Abstract
Expression of mitochondrial genomes in Kinetoplastida protists requires massive uracil insertion/deletion mRNA editing. The cascade of editing reactions is accomplished by a multiprotein complex, the 20S editosome, and is directed by trans-acting guide RNAs. Two distinct RNA terminal uridylyl transferases (TUTases), RNA Editing TUTase 1 (RET1) and RNA Editing TUTase 2 (RET2), catalyze 3′ uridylylation of guide RNAs and U-insertions into the mRNAs, respectively. RET1 is also involved in mitochondrial mRNA turnover and participates in numerous heterogeneous complexes; RET2 is an integral part of the 20S editosome, in which it forms a U-insertion subcomplex with zinc finger protein MP81 and RNA editing ligase REL2. Here we report the identification of a third mitochondrial TUTase from Trypanosoma brucei. The mitochondrial editosome-like complex associated TUTase (MEAT1) interacts with a 20S editosome-like particle, effectively substituting the U-insertion subcomplex. MEAT1 and RET2 are mutually exclusive in their respective complexes, which otherwise share several components. Similarly to RET2, MEAT1 is exclusively U-specific in vitro and is active on gapped double-stranded RNA resembling editing substrates. However, MEAT1 does not require a 5′ phosphate group on the 3′ mRNA cleavage fragment produced by editing endonucleases. The functional RNAi complementation experiments showed that MEAT1 is essential for viability of bloodstream and insect parasite forms. The growth inhibition phenotype in the latter can be rescued by coexpressing an RNAi-resistant gene with double-stranded RNA targeting the endogenous transcript. However, preliminary RNA analysis revealed no gross effects on RNA editing in MEAT1-depleted cells and indicated its possible role in regulating the mitochondrial RNA stability.
Keywords: Trypanosoma, mitochondria, RNA editing, TUTase, RNAi
INTRODUCTION
Post-transcriptional processing of most trypanosomal mitochondrial mRNAs includes U-insertion/deletion editing as an essential step in producing translation-competent messenger RNAs. The cascade of mRNA cleavage, U-insertion or U-deletion, and RNA ligation is directed by trans-acting gRNAs (Blum et al. 1990) and catalyzed by multiple enzymes assembled into a stable complex of ∼20 polypeptides, the 20S editosome (for reviews, see Stuart et al. 2005; Aphasizhev and Aphasizheva 2008). Pre-mRNA substrates for editing are thought to be produced by a nucleolytic partitioning of multicistronic maxicircle transcripts. Guide RNAs are transcribed primarily from the minicircle DNA and are 3′-urdylalted prior to entering into editing reactions. In Trypanosoma brucei, three distinct forms of the 20S editosome are distinguished by the presence of RNase III-type endonucleases responsible for mRNA cleavage at U-insertion (Carnes et al. 2005), U-deletion (Trotter et al. 2005), and unique cis-guided (Carnes et al. 2008) editing sites. Two homologous U-specific exonucleases (Kang et al. 2005), two RNA ligases (Schnaufer et al. 2001; Cruz-Reyes et al. 2002), and a set of structural proteins were reported in most of the 20S editosome preparations (Panigrahi et al. 2006).
In contrast to other types of enzymatic reactions born by the 20S editosome, the U-insertion activity has been attributed to a nonredundant terminal uridylyl transferase, RNA editing TUTase 2, RET2 (Aphasizhev et al. 2003c). In addition to the 20S editosome's activities, editing depends on guide RNAs that must be 3′-uridylylated by the RNA editing TUTase 1, RET1 (Aphasizhev et al. 2002); stabilized by binding to the gRNA binding complex (GRBC) (Weng et al. 2008); and likely subjected to other operations. Finally, polyadenylation (Etheridge et al. 2008b) and, possibly, recruitment of additional RNA binding proteins (Vondruskova et al. 2005; Hashimi et al. 2008) are required for stabilization of edited mRNAs.
Insertion and deletion cascades are functionally and spatially separated within the 20S editosome: the tripartite U-insertion subcomplex is composed of MP81 zinc finger protein, REL2, and RET2; and the U-deletion subcomplex includes MP63 zinc finger protein, REX1 and/or REX2 exonucleases, and REL1 RNA ligase (Cruz-Reyes et al. 2002; Aphasizhev et al. 2003c; Ernst et al. 2003; Schnaufer et al. 2003). Inhibition of RET2 expression by RNAi in T. brucei effectively abolished U-insertion activity and decreased the abundance of MP81 and REL2, ultimately inhibiting editing in vivo. Remarkably, the rest of the 20S editosome and its U-deletion activity remained intact (Aphasizhev et al. 2003c). Conditional gene knockout in the bloodstream form reiterated the singular role of RET2 as carrier of the U-insertion mRNA editing activity (Deng et al. 2005).
Unlike RET2, RET1 is a multifunctional enzyme participating in more complex interactions (for review, see Aphasizhev and Aphasizheva 2008). Synthesis of the oligo[U] tail at the 3′ end of gRNAs (Aphasizhev et al. 2003c), and possibly ribosomal RNAs (Adler et al. 1991), and uracil incorporation into mRNA's the poly(A) tails (Decker and Sollner-Webb 1990; Etheridge et al. 2008a) are thought to be accomplished by RET1. Uridylylation is likely to have pleiotropic effects on the turnover of mitochondrial RNAs ranging from stabilizing gRNAs and rRNAs to stimulating mRNA decay.
We have previously noted that the trypanosomal RNA editing TUTases RET1 and RET2 belong to an extended family in which five predicted candidates were termed TUT3–TUT7 (Aphasizhev 2005). Indeed, the cytosolic TUT3 (Aphasizhev et al. 2004) and TUT4 (Stagno et al. 2007b) have been characterized as RNA uridylyl transferases, while TUT5 (KPAP1) (Etheridge et al. 2008a) and possibly TUT6 (KPAP2) (Kao and Read 2007), were shown to be mitochondrial poly(A) polymerases. Beyond Kinetoplastida, data mining of eukaryotic genomes identified “noncanonical” PAPs, such as animal cytoplasmic Gld-2-type and yeast nuclear “quality control” Trf4/5 poly(A) polymerases, as the proteins most closely related to TUTases. Further cross-searches of trypanosomal genomes led to identification of two nuclear noncanonical poly(A) polymerases, ncPAP1 and ncPAP2, in T. brucei (Etheridge et al. 2008b). The protein sequence of the remaining uncharacterized member of the TUTase-ncPAP family, TUT7, showed maximum similarity to cytosolic poly(A) polymerases such as Gld-2 (Wang et al. 2002).
Here we report that TUT7, renamed mitochondrial editosome-like complex associated TUTase (MEAT1), is a mitochondrial uridylyl transferase essential for T. brucei viability in insect and bloodstream developmental forms. In the mitochondrial extract, MEAT1 exists as an unassociated protein and a component of the 20S editosome-like particle. We find that the foremost difference between reported variants of the 20S editosome (Panigrahi et al. 2006) and the MEAT1-associated complex is a nearly complete replacement of the U-insertion subcomplex by MEAT1 in the latter. The recombinant MEAT1 and its purified native complex are active in U-insertion assays in vitro, although less so than RET2 and the 20S editosome. Repression of MEAT1 by RNAi did not cause a gross inhibition of editing in vivo, as was the case for most core editosome components, but had rather stabilizing effects on the abundance of mitochondrial RNAs.
RESULTS
Identification of MEAT1
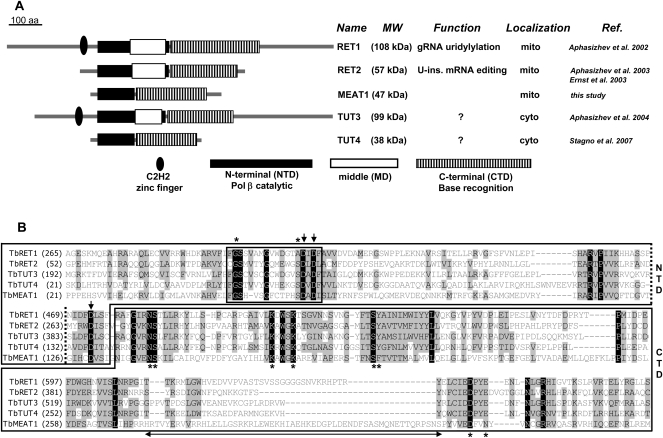
Protein sequences of RET1, RET2, and noncanonical poly(A) polymerases were used in iterative Blast searches to define a family of TUTase-like proteins in T. brucei (Aphasizhev 2005; Etheridge et al. 2008b). The domain organization of MEAT1 resembles that of the minimal TUTase TUT4 (Stagno et al. 2007b), highlighting the lack of the middle domain. This module is present in TUT3 (Aphasizhev et al. 2004) and editing TUTases and is essential for RET1 (Aphasizheva et al. 2004) and RET2 (G-E Ringpis and R Aphasizhev, unpubl.) enzymatic activities. Protein sequences of the middle domains are divergent among RET1, RET2, and TUT3, but the positioning within the N-terminal domain (Aphasizhev et al. 2002, 2004; Aphasizheva et al. 2004; Deng et al. 2005) is conserved (Fig. 1A). By analogy to RET2 and TUT4 (Deng et al. 2005; Stagno et al. 2007b), MEAT1's middle domain is seemingly replaced by a short loop connecting two β-sheets in the N-terminal domain (NTD), which is common to all members of the DNA polymerase β (Pol β) superfamily (Holm and Sander 1995). However, an ∼56-amino acids-long insertion within the C-terminal domain (CTD) may indicate a MEAT1-specific domain (Fig. 1B). A partial multiple sequence alignment illustrates that amino acid residues responsible for UTP binding and catalysis in RET1 (Aphasizheva et al. 2004), RET2 (Deng et al. 2005), and TUT4 (Stagno et al. 2007b) are mostly invariant among TUTases (Fig. 1B). The overall similarity is nonetheless limited: sequence identity between RET2 and MEAT1, for example, is only 12%.
