Abstract
Eukaryotic mRNAs are subject to quality control mechanisms that degrade defective mRNAs. In yeast, mRNAs with stalls in translation elongation are targeted for endonucleolytic cleavage by No-Go decay (NGD). The cleavage triggered by No-Go decay is dependent on Dom34p and Hbs1p, and Dom34 has been proposed to be the endonuclease responsible for mRNA cleavage. We created several Dom34 mutants and examined their effects on NGD in yeast. We identified mutations in several loops of the Dom34 structure that affect NGD. In contrast, mutations inactivating the proposed nuclease domain do not affect NGD in vivo. Moreover, we observed that overexpression of the Rps30a protein, a high copy suppressor of dom34Δ cold sensitivity, can restore some mRNA cleavage in a dom34Δ strain. These results identify important functional regions of Dom34 and suggest that the proposed endonuclease activity of Dom34 is not required for mRNA cleavage in NGD. We also provide evidence that the process of NGD is conserved in insect cells. On the basis of these results and the process of translation termination, we suggest a multistep model for the process of NGD.
INTRODUCTION
An important aspect of gene expression are quality control systems that recognize and degrade defective mRNAs (reviewed in Doma and Parker, 2007; Isken and Maquat, 2007). Some mRNA quality control systems degrade mRNAs that are defective in translation. For example, nonsense-mediated decay (NMD) rapidly degrades mRNAs with premature termination codons (Maquat, 2004). Similarly, Nonstop Decay (NSD) targets truncated mRNAs that lack termination codons to rapid 3′-5′ degradation by the exosome (Frischmeyer et al., 2002; van Hoof et al., 2002). A third cotranslational quality control pathway, No-Go decay (NGD), acts during the translation elongation step in protein synthesis (Doma and Parker, 2006).
NGD targets and degrades mRNAs with stalls in translation elongation (Doma and Parker, 2006). Decay of NGD substrate mRNAs is initiated by endonucleolytic cleavage in the vicinity of the stall site. The 5′ fragment of the mRNA is then degraded by the cytoplasmic exosome, and the 3′ fragment is degraded by the 5′ to 3′ exonuclease, Xrn1. NGD targets mRNAs with translational stalls caused by strong RNA structures, as well as rare codons and premature stop codons under some conditions. Moreover, NGD has recently been shown to degrade mRNAs that are depurinated in yeast cells (Gandhi et al., 2008), suggesting that NGD will also work on damaged mRNAs that have defects in translation elongation.
Consistent with the proposal that NGD recognizes stalled ribosomes, the Hbs1 and Dom34 proteins, which are related to translation termination factors (Inagaki and Ford Doolittle, 2000) promote endonucleolytic cleavage during NGD in Saccharomyces cerevisiae (Doma and Parker, 2006). Hbs1p is a member of the family of GTPases consisting of eEF1, which delivers the tRNA to the A site of the ribosome (Inge-Vechtomov et al., 2003); eRF3, which functions in translation termination (Jackson, 2007); and Ski7p, which has been proposed to interact with an empty A site when the ribosome reaches the 3′ end of the mRNA during nonstop decay (van Hoof et al., 2002). Dom34p binds to Hbs1p (Carr-Schmid et al., 2002; Lee et al., 2007; Graille et al., 2008) and is related to eRF1, which has a three-dimensional structure similar to a tRNA and functions with eRF3 during translation termination (Kong et al., 2004). Structural analysis of Dom34, and a related protein from archea, reveals that it is similar to eRF1 in two domains but contains a N-terminal domain that is clearly distinct from that of eRF1 (Lee et al., 2007; Graille et al., 2008). The similarity of Dom34 to eRF1 suggests that it functions it an analogous manner to eRF1, but with the Dom34 N-terminal domain serving a different role.
In eRF1, the N-terminal domain forms an extended structure that positions a conserved NIKS loop in the decoding site that is thought to participate in recognition of the stop codon (reviewed in Noble and Song, 2008). Because Dom34 functions independently of stop codons, its N-terminal domain presumably serves a different role. One possibility is that this domain interacts with the ribosome and helps to deliver Dom34 to the A-site of a stalled ribosome. Alternatively, this domain might provide the endonuclease that cleaves mRNAs during NGD. This latter possibility is based on the observation that bacterially produced preparations of both Dom34 and an archeal orthologue contained endonuclease activity (Lee et al., 2007). Moreover, a region of the N-terminal domain of Dom34 contained some acidic residues that were proposed to bind Mg2+ ions and thereby contribute to nuclease activity, which was supported by the observation that mutation of some of these residues reduced or eliminated nuclease activity in vitro (Lee et al., 2007). However, whether this proposed nuclease activity of Dom34 was required for NGD in vivo was not examined.
