbadie-McFarland and J. Abelson for anti-Prp5 antibody; K. Gould for plasmid pFA64XTAP; A. Zaug for plasmids pT7-Act; D. Bear and W. Kisiel for equipment for glycerol gradients; B. Seraphin and M. Rosbash for yeast strain BSY88; and D. Brow, A. Panganiban, D. Peabody, and J. Summers for discussions and comments on the manuscript. NSF Grants MCB9709915, MCB0220425, and MCB0404738; RAC grants C2201 and C2295; the UNM Foundation; and an ASM Undergraduate Research Fellowship to H.R.K. supported this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1065209.
REFERENCES
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Abu Dayyeh B, Quan TK, Castro MA, Ruby SW. Probing interactions between the U2 snRNP and the DEAD-box protein, Prp5. J Biol Chem. 2002;277:20221–20233. doi: 10.1074/jbc.M109553200. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Berglund N, Colot H, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes & Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody E, Abelson J. The “spliceosome”: Yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- Chang TH, Clark MW, Lustig AJ, Cusick ME, Abelson J. RNA11 protein is associated with the yeast spliceosome and is localized in the periphery of the nucleus. Mol Cell Biol. 1988;8:2379–2393. doi: 10.1128/mcb.8.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-C, Abelson J. Fractionation and characterization of a yeast mRNA splicing extract. Proc Natl Acad Sci. 1986;83:2387–2391. doi: 10.1073/pnas.83.8.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-C, Abelson J. Splicesome assembly in yeast. Genes & Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Dalbadie-McFarland G, Abelson J. PRP5: A helicase-like protein required for mRNA splicing in yeast. Proc Natl Acad Sci. 1990;87:4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Rosbash M. Yeast U1 snRNP-pre-mRNA complex formation without U1snRNA-pre-mRNA base pairing. RNA. 2001;7:133–142. doi: 10.1017/s1355838201001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- Du H, Tardiff DF, Moore MJ, Rosbash M. Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation. Proc Natl Acad Sci. 2004;101:14841–14846. doi: 10.1073/pnas.0406319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Lührmann R. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, McPheeters DS, Abelson J. In vitro assembly of yeast U6 snRNP: A functional assay. Genes & Dev. 1989;3:2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes & Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Bennett M, Xiao Y, Feld Kramer R, Wang C, Reed R. Association of U2 snRNP with the spliceosomal complex E. Nucleic Acids Res. 1997;25:354–361. doi: 10.1093/nar/25.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Vilardell J, Query CC. Pre-spliceosome formation in S. pombe requires a stable complex of SF1–U2AF59–U2AF23 . EMBO J. 2002;21:5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Garcia-Blanco MA. An ATP-independent U2 small nuclear ribonucleoprotein particle/precursor mRNA complex requires both splice sites and the polypyrimidine tract. Proc Natl Acad Sci. 1992;89:5482–5486. doi: 10.1073/pnas.89.12.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes & Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber J, Fabrizio P, Teigelkamp S, Lane WS, Hartmann E, Lührmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- Legrain P, Seraphin B, Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988;8:3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao XC, Colot HV, Wang Y, Rosbash M. Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res. 1992;20:4237–4245. doi: 10.1093/nar/20.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Lustig AJ, Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes & Dev. 1987;1:7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: A family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes & Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Michaud S, Reed R. A functional association between the 5′ and 3′ splice site is established in the earliest pre-spliceosome complex (E) in mammals. Genes & Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Nilsen TW. The spliceosome: No assembly required? Mol Cell. 2002;9:8–9. doi: 10.1016/s1097-2765(02)00430-6. [DOI] [PubMed] [Google Scholar]
- O'Day C, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae PRP5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- Perriman R, Ares MJ. ATP can be dispensable for pre-spliceosome formation in yeast. Genes & Dev. 2000;14:97–107. [PMC free article] [PubMed] [Google Scholar]
- Perriman RJ, Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes & Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Barta I, Voeltz GK, Abelson J, Ares MJ. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby SW. Dynamics of the U1 small nuclear ribonucleoprotein during yeast spliceosome assembly. J Biol Chem. 1997;272:17333–17341. doi: 10.1074/jbc.272.28.17333. [DOI] [PubMed] [Google Scholar]
- Ruby SW. A yeast spliceosome assay. In: Haynes S, editor. Methods in molecular biology. Humana; Totowa, NJ: 1999a. pp. 365–390. [DOI] [PubMed] [Google Scholar]
- Ruby SW. Yeast pre-mRNA splicing extracts. In: Haynes S, editor. Methods in molecular biology. Humana; Totowa, NJ: 1999b. pp. 323–349. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Abelson J. An early hierachic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988;242:1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes & Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Rosbash M. Identification of functional U1 sn-RNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes & Dev. 2008;22:1796–1803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RJ, Boeke J. In vitro mutagenesis and plasmid shuffling: From cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1990;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasto JJ, Carnahan RH, McDonald WH, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe . Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Umen J, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U, Parker R, Tamm J, limura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathways, particuarly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Ares M., Jr Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae . Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest D, O'Day C, Abelson J. In vitro studies of the Prp9•Prp11•Prp21 complex indicate a pwathway for U2 small nulcear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Lührmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 2004;23:376–385. doi: 10.1038/sj.emboj.7600050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavanelli MI, Ares M. Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes & Dev. 1991;5:2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]