FIGURE 1.
Domain organization of trypanosomal TUTases. (A) Domains are diagrammed based on crystal structures of RET2 (Deng et al. 2005) and TUT4 (Stagno et al. 2007b). (B) Partial multiple sequence alignment of N-terminal (NTD) and C-terminal (CTD) domains. Middle domain boundaries within NTD are shown by dotted lines. Protein sequences used in the alignment are TbRET1 (AAK38334), TbRET2 (AAO63567), TbTUT3 (XP_822966), TbTUT4 (DQ923393), and TbMEAT1 (FJ561337). Signature motif of the Pol β nucleotidyl transferase superfamily is boxed. Identical amino acids are white on black; blocks of similar amino acids are black on a gray background. Amino acids participating in UTP binding by TUTases are indicated by asterisks. Metal-coordinating catalytic carboxylate residues are shown by arrows. Double-headed arrow underlines a MEAT1-specific insertion within the CTD.
MEAT1 is a mitochondrial protein
Topological predictions of MEAT1's subcellular distribution indicated ∼70% probability of its mitochondrial targeting (MITOPROT, http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html).
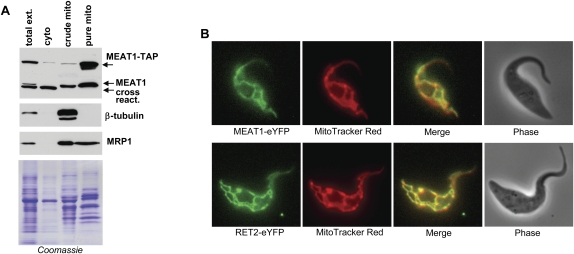
Western blotting of T. brucei subcellular fractions with antigen-purified polyclonal antibodies demonstrated that MEAT1 is indeed localized in the mitochondrial matrix. As shown in Figure 2, MEAT1 was enriched in the Renografin density gradient-purified mitochondrial fraction (Aphasizhev and Aphasizheva 2007). A subunit of the MRP1/2 mitochondrial RNA binding complex (Koller et al. 1997) showed a similar distribution, indicating that MEAT1 is localized to the mitochondrial matrix. The cytoskeleton (β-tubulin) and apparently cytosolic cross-reacting proteins were not detected in purified mitochondria. MEAT1 localization in live T. brucei cells was also examined by expressing its C-terminal fusion with TAP tag (Puig et al. 2001) from a tet-repressor controlled promoter in the 29-13 T. brucei cell line (Wirtz et al. 1999). Tagged protein cofractionated with the endogenous MEAT1, further confirming its mitochondrial location. The mitochondrial proteome project defines MEAT1 (Tb927.1.1330) as a mitochondria-enriched polypeptide (http://www.trypsproteome.org).
FIGURE 2.
Mitochondrial localization of MEAT1. (A) Subcellular fractionation of procyclic T. brucei. Approximately 10 μg of protein from hypotonic total cell extract, cytosolic (soluble) and crude membrane (pellet fraction obtained by centrifugation of the total cell lysate), and the mitochondria purified by flotation in Renografin density gradient were separated on SDS-PAGE. Western blotting was performed with antibodies against MEAT1, mitochondrial (MRP1), and cytoskeleton (β-tubulin) proteins. Coomassie-stained gel shows approximately equal protein loading in crude membrane and purified mitochondrial fractions. (B) Intracellular distribution of MEAT1. The C-terminal MEAT1- and RET2-eYFP fusion proteins were expressed in pLew79-based vector (Wirtz et al. 1999). Protein expression was induced with tetracycline for 96 h. Fluorescent images of live cells were captured in the presence of MitoTracker Red CMX ROS stain.
To assess the submitochondrial distribution of MEAT1 and the 20S editosome, the RET2 and MEAT1 TUTases were expressed in procyclic T. brucei as C-terminal fusions with the enhanced Yellow Fluorescent Protein (eYFP). The colocalization of both proteins with the membrane potential-dependent MitoTracker Red CMX-Ros shows a uniform distribution of MEAT1 and RET2-containing editing complex throughout the mitochondrial matrix (Fig. 2B). Interestingly, polyadenylation complex was previously observed at two punctate antipodal regions adjacent to the kDNA disk (Etheridge et al. 2008a), while the mitochondrial RNA binding complex 1 (MRB1) localized to discrete spots in the proximity of the kinetoplast (Hashimi et al. 2009). MRB1 is virtually identical in protein composition to GRBC, which is essential for gRNA stability (Weng et al. 2008).
MEAT1 is a U-specific transferase active on single-stranded and double-stranded RNAs
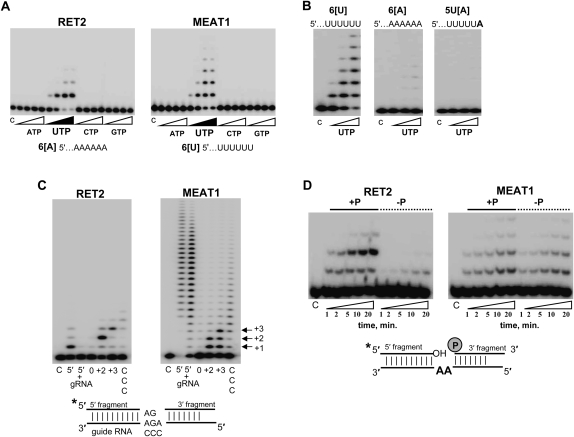
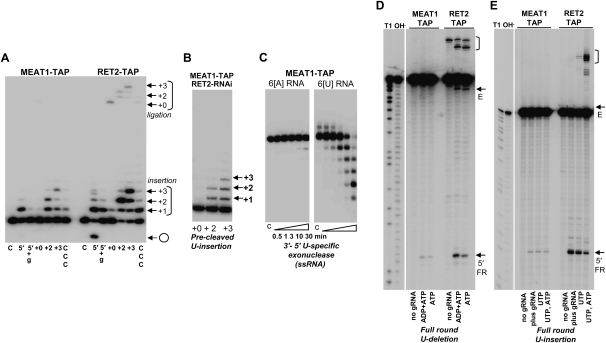
RNA uridylyl transferases display a range of nucleotide triphosphate specificities in vitro: from RET2's virtually absolute specificity for UTP (Deng et al. 2005; Aphasizhev and Aphasizheva 2007) to moderate C-incorporation by TUT4 (Stagno et al. 2007b) to equally efficient ATP/UTP polymerization by Cid1 (Read et al. 2002). The recombinant MEAT1 purified from E. coli showed an exclusive UTP specificity in the presence of Mg2+ ions (Fig. 3A), but distinctly different elongation patterns and requirements for single-stranded RNA (ssRNA) substrates. At UTP concentrations above 10 μM, MEAT1 added multiple Us to ssRNA, while RET2's activity was limited to a single nucleoside incorporation. Mutation of the metal-binding acidic residue in the catalytic site (D67A) inactivated UTP polymerization, confirming the enzymatic identity of MEAT1 (data not shown). We have reported previously that RET2 prefers ssRNA substrates ending with purines and is virtually inactive on RNA with six uridines at the 3′ end. The replacement of the terminal uracil in 6[U] RNA (Fig. 3) with adenosine fully restored RET2 activity to that observed on 6[A] RNA (Stagno et al. 2007a). The opposite effects were observed for MEAT1: virtually no UMP incorporation into 6[A] RNA compared with 6[U] RNA. Replacement of the terminal uracil with an adenosine inhibited activity, indicating that the terminal base's interactions are critical for the productive RNA binding (Fig. 3B).