To test the in vivo significance of various features of Dom34, including its nuclease activity, we created several Dom34 mutants and examined their effects on NGD in Saccharomyces cerevisiae. We observed that three regions, all located in extended loops of the Dom34 structure, affect the efficiency of NGD. In contrast, mutations predicted to inactivate the nuclease activity do not affect NGD in vivo suggesting that any nuclease activity of Dom34 is not required for mRNA cleavage during NGD. Consistent with this interpretation, we observed that overexpression of the Rps30a protein, a high copy suppressor of dom34Δ cold sensitivity, can restore some mRNA cleavage in a dom34Δ strain. On the basis of these results, and by drawing analogy to the process of translation termination, we propose a multistep model for the process of NGD.
MATERIALS AND METHODS
Yeast Strains and Plasmids
All yeast strains used in this study were in the genetic background of yRP1674 (Mat a his3Δ1 leu2Δ met15Δ ura3Δ). yRP2578 (Mat a his3Δ1 lys2Δ leu2Δ ura3Δ dom34Δ:NEO ski7Δ:NEO) was used for the analysis of Dom34 mutants. Double mutants for rps30aΔski7Δ, rps30aΔxrn1Δ strains were generated by standard genetic crosses and verified by PCR. Plasmids GAL-RPS30A was made by amplifying the RPS30A open reading frame from B1116 plasmid (Davis and Engebrecht, 1998) using primers with BamHI and HindIII ends and cloning the insert into the YEP351 plasmid cut with BamHI and HindIII generating pRP1790. The GAL-PELOTA Plasmid was released from the B806 plasmid (Davis and Engebrecht, 1998) using SacI and XbaI enzymes and cloned into YEP351 with same ends generating pRP1793. The reporter mRNAs, PGK1 and PGK1-SL, are described in (Doma and Parker, 2006). Mutants in Dom34 were made by Quick-Change and verified by sequencing. Plasmids were introduced into yeast by standard lithium acetate transformation.
RNA Analysis
All RNA analyses including yeast total RNA extractions and Northern analysis were performed as described previously. Steady-state cultures were grown in SC-Ura containing 2% galactose until OD = 0.35–0.40. All experiments were performed at 30°C in galactose-containing medium with midlog cultures. Total RNA (20 μg) was analyzed by electrophoresis through 1.25% formaldehyde agarose gel. For the PGK1 reporter mRNAs, the oligonucleotide oRP132 was used for detection of the 5′ fragment and oligonucleotide oRP70 for the detection of the 3′ fragment. All oligo sequences are available upon request. Loading corrections for quantification were determined by hybridization with oRP100, an oligonucleotide directed to SCR1 RNA, a stable RNA polymerase III transcript.
Dom34 Purification and Nuclease Analysis
Protein Expression and Purification.
Amplification of Dom34 (FOR: 5′-CATGCCATGGAGATGAAGGTTATTAGTCTGAAAAA-3′; REV: 5′-CGGGATCCTCAGTGATGGTGATGGTGATGCTCCTCACCATCGTCTTC-3′) and Hbs1 (FOR: 5′-CATGCCATGGAGATGGCTTACAGTGACTACAG-3′; REV: 5′-CGGGATCCTCAGTGATGGTGATGGTGATGCTGAGTTATTTCGGATATTTTACC-3′) was performed using the indicated primers. These primers added a 5′ NcoI site, a C-terminal hexahistidine tag, and a 3′ BamHI site to each product (added sequences underlined). The restriction sites were used to subclone each DNA fragment into a pET28 expression vector. Rosetta2(DE3)pLysS cells (EMD Biosciences, San Diego, CA) containing pET28:Dom (pCS37) or pET28:Hbs1 (pCS39) were induced at OD600 0.6 for 18 h at 16°C.
Dom34 domain 1 (FOR: 5′-GCTCTAGAATGAAGGTTATTAGTCTGAAAAA-3′; REV: 5′-ATAAGAATGCGGCCGCTCATTTATACTCGATATTACAGGCTT-3′) and domain 2 (FOR: 5′-GCTCTAGATCAGACACCGCGGCGGTC-3′; REV: 5′-ATAAGAATGCGGCCGCTCAATTTTTCAAAACTTCATTAATGC-3′) were amplified with primers containing 5′ XbaI and 3′ NotI sites (underlined). Digested fragments were cloned into an MBP-HTSHP expression vector. Rosetta2(DE3)pLysS cells (OD600 0.6) containing the expression vector with MBP-domain 1 (pCS28) or MBP-domain 2 (pCS29) were induced with 1 mM IPTG for 1 h at 23°C.
Cells were pelleted after the indicated times, lysed via sonication, and purified over Ni-NTA agarose beads (Qiagen, Chatsworth, CA). Dom34/Hbs1 eluates were further purified over a HiPrep 26/60 Sephacryl S-200 column (GE Healthcare, Waukesha, WI) and, in the case of Dom34, a HiTrap Q FF column (GE Healthcare). MBP-domain 1 and MBP-domain 2 eluates were further purified over amylose resin (New England Biolabs, Beverly, MA).
RNA Substrate Formation and Ribonuclease Reactions.
Hairpin cleavage assays were performed using a 188-nt RNA substrate identical to that described previously (Lee et al., 2007). Synthetic oligos (5′-GCCTTTCTTTATGTTTTTGGCGTCTTCGAGCTCAGGGAAGTTGAAGGATCCGATATCCCGTGGAGGGGCGCGTGGTGGCGGCTGCAGCCGCCACCACGCGCCCCTCCACGGGATATCGTCGACATTTGTTCATTTTTGAGAACTCGCTCAACGAACGATTTGATATAGACGTCGGGCCCAATTCGCCC-3′; 5′- TTAATACGACTCACTATAGGGCGAATTGGGCCCGA-3′) were annealed and extended to create a double-stranded DNA (dsDNA) template. The resulting template was cloned into a pCR-BluntII-TOPO vector (Invitrogen, Carlsbad, CA) and subcloned into SmaI-digested pUC19 using hairpin-specific primers (FOR: 5′-TTAATACGACTCACTATAGGG-3′ and REV: 5′-GCCTTTCTTTATGTTTTTGGC-3′). Sequencing confirmed the integrity of the hairpin construct sequence. PCR amplification of the hairpin region of the pUC19 construct (pCS44) was conducted using the above primers. In vitro transcription of the PCR product using T7 polymerase produced the following transcript: 5′-GGGCGAAUUGGGCCCGACGUCUAUAUCAAAUCGUUCGUUGAGCGAGUUCUCAAAAAUGAACAAAUGUCGACGAUAUCCCGUGGAGGGGCGCGUGGUGGCGGCUGCAGCCGCCACCACGCGCCCCUCCACGGGAUAUCGGAUCCUUCAACUUCCCUGAGCUCGAAGACGCCAAAAACAUAAAGAAAGGC-3′ (34-base pair hairpin underlined). The RNA product was subsequently purified via gel extraction from a 7 M urea/5% polyacrylamide gel. Purified RNA substrate was 3′-end labeled with 32P-pCp using T4 RNA ligase (Promega, Madison, WI) for 4 h at 22°C and labeled product was gel-purified again to ensure a homogenous substrate composition.
Hairpin cleavage reactions were performed in 1× Buffer A (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 20 U RNasin Plus inhibitor [Promega] and 2 mM MgCl2) for up to 30 min at 37°C. RNA substrate was present in twofold molar excess over protein (75 nM). Completed reactions were extracted with TRIzol (Invitrogen) and precipitated with isopropanol. Samples were separated in a 7 M urea/6% polyacrylamide gel and analyzed using a Typhoon phosphorimager system (GE Healthcare).
Bulk RNA cleavage assays were performed in 1× buffer A where 20 μg bulk RNA substrate was incubated with 1 μM protein at 40°C for 1 h, following previously published methods (Lee et al., 2007). Products were loaded directly onto 1.2% formaldehyde-agarose gels and inspected for cleavage.
Description of S2 Cell Experiments
Drosophila Schneider 2 (S2) cells were grown at 25°C in Schneider's medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. For RNA interference, we synthesized double-stranded RNA (dsRNA) corresponding to nucleotides 469-953 of the Pelota (CG3959) transcript and nucleotides 231-874 of the Ski2 (CG10210) transcript. Cells were incubated with dsRNA in serum-free media for 60 min and then allowed to recover in media containing 10% serum for 4 d. Cells were then passaged and retreated with dsRNA to maintain knockdown. One day after retreatment, we transiently transfected the cells with reporters expressing yPGK1 or yPGK1-SL under the control of the Drosophila metallothionein promoter (pMT/V5-His, Invitrogen). After a 1-d recovery, we induced reporter expression with 500 μM CuSO4 for 24 h, harvested the cells in Trizol reagent, and purified total RNA. We analyzed 10 μg of total RNA by Northern blotting using a digoxigenin-labeled probe complementary to the 5-prime 952 nucleotides of the yPGK reading frame, upstream of the stem-loop insertion site. Intensities for the full-length and fragment bands were quantified in NIH ImageJ (http://rsb.info.nih.gov/ij/).