FIGURE 3.
Substrate specificity of the purified MEAT1. (A) Nucleotide triphosphate substrate specificity. C indicates control RNA. Reactions were carried out for 30 min in the presence of 1, 10, 100, and 500 μM of respective NTPs; 10 nM of each enzyme; and 100 nM of chemically synthesized RNAs. Products were separated on 15% acrylamide/8 M urea gels. (B) Effect of the terminal ssRNA nucleotides on MEAT1 polymerization activity. Reactions were set up as in A at 1, 10, and 100 μM of UTP. (C) Precleaved U-insertion editing assay. RNA substrate was assembled by heating and slow cooling of the radiolabeled 5′ fragment, guide RNA, and 3′ fragment and at 1:1.5:2 molar ratios, respectively. Guide RNAs were designed to direct zero, two, or three U-insertions or to have three cytidines in guiding positions. Reactions containing 100 μM of UTP, 100 nM of 5′ fragment, and 50 nM of enzyme were incubated for 30 min. 5′ indicates 5′ fragment; 5′+g, only 5′ fragment and gRNA were assembled; 0, +2, and +3, number of guiding nucleotides; and CCC, cytidines instead of guiding purines. (D) Reactions were done as in C for +2 guiding, except with the dephosphorylated 3′ fragment in the “–P” panels.
Editing substrates that resemble 5′ and 3′ mRNA cleavage fragments tethered by gRNA after the endonucleolytic cleavage (Fig. 3C; Igo et al. 2000) have been instrumental in dissecting enzymatic activities of the 20S editosome and studies of individual editing enzymes. Recombinant RET1 is active to some extent on dsRNA and is highly processive on ssRNAs (Aphasizhev et al. 2002). Purified RET2 adds Us according to the number of guiding nucleotides and requires base-pairing of the newly added uracil with purine on the opposite strand for incorporation of the next nucleoside. Substitution of the guiding purines to cytidines or omission of the 3′ cleavage fragment inhibited RET2 reaction (Fig. 3C, left panel). Under the same conditions, U-insertion by MEAT1 efficiently terminated at positions specified by the gap size, +2 or +3 as shown in Figure 3C, right panel, but intermediate extension products accumulated at higher levels than in RET2-catalyzed reaction.
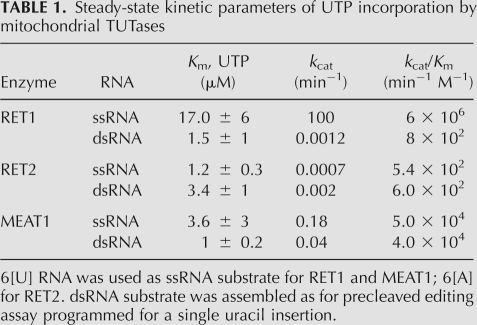
To compare catalytic properties of mitochondrial TUTases, we have determined steady-state kinetic parameters for RET1 and MEAT1 on 6[U], and for RET2 on 6[A] ssRNAs. In addition, the apparent Km and rate constants for UMP incorporation into dsRNA (+1 insertion into precleaved substrate) were examined for all three TUTases (Table 1). The Km values for UTP were confined to ∼1–10 μM range in all cases. The catalytic rate constant, and therefore, catalytic efficiency of RET2 was found to be similar for ssRNAs and dsRNAs, as was the case for MEAT1. However, the absolute values were higher by ∼100-fold for MEAT1. As expected, catalytic efficiency of RET1 on ssRNA exceeded that for dsRNA by approximate four orders of magnitude. To conclude, NTP and RNA substrate specificities of MEAT1 are more consistent with that of TUTase acting on double-stranded RNA (dsRNA) substrates.
TABLE 1.
Steady-state kinetic parameters of UTP incorporation by mitochondrial TUTases
RET2 and MEAT1 are both active on dsRNA and belong to the Pol β superfamily (Holm and Sander 1995). Gapped dsRNA editing substrate resembles an intermediate in abasic DNA site repair that is recognized by the DNA Pol β (Beard and Wilson 2006). In the DNA substrate, the 5′ phosphate group at the 3′ fragment is required for enzyme binding (Prasad et al. 1994). The phosphate group reportedly stimulated U-insertion by partially enriched editing complex (Igo et al. 2000), raising the question of whether individual RET2 or MEAT1 requires phosphate at the 3′ fragment for activity. As shown in Figure 3D, removal of the phosphate abrogated +2 guided additions by RET2 and also inhibited the rate of nonguided +1 insertion by approximately fivefold. In contrast, lack of a phosphate had no discernable effect on MEAT1-catalyzed reaction. Hence, it is plausible that the effects observed by Igo et al. (2000) were produced by the overlapping activities of RET2 and MEAT1 in partially purified editing complexes. Alternatively, other editosome subunits may compensate for poor binding of RET2 to the gapped dsRNA lacking a phosphate group.
MEAT1 interacts with RNA editing ligase 1-containing complexes
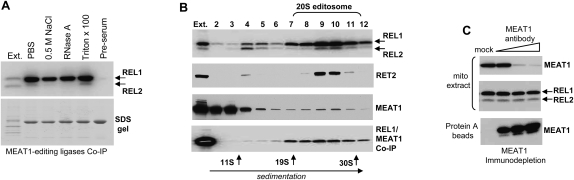
RNA editing ligases are capable of covalent self-adenylation in the presence of [α-32P]ATP, which provides a method for highly-sensitive detection of editing complexes. To assess interaction of MEAT1 with RNA ligase-containing complexes, immunoprecipitations (IPs) with antigen-purified anti-MEAT1 antibody were performed in an extract from highly enriched mitochondrial fraction (Fig. 4A). RNA ligases in the IP material were de-adenylated by incubation with 10 mM of inorganic pyrophosphate during IP and were self-adenylated on the beads. Co-IP of MEAT1 under high-salt conditions and in the presence of RNase A demonstrated a stable, RNA-independent association of MEAT1 with RNA editing ligase 1; only traces of REL2 were detected in the same material.
FIGURE 4.
MEAT1 interactions in mitochondrial extract. (A) Coimmunoprecipitation in the mitochondrial extract. Washes were for 30 min at room temperature in phosphate-buffered saline with 0.1 mg/mL of BSA + 0.1% of Triton X 100 (PBS), or with addition of RNase A to 0.1 mg/mL. For high-salt or Triton X 100 washes, the sodium chloride and detergent were adjusted to 0.5M and 1%, respectively. RNA ligases were detected by exposing the nitrocellulose membrane to a phosphor storage screen. SDS-PAGE was stained with Coomassie to show equal antibody load in immunoprecipitations. Approximately 5 μg of protein from mitochondrial extract was loaded as positive control; “Ext.” Protein A beads saturated with pre-immune serum were used as negative control. (B) Glycerol gradient sedimentation analysis of MEAT1 complexes. Mitochondrial extract was loaded on 10%–30% glycerol gradient formed in SW41 (Beckman) tubes and centrifuged at 50,000g for 20 h. Each fraction was pre-adenylated and tested for RNA ligases. MEAT1 and RET2 were detected by Western blotting. Half of each fraction (300 μL) was used for immunoprecipitation under conditions designated “PBS” in A. (C) MEAT1 immunodepletion. Protein A beads were saturated with purified MEAT1 antibodies and added to mitochondrial extract at increasing amounts and removed by centrifugation after 1 h of incubation. REL1 and REL2 were monitored in the extract by adenylation; MEAT1 was probed in the extract and in the SDS elution off the Protein A beads. See Materials and Methods for details.
The core editing complex sediments in 10%–30% glycerol gradient as ∼20S particle and has an estimated molecular mass of ∼1.6 MDa (Rusche et al. 1997). The apparent molecular mass of MEAT1, as determined by gel-filtration on Sepharose 12 column closely matched the predicted value for the His-tagged polypeptide (∼50 kDa), indicating that the recombinant protein is a monomer (data not shown). We next analyzed size distribution of MEAT1-associated complexes. Mitochondrial extract was separated on 10%–30% glycerol gradients and each fraction pre-adenylated, separated on SDS-PAGE, transferred to membrane, and probed for RET2 and MEAT1. In addition, IPs with anti-MEAT1 antibody were performed in each fraction, and coprecipitated material was assayed for RNA editing ligases (Fig. 4B). In contrast to the 20S editosome-confined RET2, the major fraction of MEAT1 protein sedimented at the 5–10S region while ∼3%–5% of the protein, as determined by densitometry, co-sedimented with the 20S editosome. Co-IP of MEAT1-REL1 was also detected in the 20–30S range. Thus, in mitochondrial extract, only a minor portion of the total MEAT1 pool interacts with REL1-containing complexes.
In the reciprocal experiment, we asked if a significant portion of REL1-containing complexes interacts with MEAT1. As seen from Figure 4C, no appreciable loss of REL1 or REL2 self-adenylation signal was detected in mitochondrial extract upon nearly complete immunodepletion of MEAT1. To conclude, the major fraction of MEAT1 protein is not associated with high molecular mass particles, while a minor portion stably interacts with REL1-containing complex.
MEAT1 and RET2 belong to distinct protein complexes that share several subunits
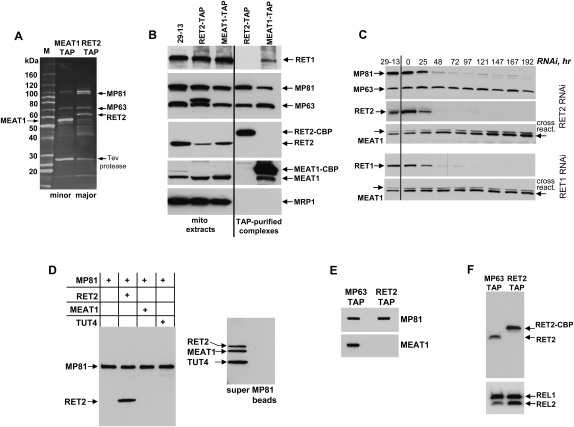
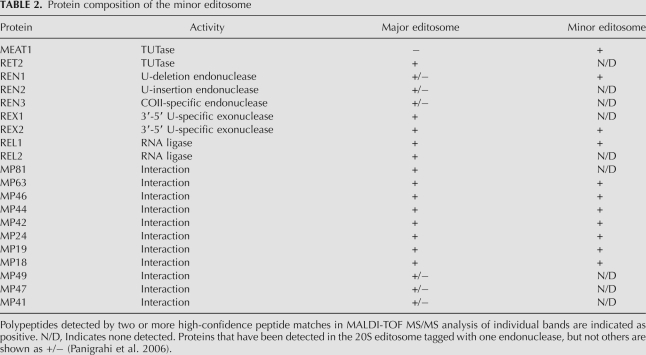
To identify protein components of the MEAT1-associated complex and to compare its composition with the 20S editosome, TAP-tagged MEAT1 and RET2 were overexpressed under control of an inducible promoter in procyclic T. brucei. Tandem affinity purifications were performed from highly enriched mitochondria, and the final fractions were separated on SDS-PAGE (Fig. 5A). Individual bands were excised from the MEAT1 lane, digested with trypsin, and subjected to MALDI-TOF MS/MS analysis (Table 2). In addition, the entire complex digest was analyzed by nanoLC-MS/MS analysis. Work by Panigrahi et al. (2006) demonstrated that depending on which endonuclease is TAP-tagged for purification, certain proteins are not present in the resultant complex. For example, REN1-tagged 20S editosome did not contain REN2, MP67, MP 49, and MP47, which was interpreted as existence of three forms of the 20S editosome specific for U-insertion, U-deletion, and cis-guided U-insertion sites (Carnes et al. 2008). Subunits of the U-insertion subcomplex (MP81, RET2, and REL2) have been reported in all affinity purifications of the 20S editosome. These proteins, however, were not detected in the MEAT1-associated complex, while several other editosome subunits have been identified with high confidence. This is consistent with the absence of REL2 in the MEAT1-coimmunoprecipated material (Fig. 4A).
FIGURE 5.
Purification and analysis of the MEAT1-associated complex. (A) Peak fractions from MEAT1 and RET2 tandem affinity purifications were separated on 8%–16% SDS acrylamide gel and stained with Sypro Ruby. (B) Approximately 10 μg of protein from mitochondrial extracts used for TAP procedure and from the parental cell line (29-13) were separated beside the same fractions as in A and probed on immunoblots with monoclonal antibodies against MP63 and MP81 simultaneously, and then sequentially with polyclonal antibodies against MEAT1, RET2, RET1, and MRP1. The duplet band observed in RET2-TAP was most likely produced by secondary antibody binding to Protein A moiety in the TAP tag. (C) T. brucei cells collected at indicated time points after RET1 and RET2 RNAi induction were analyzed by Western blotting for changes in MEAT1 abundance. (D) In vitro binding of purified RET2, MEAT1, and TUT4 to MP81 synthesized in TNT system. Protein A beads were precoated with monoclonal anti-MP81 antibody and incubated with mixtures containing recombinant proteins and the reticulocyte extract with MP81 expression plasmid (left). No MP81 added to the mixture of all three TUTaes (right). Bound proteins were eluted off the beads with SDS loading buffer and analyzed by Western blotting with anti-6His tag antibody. (E) TAP-tagged MP63 was expressed and purified from T. brucei under the same conditions as RET2 and MEAT1 complexes and probed with anti-MEAT1 antibody in reference to purified RET2 complex. Protein samples were normalized by immunoblotting with anti-MP81 antibody. (F) Same samples as in E were incubated with [α-32P]ATP, separated on SDS-PAGE, transferred to nitrocellulose membrane, exposed to phosphor storage screen, and treated with anti-RET2 antibody.
TABLE 2.
Protein composition of the minor editosome
To corroborate mass-spectrometry data, purified complexes were probed by Western blotting alongside mitochondrial extracts from the parental cell line and cells expressing either RET2-TAP or MEAT1-TAP (Fig. 5B). Remarkably, no traces of RET2 were detected in the MEAT1 complex and vice versa, suggesting that these proteins are associated with their respective complexes in a mutually exclusive manner. MP81 was detected in the MEAT1 complex by Western blotting. In comparison to the 20S editosome, however, its abundance was decreased by approximately fourfold, as determined by densitometry relative to MP63 (data not shown). The MRP1 subunit of the MRP1/2 RNA binding complex, which is loosely associated with the 20S editosome (Aphasizhev et al. 2003b), was not present in either complex, attesting to the stringency of purification conditions. RET1, however, was selectively present as a substoichiometric band in the MEAT1-TAP isolated fraction.
Repression of individual subunits often results in loss of the entire 20S editosome (Babbarwal et al. 2007; Tarun et al. 2008) or subcomplexes within the editosome (Gao and Simpson 2003). To determine whether MEAT1, RET1, and RET2 complexes are interdependent, cells expressing dsRNA fragments targeting RET1 or RET2 transcripts were collected at ∼24 h intervals after induction with tetracycline, and analyzed for changes in MEAT1 protein level (Fig. 5C). Consistent with a previous report, inhibition of RET2 expression led to depletion of MP81 but had no effect on MP63 (Aphasizhev et al. 2003c). However, neither RET1 RNAi nor RET2 RNAi affected MEAT1's abundance, in reference to cross-reacting band or β-tubulin (data not shown). This provides further evidence that all three mitochondrial TUTases belong to distinct particles.
Yeast-two hybrid screening and in vitro binding studies established that RET2 forms a stable complex with MP81 (Ernst et al. 2003; Schnaufer et al. 2003), which is the most probable explanation for MP81 loss in T. brucei cells depleted of RET2. MP81 is also critical for the entire U-insertion editing cascade and, to some extent, for the integrity of the 20S editosome (O'Hearn et al. 2003; Law et al. 2005). Immunodetection of MP81, although at reduced level, in the MEAT1 affinity-purified fraction prompted us to inquire whether MEAT1 association with its complex occurs via MP81. Recombinant 6His-tagged RET2, MEAT1, and cytosolic TUTase TUT4 as a control (Stagno et al. 2007b) were purified from Escherichia coli and incubated with 6His-tagged MP81 produced in vitro by coupled transcription-translation. Following IP with immobilized anti-MP81 monoclonal antibody, co-IP material was separated on SDS-PAGE and probed with Penta-His antibody (Fig. 5D, left panel). As a specificity control, antibody-coated beads were incubated in the absence of MP81 with combined RET2, MEAT1, and TUT4 and subjected to the same procedure (Fig. 5D, right panel). As expected, MP81 and RET2 formed a stoichiometric complex, while no interaction was observed between MP81 and MEAT1.
If MEAT1 and RET2 belong to distinct particles that share several components, we hypothesized that MEAT1 should be present in the complex purified by tagging a shared polypeptide, such as MP63. As seen from Figure 5E, indeed, MEAT1 can be detected by Western blotting in the MP63-TAP pull-down but not in the RET2-TAP purified complex (Fig. 5E). Because relative abundance of the 20S editosome's subunits changes depending on which protein is used for TAP-tagging, the same samples were subjected to self-adenylation and immunoblotting with anti-RET2 antibody (Fig. 5F). Under these conditions, RET2's signal intensity in the RET2-TAP exceeded that in the MP63-TAP by about 2.5-fold if normalized by REL1; RET2 signals are virtually identical if adjusted by REL2 adenylation. Collectively, these results demonstrate that MEAT1 and RET2 belong to distinct protein complexes that share several subunits.