RESULTS AND DISCUSSION
Comparison of eRF1 and Dom34 to Identify Residues for Mutagenesis
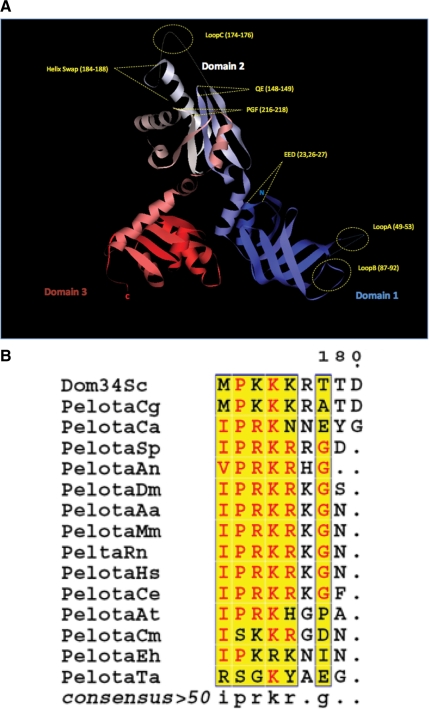
To predict important functional regions of Dom34, we first compared the structures of Dom34 and eRF1. Both Dom34 and eRF1 are three domain proteins, with the middle and C-terminal domains showing strong sequence and structural similarity (Song et al., 2000; Lee et al., 2007; Graille et al., 2008). As described below for each domain, we took advantage of this relationship in choosing residues of Dom34 to examine by mutagenesis. We made several substitution mutations in Dom34, each of which is shown on the structure of Dom34 (Figure 1A) and listed in Table 1. The effects of these mutations were then assessed by introducing the Dom34 variants into a dom34Δ ski7Δ strain carrying a PGK1 mRNA with a strong stem-loop in the coding region, termed PGK1-SL, which triggers NGD (Doma and Parker, 2006). The lack of Ski7 inactivates the cytoplasmic exosome and allows for the accumulation of the 5′ cleavage product of NGD, which is absent in a dom34Δ strain (Doma and Parker, 2006), but can be restored when wild-type Dom34 is provided on a plasmid (Figure 2A, lanes 1 and 2).
Figure 1.
(A) Ribbon diagram of the crystal structure of S. cerevisiae Dom34, highlighting the location of each mutant used in this study. Domain 1 (blue) contains the mutants loop A, loop B, and EED. Domain 2 (white) includes the mutants QE, NLS, loop C and PGF. Domain 3 is shown in red. The N- and C- termini are also represented as N and C. (B) Alignment of dom34 homology proteins from different organisms shows a cluster with highly conserved basic residues (NLS). The alignment was processed using the network protein sequence analysis (NPS) at http://npsa-pbil.ibcp.fr/ (Combet et al., 2000). The following organisms were used: S. cerevisiae (Sc); Candida glabrata (Cg); Candida albicans (Ca); Drosophila melanogaster (Dm); Aedes aegypti (Aa); Homo sapiens (Hs); Mus musculus (Mm); Rattus norvegicus (Rn); Caenorhabditis elegans (Ce); Arabidopsis thaliana (At); Schizosaccharomyces pombe (Sp); Aspergillus niger (An); Plasmodium falciparum (Pf); Toxoplasma gondii (Tg); Cryptosporidium muris (Cm); Entamoeba histolytica (Eh); Thermoplasma acidophilum (Ta).
Table 1.