Editosome-like particle catalyzes guided U-insertion and mRNA cleavage in vitro
Guided U-insertion activities of the MEAT1-associated editosome-like particle and the 20S editosome were compared in a precleaved assay (Fig. 6A). Protein samples shown in Figure 5A were adjusted for equal amounts of MP63 and used in a precleaved insertion assay with substrates diagrammed in Figure 3C. Consistent with the properties of the individual MEAT1 and RET2, U-insertions by the MEAT1 complex were effectively terminated by the gap size. The 20S editosome-catalyzed reaction was more efficient: ∼35% of the 5′ fragment was correctly extended into +3 product versus less than 5% conversion achieved in the presence of the MEAT1 complex. Catalytic efficiency of the recombinant MEAT1 on dsRNA exceeds that of RET2 by almost a 100-fold, yet the RET2-containing editosome appears to be more efficient and precise in filling gaps in precleaved substrates. Although ATP was not present in the reaction, minor ligation products were seen in RET2-TAP likely due to REL2 activity (Cruz-Reyes et al. 2002).
FIGURE 6.
Enzymatic activities of the MEAT1 complex. (A) Precleaved U-insertion reaction. RNA substrates for +0, +2, and +3 insertion were assembled as in Figure 3C. Extended and circularized forms of the 5′ fragment and the ligated products are indicated. UTP was added to 100 μM. C indicates labeled 5′ fragment incubated without protein; 5′ +g: 3′ fragment was omitted. (B) MEAT1-TAP expression and RET2 RNAi were induced simultaneously, and MEAT1 complex was purified after 72 h via standard procedure (see Materials and Methods for details). Precleaved insertion assays were performed with fully assembled substrates with indicated gap sizes. (C) Single-stranded 5′-labeled 24-mer RNAs ending with six adenosines or uracils were incubated with MEAT1-TAP purified complex in the absence of NTPs for specified time intervals. Products were resolved on 15% acrylamide/8M urea gel. (D) For the full-round U-deletion editing assay, a 5′-labeled mRNA fragment was annealed with a twofold molar excess of gRNA by heating and slow cooling and incubated with affinity purified fractions for 1 h. ADP was added to stimulate cleavage reaction (Cruz-Reyes et al. 1998). Positions of 5′ cleavage fragment and -3 edited products are shown by arrows; gRNA-mRNA chimeras are in brackets. Fragment sizes were determined by comparison to partial RNase T1 and alkaline hydrolysis. (E) Full-round U-insertion reactions were performed in the presence of UTP or UTP/ATP. The fully edited +2 product accumulated at a lower level than in the deletion reaction and is partially overshadowed by the input mRNA signal.
To exclude even a slight possibility that MEAT1-TAP was contaminated with RET2 at levels not detectable by mass spectrometry or Western blotting, we have coexpressed TAP-tagged MEAT1 and dsRNA targeting RET2's mRNA. MEAT1 complex was purified from T. brucei cells depleted of RET2 (data not shown) and tested in a precleaved assay. As seen from Figure 6B, the extension profile generated with the MEAT1-TAP/RET2 RNAi fraction resembles those observed for the recombinant protein (Fig. 3C) and MEAT1-TAP pull-down (Fig. 6A).
Two U-specific exonucleases have been identified in the 20S editosome from T. brucei (Panigrahi et al. 2003): REX1, which is essential for parasite viability (Kang et al. 2005) and has an ortholog in Leishmania (Aphasizhev et al. 2003a), and apparently nonessential REX2 (Rogers et al. 2007). Because only the latter was detected in the MEAT1 complex (Table 2), we next tested the TAP-purified fraction for a 3′-5′ exonuclease activity that selectively degrades synthetic ssRNA ending with Us (Aphasizhev and Simpson 2001). In agreement with the report that both REX1 and REX2 are active in vitro as U-specific exonucleases (Rogers et al. 2007), this activity was indeed readily detectable in the MEAT1 complex (Fig. 6C).
The heterogeneity of the major 20S editosome is thought to be determined by the mutually exclusive presence of three endonucleases required for cleavage at U-insertion, U-deletion, and cis-edited sites and a few associated proteins (Panigrahi et al. 2006; Carnes et al. 2008). Mass spectrometry analysis of the editosome-like particle identified only U-deletion-specific REN1, which raised the question if the lack of U-insertion-specific REN2 is caused by the loss of U-insertion subcomplex. To assess the endonuclease activities in the MEAT1-TAP-purified fraction, the in vitro synthesized A6 mRNA fragments and gRNAs (see Materials and Methods for sequence modifications) directing deletion of three Us (Fig. 6D) or insertion of two Us were used in a full-round editing assay (Fig. 6E). Although the relative amounts of REN1 and REN2 could not be determined from this assay, both endonuclease activities were present in the MEAT1 complex. However, in comparison with the 20S editosome purified by RET2 tagging and adjusted by Western blotting with anti-MP63 antibody, formation of the 5′ cleavage product was decreased by more than 10-fold.
To conclude, the data presented in Figure 6 demonstrate that the MEAT1-associated complex possesses individual enzymatic activities that have been shown to be associated with the 20S editosome. The U-insertion activity of the editosome-like particle in vitro is significantly lower than that of the 20S editosome.
MEAT1 repression by RNAi causes cell growth inhibition, which can be rescued by a functional RNAi-resistant knock-in
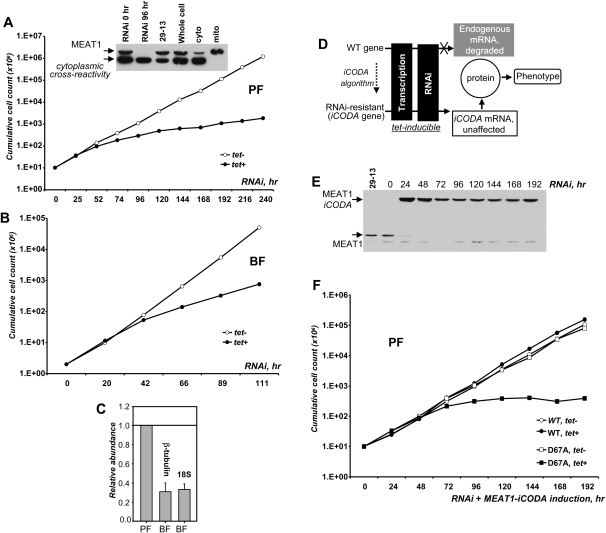
To inhibit gene expression, two fragments corresponding to positions 65–576 (U-fragment) and 360–926 (M-fragment) of the MEAT1 gene were cloned between opposing tet-repressor binding sites and T7 RNA polymerase promoters into a p2T7-177 vector (Wickstead et al. 2002). In addition, a fragment covering the 3′ end of the gene (positions 870–1221) and extending into the 3′ untranslated region (3′ UTR) by 140 nucleotides (nt) (D-fragment) was cloned likewise. Upon transfection into 29–13 procyclic (PF) or a “single” marker bloodstream (BF) strains of T. brucei (Wirtz et al. 1999), stable clonal cell lines for conditional expression of dsRNAs were established by limiting dilution. RNAi was induced by addition of tetracycline in three clones for each cell line, and growth rates were monitored over 10 d. All PF and BF clones generated with U and M gene fragments showed insignificant growth rate variations after RNAi induction (data not shown). However, targeting the 3′ portion of the gene and 3′ UTR (D fragment) caused growth inhibition after ∼60 h post-induction in PF (Fig. 7A) and ∼40 h in BF (Fig. 7B) parasites, suggesting that MEAT1 expression is essential for viability of both forms. Efficient depletion of MEAT1 protein in procyclic trypanosomes was confirmed by Western blotting (Fig. 7A, inserted panel). Sensitivity of immunoblotting was insufficient for a reliable detection of MEAT1 in total cell extracts of bloodstream trypanosomes, most likely reflecting a lower abundance of this mitochondrial protein. This conclusion is consistent with an approximately fourfold decrease in relative abundance of the MEAT1 transcripts in BF (Fig. 7C).
FIGURE 7.