Substitution mutations in Dom34
| Mutation | Name | Plasmid no. |
|---|---|---|
| E23A | E23A | pRP1732 |
| E26A | E256A | pRP1733 |
| D27A | D27A | pRP1734 |
| E23A, E25A, D27A | EED triple | pRP1736 |
| 49 SKLDf53 to 49AAAAA53 | Loop A | pRP1737 |
| 87TVTDES92 to 87AAAAAA92 | Loop B | pRP1738 |
| Q148AE149A | QE | pRP1739 |
| 174KKK176 to 174AAA176 | Loop C | pRP1740 |
| 184FDEKT188 to 184AAAAA188 | Helix swap | pRP1741 |
| 213PGF215 to 213AAA215 | PGF triple | pRP1744 |
| P213A | P213A | pRP1742 |
| G214A | G214A | pRP1743 |
| Loop A + loop B | Loop A+B | pRP1771 |
| Loop B + loop C | Loop B+C | pRP1772 |
| Loop A + loop C | Loop A+C | pRP1773 |
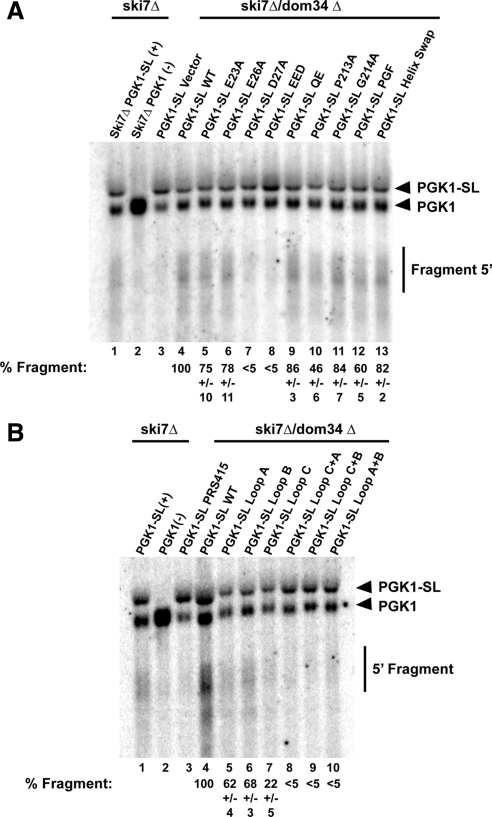
Figure 2.
Affects of Dom34 Mutations on NGD. (A) Northern analysis of the levels of the mRNA fragment produced from the PGK1-SL reporter mRNA in strains double deleted for dom34 and ski7 (dom34Δski7Δ) transformed with different Dom34 point mutants. The % fragment represents the relative ratio of fragment to full-length PGK1-SL from multiple experiments standardized to a percentage of 100% for strains expressing the wild-type Dom34. (B) Northern analysis of the levels of the mRNA fragment produced from the PGK1-SL reporter mRNA in strains double-deleted for dom34 and ski7 (dom34ski7Δ) transformed with Dom34 variants with alterations in the three loops of the Dom34 structure. The % Fragment, the relative ratio of fragment to full length PGK1-SL from multiple experiments standardized to a percentage of 100% for strains expressing the wild-type Dom34.
The Central Domain
The central domain of eRF1 is thought to interact with the ribosome and to position a conserved GGQ sequence in the peptidyl transferase center to trigger hydrolysis of the peptide-tRNA bond, thereby allowing release of the nascent peptide (reviewed in Noble and Song, 2008). In Dom34, the extended helix that contains this GGQ motif in eRF1 is not as long and does not contain a GGQ in its loop sequence. Instead, in eukaryotic Dom34s, the corresponding loop, which we refer to as loop C, is disordered in the structure but is very rich in basic residues (KKKR), with three or four basic residues preceded by a proline conserved in all eukaryotic Dom34 orthologues (Figure 1B). This suggests that loop C has an important role in Dom34 function, which we tested by substituting the four basic residues with alanine.
We also constructed additional mutations in the central domain (Figure 1A). Specifically, there is a universally conserved PGF tripeptide (residues 213-215) that we mutated to alanine. We also mutated a conserved QE pair (residues 148-149) found in the turn between beta8 and beta 9 in the structure, which is located near the conserved PGF tripeptide. Finally, we mutated a stretch of amino acids that comprise part of the helix that extends the loop C away from the central domain, thus testing if the specific amino acids in this region were required for Dom34 function in NGD. We expressed these variants from a centromere plasmid under the control of the Dom34 promoter in a dom34Δ ski7Δ strain and tested whether they complemented the function of Dom34 in NGD as judged by production of the mRNA cleavage product.
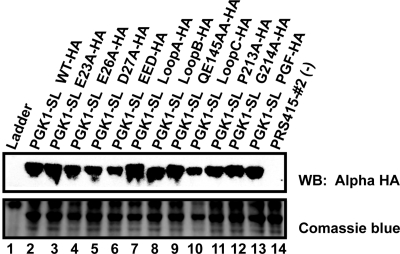
We observed that mutations in the PGF tripeptide, either singly (lanes 10 and 11) or a triple mutant in combination (PGF triple, lane 12), mutations in the QE domain (QE, lane 9), or changes to residues 184-188 (Helix Swap, lane 13) still exhibited NGD (Figure 2A). However, mutation of Proline213 or the entire PGF tripeptide showed a reduction in the efficiency of NGD (Figure 2A). In contrast, we observed that mutation of the basic loop region (loop C mutant) strongly reduced, but did not abolish the process of NGD (Figure 2B, lane 7). Moreover, the mutations in the PGF tripeptide or loop C did not lead to a significant reduction in the levels of the Dom34 protein as assessed by Western blots with a hemagglutinin (HA)-tagged version of Dom34 (Figure 3). These observations suggest that the PGF tripeptide and loop C have functional roles in NGD.
Figure 3.
Western analysis of Dom34 variants using a HA-tagged construct. Dom34 variants were tagged with a 3XHA tag on their C-terminus and their levels analyzed by Western analysis.
There are two likely functions for the basic region of Dom34. First, analogous to the GGQ motif in eRF1, this region may interact with the ribosome and influence some aspect of translation. Alternatively, as proposed previously (Davis and Engebrecht, 1998), this region may act as a nuclear localization signal. To test if the basic region affected Dom34 localization, we tagged Dom34 at its C-terminus with mCherry and examined its localization with and without mutation of loop C. We observed that both the wild-type Dom34 and the loop C mutant showed a similar, and predominantly cytosolic distribution, although we did not observe Dom34 being excluded from the nucleus (data not shown). We interpret this observation to indicate that loop C does not influence Dom34 subcellular location. This implies that this region may play a direct role in the process of NGD, perhaps by interacting with the ribosome in a manner analogous to the GGQ motif of eRF1.
The C-Terminal Domain
The C-terminal domain of eRF1 is thought to interact with eRF3 and to form a complex that allows for efficient translation termination (Merkulova et al., 1999; Eurwilaichitr et al., 1999). By this analogy, the C-terminal domain of Dom34 may interact with Hbs1 to form a Hbs1-Dom34 complex that functions most efficiently in NGD. Indeed, comparison of the C-terminal domains of eRF1 and Dom34 has led to the proposal that three regions (residues 278-302, 370-374, and an acidic tail) of the Dom34 C-terminal domain may mediate interaction with Hbs1 (Graille et al., 2008). However, since Hbs1 is not absolutely required for NGD (Doma and Parker, 2006), we have not yet investigated the effects of mutations in this C-terminal domain in vivo.
The N-Terminal Domain
The N-terminal domain of eRF1 contains an extended structure thought to position a conserved NIKS sequence in the decoding site to recognize stop codons (reviewed in Noble and Song, 2008). In contrast, Dom34 has a distinctly different N-terminal domain with some similarity to an Sm fold (Lee et al., 2007; Graille et al., 2008). Because the spatially corresponding domain of eRF1 interacts with the ribosome and the A site, one possibility is that the Dom34 N-terminal domain interacts with the ribosome, but does so it a manner independent of the nature of the stop codon. If this is the case then we would expect that mutations in this domain would decrease NGD. By analogy to the NIKS loop sequence in eRF1, we hypothesized that the two loops (loop A, residues 49-53; loop B, residues 87-92) seen in the N-terminal domain of Dom34 might be important in NGD, and therefore we mutated residues in these regions to alanine. We observed that mutations in the both loop A and loop B led to a modest but reproducible decrease in the amount of mRNA fragment produced (Figure 2B, lanes 5 and 6), despite being well expressed as assessed by Western analysis (Figure 3). Moreover, we observed that double mutants containing substitutions in both loop A and loop B, in loop A and loop C, or in loop B and loop C essentially reduced NGD to the levels seen in a dom34Δ strain (Figure 2B, lanes 8–10). From these observations we conclude that both loop A and loop B in the N-terminal domain are required for fully efficient NGD and that Dom34 variants with lesions in any two of the three major loops of the Dom34 structure are nonfunctional.