Inhibition of MEAT1 expression by RNAi. (A) Growth kinetics of procyclic T. brucei RNAi cell line (D fragment) after mock induction (open circles) or addition of tetracycline (closed circles). (Inserted panel) Parasites were collected at ∼24-h intervals; 5 × 106 cells were separated on denaturing gel and probed with anti-MEAT1 antibody on immunoblots. Subcellular fractions were analyzed alongside to indicate migration of MEAT1 and cytoplasmic cross-reactivity. (B) Growth kinetics of bloodstream form T. brucei RNAi cell line (D fragment). (C) Relative abundance of MEAT1 mRNA in PF and BF was normalized to β-tubulin and 18S ribosomal RNA. (D) Schematic diagram of the iCODA RNAi rescue strategy. (E) Cells were collected at indicated time points and cell lysates analyzed by immunoblotting with anti-MEAT1 antibody. (F) Growth kinetics of MEAT1-iCODA knock-in T. brucei RNAi cell line (D fragment) after mock induction (open circles) or addition of tetracycline (closed circles). Cell counts for knock-in cell line expressing catalytically-inactive MEAT1 are represented by open squares (mock induction) and closed squares (tet added).
Targeting of the 3′ portion of the gene and a short (100–150 nt) region of the 3′ UTR may provide a general approach for a more efficient inhibition of expression by RNAi in T. brucei. However, this raises two questions: off-targeting and possible effects on trans-splicing for the downstream gene. To address these potential problems, we have developed a new functional RNAi complementation system, the iCODA design (RNAi resistant genes via computationally optimized DNA assembly) diagrammed in Figure 7D. Briefly, the gene fragment targeted by RNAi (∼500 base pairs [bp]) was modified to contain at least one silent mutation per 10 bp and assembled from oligonucleotides (Larsen et al. 2008). Codon context and other considerations were taken into account to minimize changes in translational efficiency (Hatfield and Roth 2007). The C-terminal TAP tag was also introduced to allow purification of the MEAT1-iCODA protein from cells depleted of the endogenous protein by RNAi, if necessary. Finally, a D67A mutation in the catalytic site was introduced to inactivate the enzyme (see Materials and Methods for details).
Genetic constructs for tetracycline-inducible expression of dsRNA and MEAT1-iCODA bearing phleomycin and blastocidin resistance markers, respectively, were introduced into the 29-13 transgenic T. brucei strain, which constitutively expresses tet-repressor and T7 RNA polymerase (Wirtz et al. 1999). RNAi and MEAT1-iCODA coexpression was induced in a clonal cell line resistant to phleomycin, blasticidin, neomycin, and hygromycin. Western blotting of total cell lysates obtained at ∼24-h time intervals showed rapid depletion of the endogenous MEAT1 and stable expression of the TAP-tagged MEAT1 (Fig. 7E). Growth rates of uninduced (RNAi off, MEAT1-iCODA off) and induced (RNAi on, MEAT1-iCODA on) cultures were virtually identical (Fig. 7F). Combined with the immunoblotting in Figure 7E and growth curves in Figure 7A, these results demonstrate that expression of the RNAi-resistant MEAT1 gene alleviates inhibition of cell growth by MEAT1 RNAi. To confirm that the enzymatic activity of MEAT1 is required for the phenotype rescue, a catalytically-inactive version of the MEAT1-iCODA was coexpressed with RNAi. Although the RNAi efficiency and TAP-tagged protein expression level were similar to those observed for the wild-type polypeptide (data not shown), a severe growth inhibition was observed after ∼70 h of induction (Fig. 7F). Overexpression of the mutant protein alone did not produce an appreciable dominant-negative phenotype (data not shown). In summary, conditional RNAi knockdown and knock-in of the RNAi-resistant gene demonstrated that MEAT1 expression is essential for T. brucei viability.
MEAT1 repression leads to increase in abundance of several mitochondrial RNAs
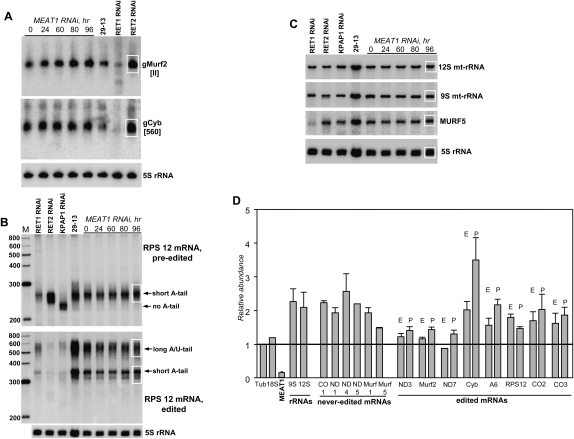
RNAi-mediated depletion of RET1 (Aphasizhev et al. 2002) or RET2 (Aphasizhev et al. 2003c) resulted in the inhibition of RNA editing, which was likely set off by the loss of functional gRNAs and U-insertion mRNA editing activity, respectively. To investigate the consequences of MEAT1 repression on guide RNAs, total RNA was isolated at indicated time periods post-induction and analyzed by Northern blotting in reference to cytoplasmic 5S rRNA (Fig. 8A). RNA isolated from RET1 and RET2 RNAi cell lines after 72 h of induction was included to illustrate a range of possible effects on the gRNA population. Assuming the gRNA/5S rRNA ratio in the uninduced cells as 100%, abundance of the maxicircle encoded gMURF-II increased to ∼160%, while the level of the mini-circle encoded gCyb[560] did not change.
FIGURE 8.
Analysis of mitochondrial RNAs from MEAT1-depleted T. brucei. (A) Northern blotting of individual gRNAs. RNAs from the parental (29-13) and cell lines with induced RNAi were separated on 12% polyacrylamide/8 M urea gels and sequentially hybridized with oligonucleotide probes. 5S ribosomal RNA was used as a loading control. Areas used for quantitation are shown by a white rectangle. (B) Northern blotting of pre-edited and fully edited RPS12 mRNAs. RNA was separated on 6% acrylamide/urea gel. (C) Northern blotting of never-edited MURF5 transcript and mitochondrial ribosomal RNAs (mt-rRNAs). The same membrane as in B was sequentially hybridized with ssDNA probes. (D) Quantitative RT-PCR analysis of mitochondrial mRNAs and RNAi-targeted transcript. The RNA levels were normalized to β-tubulin mRNA. P indicates pre-edited mRNA; E, edited form. Error bars, SD from at least three replicates. The thick line at “1” stands for no change in mRNA's relative abundance with bars above or below representing an increase or decrease, respectively.
High-resolution Northern blotting was employed to assess changes in editing and polyadenylation status of the small ribosomal subunit 12 (RPS12) mRNA. Single-stranded probes were designed to detect pre-edited and fully edited RPS12 mRNA species in RNA isolated from RNAi cell lines indicated in Figure 8B. RNA isolated from cells with repressed mitochondrial poly(A) polymerase (KPAP1) served as control for the loss of short (A) tail in the pre-edited mRNA and lack of the edited forms with short (A) tail and long (A/U) tail (Etheridge et al. 2008a); RET1 and RET2 RNAi samples were included to demonstrate gross effects of the disabled editing process on the abundance of respective species. Sequential hybridizations were performed with probes for mitochondrial 9S and 12S ribosomal RNAs and with never-edited mitochondrial unidentified open reading frame 5 (MURF5) (Fig. 8C). In contrast to RET1 and RET2 RNAi, we did not observe deleterious effects on edited mRNAs as a consequence of MEAT1 repression.
To extend the analysis of mitochondrial transcripts, the RNAi-dependent changes in relative abundance of five never-edited and eight edited transcripts have been determined by real-time PCR at 96-h post-induction. In close agreement with Northern blotting, no major changes were observed for MURF5 and for pre-edited and edited forms of RPS12 mRNA (Fig. 8D). Relative abundance of most other transcripts remained unchanged or increased up to about threefold in the case of pre-edited cytochrome b mRNA. To conclude, a preliminary analysis demonstrated a moderate increase in the steady-state level of mitochondrial mRNAs and rRNAs caused by MEAT1 repression but produced no immediate cues to the specific MEAT1 function. The apparent integrity of editing and polyadenylation patterns in RNA species analyzed in this work suggests that either MEAT1 is not involved in these processes or different approaches such as deep sequencing are required to elucidate its role.
DISCUSSION
RNA uridylyl transferases have been discovered in biochemical studies of U-insertion/deletion RNA editing in the Kinetoplastida protozoans Leishmania tarentolae and T.a brucei (for review, see Aphasizhev and Aphasizheva 2008). The editing events are most often found in organelles, and it is thought that the divergent editing mechanisms evolved independently as a means to correct mutations and modulate gene expression. Known TUTase functions in trypanosomal RNA editing provided little insight into the broader biological roles of UTP-specific terminal RNA transferases in eukaryotic cells. Recent reports indicated that TUTases may participate in processes as diverse as U6 snRNA 3′ processing (Trippe et al. 2006), cell cycle-dependent regulation of histone mRNA stability in human cells (Mullen and Marzluff 2008), and the 3′ uridylylation of actin mRNAs after S-phase arrest in Schizosaccharomyces pombe (Rissland and Norbury 2008). Crystallographic studies of RET2 (Deng et al. 2005) and TUT4 (Stagno et al. 2007a,b) provided insights into the structural basis of UTP recognition and domain organization for TUTases underscoring conservation of the key UTP binding residues in otherwise divergent proteins.
Here we report identification and characterization of a third mitochondrial TUTase in trypanosomes, MEAT1. Notwithstanding low sequence similarity between RET2 and MEAT1, both enzymes are characterized by exclusive UTP specificity and the capacity to utilize ssRNA and dsRNA substrates. There are, however, opposing preferences for either purines or pyrimidines as terminal nucleotides in ssRNA. Recombinant RET2 is more precise in guided U-insertions on gapped dsRNA substrates and requires a phosphorylated 3′ fragment presumably produced by the endonucleolytic mRNA cleavage. Under the same conditions, MEAT1 generates intermediate products almost as efficiently as correctly edited products, and its activity is not affected by the lack of a phosphate group.
A possibility that MEAT1 may have a role in editing was raised by experiments demonstrating its association with RNA editing ligase 1-containing complex. In a contrast to RET2, which is confined to the 20S editosome (Aphasizhev et al. 2003c), only ∼5% of MEAT1 is associated with this high molecular mass complex. Mass spectrometry and immunochemical analyses established that MEAT1 is associated with a 20S editosome-like complex. We provide several lines of evidence that MEAT1 and RET2 interact with their respective complexes in a mutually exclusive manner. First, MEAT1 was not detected in a complex purified via affinity tagging of RET2 and vice versa. Second, MEAT1 was readily detectable in a complex purified by tagging a shared component of the editosome-like complex and the 20S editosome, the zinc finger protein MP63. Third, RET2 RNAi led to a loss of the U-insertion subcomplex in the 20S editosome but had no effect on MEAT1 protein level. Fourth, MEAT1 does not interact with MP81, which binds RET2 and is required for U-insertion subcomplex assembly. MEAT1 complex displayed a robust 3′-5′ U-specific exonuclease activity. However, the endonucleolytic activities specific for U-insertion and U-deletion sites were significantly reduced in comparison to the 20S editosome; the RNA ligase activity was undetectable under the experimental conditions used.
The RNA analysis methods that are routinely employed to assess inhibition of RNA editing in vivo produced no obvious indications that this process is affected by MEAT1 repression. On the contrary, a moderate increase in abundance has been detected for several mitochondrial RNAs. Speculating on the MEAT1's possible regulatory role, random uracils found in the short A-tails, one or two at a time, in most never edited, pre-edited, and edited mRNAs (Etheridge et al. 2008a) may be added by MEAT1, thereby promoting RNA decay. In this scenario, repression of MEAT1 would stabilize short A-tail-containing species although it is not immediately clear how this would affect cell viability.
Most of the editosome components have been studied by conditional gene knockouts or RNAi-induced knockdowns. Although the resultant phenotypes and effects on editing in vivo varied considerably, to our knowledge only REL2 (Gao and Simpson 2003; O'Hearn et al. 2003) and REX2 (Rogers et al. 2007) knockdowns had no effect on T. brucei viability. Puzzled by the apparently intact editing process but inhibited cell growth in MEAT1-depleted cells, we developed a functional RNAi knock-in system to address the questions of possible RNAi off-targeting and whether MEAT1's enzymatic activity is responsible for the observed phenotype. This was achieved by coexpressing a dsRNA targeting the MEAT1 transcript with a synthetic RNAi-resistant gene. Rescue of the RNAi-induced phenotype by coexpression of catalytically active MEAT1, but not by expression of the mutated protein, confirmed that this TUTase is essential for cell viability. Further work is needed to determine its exact function.
For reasons that are not clear, conditional gene knockouts and knock-ins of essential mitochondrial genes (Schnaufer et al. 2001; Trotter et al. 2005) have been possible only in bloodstream form T. brucei, which is deficient in oxidative phosphorylation. This approach requires sequential generation of four clonal cell lines, and more importantly, the scale of biochemical experiments is limited because of low cell density (<106 cells/mL) in BF cell cultures. The iCODA methodology presented in this work is more expeditious and allows not only phenotype analysis but also purification of a protein of interest from insect form parasites that possess functional mitochondria and can be readily cultured at >107 cells/mL.
MATERIALS AND METHODS
Cell cultures and RNAi
RNAi expression plasmids were generated by cloning MEAT1 fragments indicated in the Results section and RET2 fragment (41–545) into a p2T7-177 vector (Wickstead et al. 2002). DNA oligonucleotides used in this study are listed in the Supplemental Materials. Clonal tet-inducible RNAi cell lines were obtained by transfecting these plasmids into procyclic 29–13 or bloodstream “single marker” T. brucei strains (Wirtz et al. 1999) followed by limiting dilution. RNAi was induced with 5 mg/L tetracycline, and cells were diluted to 106/mL every 24 h.
Protein expression and purification
The full-length MEAT1 gene was cloned into a pET15b vector to generate an N-terminal 6His-fusion protein. E. coli cultures were grown in 2YT media at 37°C to 0.2 OD600, cooled to 10°C and induced with 0.5 mM IPTG for 3 h. His-tagged protein was purified by sequential metal affinity chromatography on Talon resin (Clontech) and anion exchange Sepharose Q HP column (GE Healthcare), as recommended by the manufacturers. More than 95% homogeneity, as determined by Sypro Ruby staining of SDS-PAGE, was achieved routinely. Recombinant RET2 (Aphasizhev and Aphasizheva 2007) and TUT4 (Stagno et al. 2007b) were purified as described previously. For expression in T. brucei, MEAT1, RET2, and MP63, we cloned into mhTAP plasmid (kind gift from Marilyn Parsons, Seattle Biomedical Research Institute) containing a phleomycin resistance gene. For coexpression with RET2 RNAi, a MEAT1-TAP fusion was subcloned from mhTAP vector into a pLEW100-based vector with blasticidin-resistant gene (kind gift of Elisabetta Ullu, Yale University). Clonal cell lines of MEAT1-TAP/RET2-RNAi were generated by sequential stable transfection of p2T7-177-RET2 and pLEW100-MEAT1-TAP-BSR into procyclic T. brucei strain 29-13 (Wirtz et al. 1999). TAP fusion protein expression and RNAi was concurrently induced with 1 μg/mL tetracycline.
iCODA
To generate the iCODA cell line, the following sequence was assembled from oligonucleotides to replace the XhoI/XbaI fragment (positions 874–1219 of the gene) in the pLEW100-MEAT1-TAP-BSR: 5′-gaagcgtcttgagtgggagaagcacattgcagagcacaaagaggacggtccgcttgatgagaacgattttagtgcgagtatgcagaacgagacaacacagagaccaagtaatagtccatatgtggttgaagattttgtgaactacgttaactgtggtcgtcgcgttcaggcaagtcgtgttcgtcatattcaacaggagtttaaccgtcttcgtgagatgttgattgataaagagagtgagcttaagttcgatgaggtatttcgtgagagtgatacggtgccacgttttcagggttttgaaggtgtgggtacacgtgatcaccgggttaagacatttcgtccacag-3′. DNA design, assembly, and sequencing were made by CODA Genomics, Inc.
Clonal cell lines for MEAT1-iCODA/MEAT1-RNAi were generated by cotransfecting p2T7-177-MEAT1 and pLEW100-MEAT1-iCODA-BSR into procyclic T. brucei strain 29-13 and cloning parasites resistant to phleomycin, blasticidin, neomycin, and hygromycin.
Subcellular fractionation, mitochondria purification, extracts preparation, IP, and mass spectrometry
Mitochondria isolation, extract preparation, glycerol gradient fractionation, and self-adenylation reactions and tandem affinity purification were performed according to the method of Aphasizhev and Aphasizheva (2007) and Pelletier et al. (2007). Mass spectrometry was carried out as described earlier (Etheridge et al. 2008a). For IP, 0.3 g (wet weight, ∼15 mg total protein) of purified mitochondria were extracted in 1 mL of 50 mM HEPES (pH 7.6), 150 mM KCl, 1 mM EDTA, 0.4% of NP-40 by sonication (three times 30 sec at 12W). The membrane fraction was removed by centrifugation for 10 min at 100,000g. Antigen-purified antibody (100 μg), total IgG from pre-immune serum, or 200 μL of ascitic liquid with MP81 monoclonal antibody (kind gift of Ken Stuart, Seattle Biomedical Research Institute) were pre-incubated with 100 μL of rProtein A Sepharose (Invitrogen) and saturated in TBS plus 10 mg/mL of BSA. Protein A beads were incubated with mitochondrial extract or gradient fractions (10 μg of beads per 100 μL or 400 μL, respectively) for 1 h, and washed three times for 30 min with 1 mL of 1 mM EDTA, 150 mM NaCl, 25 mM (Tris at pH 8.0) buffer. Following incubation with 0.5 M NaCl, 0.1 mg/mL RNaseA-containing or 1% Triton X 100-containing TBS, beads were transferred into 25 mM Tris (pH 7.6), 10 mM MgCl2, 60 mM KCl for self-adenylation reaction with [α-32P]ATP. Proteins were eluted off the beads by boiling in SDS loading buffer, separated on 8%–16% SDS-PAGE, transferred onto nitrocellulose membrane, exposed to phosphor storage screen, and subjected to immunoblotting.