A second interesting feature of the N-terminal domain is a set of aspartic and glutamic acid residues that have been proposed to function as part of a nuclease domain (Lee et al., 2007). We substituted alanine for each of the acidic residues (residues Glu23, Glu26, and Asp27), either separately or in combination. We observed that mutations of these acidic residues had varying effects. Substitution of E23 or E26 by alanine, which are both residues required for proposed nuclease activity (Lee et al., 2007), led to little or no difference in mRNA fragment production, indicating these residues are not required for nucleolytic cleavage of the mRNA (Figure 2A, lanes 5 and 6). In contrast, mutation of D27, which is not expected to affect the proposed nuclease (Lee et al., 2007), led to a dramatic reduction in NGD (Figure 2A, lane 7). A triple mutant in which all three acidic residues were substituted (EED triple) was also defective in NGD, which is likely to a consequence of the D27A mutation (Figure 2A, lane 8). We interpret these results to suggest that disruption of the previously proposed nucleolytic site does not inhibit NGD, although D27 does play some important role in Dom34 function. Based on the structure of Dom34, one possibility is that D27 stabilizes the fold of the N-terminal domain and that in the absence of this residue the protein folds poorly (Graille et al., 2008).
Analysis of the Nuclease Activity of Dom34
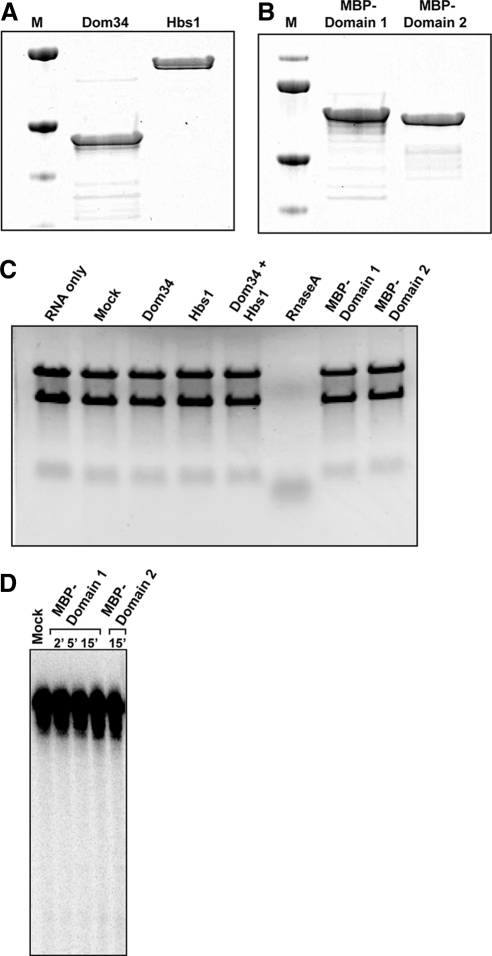
Previous work has described Dom34 and a Dom34 orthologue from archea as having nuclease activity (Lee et al., 2007). To examine the properties of this nuclease activity, we purified recombinant Dom34, and Hbs1 to high purity (Figure 4, A and B) and examined their ability to degrade RNA. We first checked the ability of these polypeptides to degrade RNA using the previously described assay of the degradation of total RNA (Lee et al., 2007). Surprisingly, we were unable to observe any RNA degradation by Dom34, Hbs1, or a Dom34/Hbs1 mixture (Figure 4C).
Figure 4.
Nuclease activity assays for purified Dom34 (and its domains). (A) Coomassie-stained protein gel of purified Dom34 and Hbs1. (B) Coomassie-stained protein gel of purified Dom 34 domains (MBP-domain 1 and MBP-domain 2 conjugates). (C) RNA cleavage assay with various expressed proteins. Bulk RNA (from yeast) was incubated with 1 μM of the indicated protein for 1 h at 40°C. Reactions were analyzed on a 1.2% formaldehyde-agarose gel. (D) Analysis of cleavage products from 3′ end-labeled hairpin RNA. Radiolabeled RNA containing a stable 34-base pair hairpin was incubated (alone, with MBP-domain 1 or with MBP-domain 2) for indicated times and cleavage products were analyzed on a 7 M urea/6% polyacrylamide gel.
Our inability to see nuclease activity of purified Dom34 or the Dom34/Hbs1 mixture was inconsistent with earlier work (Lee et al., 2007). However in that earlier work, the most robust nuclease activity was observed when domain 1 of Dom34 was purified as an isolated domain and tested either on total RNA or on a stem-loop substrate (Lee et al., 2007). Given this, we individually purified domain 1 and domain 2 of Dom34 as a maltose-binding protein fusion and tested their nuclease activity (Figure 4B). We observed no degradation of total RNA with these purified domains of Dom34 (Figure 4C). In addition, we were also unable to observe any cleavage of a stem-loop substrate as described previously (Figure 4D). Taken together, these results are consistent with Dom34 not being a nuclease for NGD in vivo and suggest that any nuclease activity of Dom34 might be highly sensitive to experimental conditions.