Enzymatic assays
Nucleotide specificity assay reactions were performed in the buffer containing 50 mM Tris-HCl (pH 8.0), 1 mM DTT, 10 mM KCl, 10 mM MgAc, and 0.5 μM RNA substrate for 30 min at 27°C. Reactions were started by adding recombinant proteins to 50 nM and stopped by adding 1.5 V of 10 mM EDTA, 95% formamide solution. Products were separated on 15% acrylamide/8 M urea gel. RNA substrates were as follows:
6[A] GCUAUGUCUGUCAACUUGAAAAAA;
6[U] GCUAUGUCUGUCAACUUGUUUUUU; and
5U[A] GCUAUGUCUGUCAACUUGUUUUUA.
Precleaved editing assays were performed with 3 μL of TAP-purified RET2 and MEAT1 fraction as described by Aphasizhev and Aphasizheva (2007). For the full-round assays, RNA substrates were derived from those described by Carnes and Stuart (2007) and synthesized by in vitro transcription:
A6-short mRNA (−3 deletion): GGGAGGAGAGAAGAAAGGGAAAGUUGUGAUUUUGGAGUUAUAGAAUACUUACCUGGCAUC3;
gRNA D34: GGGUAUACUAUAACUCCACCCUCACAACUUCUU;
A6-ES1-mRNA (+2 insertion): GGGAGGAGAGAAGAAAGGGAAAGUUGUGAUUGGAGUUAUAGAAUACUUACCUGG; and
gA6ES1: GGGAUACUAUAACUCCAAUAACGAAUCAGAUUUUGACAGUGAUAUGAUAA.
Steady-state kinetics parameters of UMP incorporation into ssRNA were measured using a filter assay at 2.5 nM of RET1 and 50 nM of MEAT1. Reactions were performed in a 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1mM DTT buffer containing 100 nM of 6[U] ssRNA, and 20 nM of [α-32P] UTP. Reaction time and UTP concentrations varied from 30 sec to 45 min and from 0.1 to 400 μM, respectively. Reactions were stopped with 2 volumes of 1% SDS, 0.5 M sodium phosphate (pH 8.0). Samples were transferred on DE81 filters (Whatman); washed three times with 0.5 L of 0.5 sodium phosphate (pH 8.0), 30 mM sodium pyrophosphate; and dried. Filters were counted in a scintillation cocktail. Initial velocities for insertion of a single uracil into precleaved editing dsRNA substrate by RET1, MEAT1, and RET2 and into ssRNA by RET2 were determined by gel-based assay. Reactions were performed in the same buffer containing 100 nM of 5′-labeled ssRNA or precleaved dsRNA substrates assembled with [γ-32P]ATP labeled 5′ fragment. UTP concentration and reaction time were varied from 0.5 to 150 μM and from 1 to 20 min, respectively. Reactions were stopped by addition of 1.5 volume of 95% formamide, 20 mM EDTA and were separated on 10% 8 M urea/acrylamide gel. Quantity One software was used to calculate intensity of the input and product bands. Steady-state kinetics parameters were obtained using the enzyme kinetics module in the Sigma Plot package.
RNA analysis
RNA isolation, real-time PCR, and Northern blotting were done as described previously (Etheridge et al. 2008a; Weng et al. 2008).
Accession numbers
GenBank accession numbers for MEAT1 genes reported in this paper are FJ561337 (T. brucei) and FJ561338 (L. major).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Marilyn Parsons and Elisabetta Ullu for kind gifts of plasmids, and Ken Stuart and Uli Göringer for generously providing antibodies. We thank Ronald Etheridge and Ruth Chang for help with fluorescent microscopy and acknowledge fruitful discussions with members of our laboratory. G.-E.R. was supported by the UCI Biomedical Informatics Training Program. This work was supported by NIH grant AI064653 to R.A.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1538809.
REFERENCES
- Adler BK, Harris ME, Bertrand KI, Hajduk SL. Modification of Trypanosoma brucei mitochondrial rRNA by post-transcriptional 3′ polyuridine tail formation. Mol Cell Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R. RNA uridylyltransferases. Cell Mol Life Sci. 2005;62:2194–2203. doi: 10.1007/s00018-005-5198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. RNA editing uridylyltransferases of trypanosomatids. Methods Enzymol. 2007;424:51–67. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim Biophys Acta. 2008;1779:270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Simpson L. Isolation and characterization of a U-specific 3′-5′ exonuclease from mitochondria of Leishmania tarentolae. J Biol Chem. 2001;276:21280–21284. doi: 10.1074/jbc.M100297200. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003a;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003b;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad Sci. 2003c;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L. Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett. 2004;572:15–18. doi: 10.1016/j.febslet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: Identification of functional domains by mutational analysis. J Biol Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- Babbarwal VK, Fleck M, Ernst NL, Schnaufer A, Stuart K. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei. RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Carnes J, Stuart KD. Uridine insertion/deletion editing activities. Methods Enzymol. 2007;424:25–54. doi: 10.1016/S0076-6879(07)24002-9. [DOI] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Rusche L, Piller KJ, Sollner-Webb B. T. brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol Cell. 1998;1:401–409. doi: 10.1016/s1097-2765(00)80040-4. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J, Zhelonkina AG, Huang CE, Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol Cell Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Sollner-Webb B. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell. 1990;61:1001–1011. doi: 10.1016/0092-8674(90)90065-m. [DOI] [PubMed] [Google Scholar]
- Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst NL, Panicucci B, Igo RP, Jr, Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008a;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge RD, Clemens DM, Aphasizhev R. Identification and characterization of nuclear noncanonical poly(A) polymerases from Trypanosoma brucei. Mol Biochem Parasitol. 2008b;164:66–73. doi: 10.1016/j.molbiopara.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J Biol Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield GW, Roth DA. Optimizing scaleup yield for protein production: Computationally optimized DNA assembly (CODA) and translation engineering. Biotechnol Annu Rev. 2007;13:27–42. doi: 10.1016/S1387-2656(07)13002-7. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- Igo RP, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc Natl Acad Sci. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CY, Read LK. Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3′ tails in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 2007;154:158–169. doi: 10.1016/j.molbiopara.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller J, Muller UF, Schmid B, Missel A, Kruft V, Stuart K, Goringer HU. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J Biol Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- Larsen LS, Wassman CD, Hatfield GW, Lathrop RH. Computationally optimised DNA assembly of synthetic genes. Int J Bioinform Res Appl. 2008;4:324–336. doi: 10.1504/IJBRA.2008.019578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Huang CE, O'Hearn SF, Sollner-Webb B. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol Cell Biol. 2005;25:2785–2794. doi: 10.1128/MCB.25.7.2785-2794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hearn SF, Huang CE, Hemann M, Zhelonkina A, Sollner-Webb B. Trypanosoma brucei RNA editing complex: Band II is structurally critical and maintains band V ligase, which is nonessential. Mol Cell Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M, Read LK, Aphasizhev R. Isolation of RNA binding proteins involved in insertion/deletion editing. Methods Enzymol. 2007;424:69–96. doi: 10.1016/S0076-6879(07)24004-2. [DOI] [PubMed] [Google Scholar]
- Prasad R, Beard WA, Wilson SH. Studies of gapped DNA substrate binding by mammalian DNA polymerase β. Dependence on 5′-phosphate group. J Biol Chem. 1994;269:18096–18101. [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Read RL, Martinho RG, Wang SW, Carr AM, Norbury CJ. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc Natl Acad Sci. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. The Cid1 poly(U) polymerase. Biochim Biophys Acta. 2008;1779:286–294. doi: 10.1016/j.bbagrm.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Rogers K, Gao G, Simpson L. Uridylate-specific 3′-5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J Biol Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2161. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc Natl Acad Sci. 2007a;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007b;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase 1. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Vondruskova E, van den Burg J, Zikova A, Ernst NL, Stuart K, Benne R, Lukes J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]