The Rps30a Protein Partially Complements the Endonuclease Defect in NGD in dom34Δ Strains
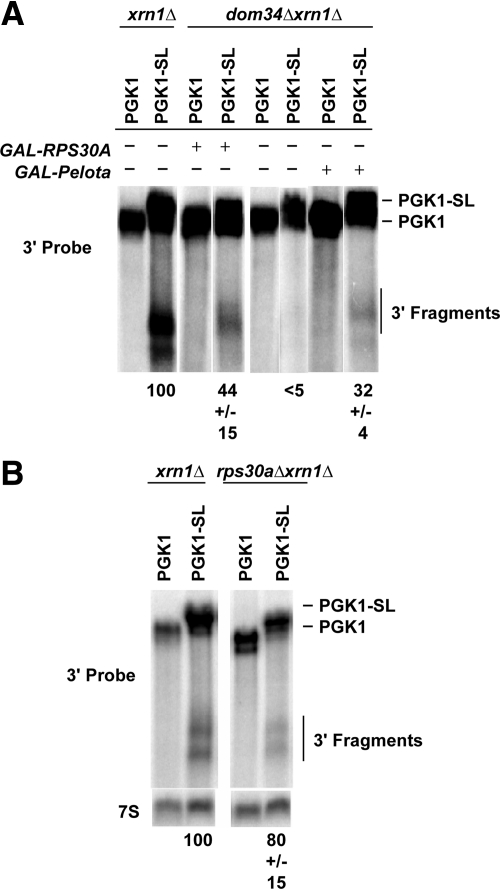
The above results suggested that Dom34 may not be the nuclease responsible for NGD. If this were true, then under some conditions cleavage of NGD substrates might be observed even in the absence of Dom34. Interestingly, RPS30a, a ribosomal protein, has previously been identified as a high copy suppressor of the dom34Δ growth defect (Davis and Engebrecht, 1998). This raises the possibility that overexpression of Rps30a might allow some cleavage of NGD substrates in a dom34Δ strain. To test this possibility, we analyzed the effects of Rps30a overexpression from the GAL promoter on NGD. We observed that overexpression of Rps30a complements the NGD defects in dom34Δ xrn1Δ mutant and restores cleavage of PGK1-SL at low levels (Figure 5). This suppression by Rps30a is consistent with observations that overexpression of this protein rescues sporulation, pseudohypheal growth defects, and the slow growth at low temperatures in dom34Δ mutants (Davis and Engebrecht, 1998) and also provides a second line of evidence that Dom34 is not absolutely required for the endonuclease cleavage of mRNA.
Figure 5.
Rps30A or Pelota overexpression complements the dom34Δ defect in endonucleolytic cleavage during NGD. (A) Northern analysis of steady-state PGK1 and PGK1-SL reporter mRNA in dom34 Δxrn1 Δ mutant strains with or without the presence of the GAL-RPS30A or GAL-Pelota plasmid. Probes were specific for mRNA regions 3′ of the stall site as illustrated. (B) Northern analysis of steady-state PGK1 and PGK1-SL reporter mRNA in rps30 Δxrn1 Δ mutant strains. Probes were specific for mRNA regions 3′ of the stall site.
To test if Rps30a was required for NGD, we examined the NGD-associated endonucleolytic cleavage of PGK1-SL reporter mRNA in rps30AΔ ski7Δ and rps30AΔ xrn1Δ strains. We observed that Rps30a was not required for NGD (Figure 5). Thus, Rps30a can be a high copy suppressor of the Dom34 role in NGD, but is not required for NGD normally.
It is unclear why overexpression of Rps30a would suppress the defect in NGD in the dom34Δ strains. One possibility is that extra Rps30a leads to prolonged elongation pauses or peptide release, thereby committing ribosomes to a mRNA cleavage event (see below for kinetic model of NGD). Such a role of Rps30a could be due to it having additional abnormal interactions with the ribosome when overexpressed, or by indirect effects on gene expression of other factors. Future experiments will be required to understand the mechanism by which Rps30a can activate Dom34 independent NGD.
Pelota, a Drosophila Homolog of Dom34p, Functions in NGD in Insect Cells and Can Complement a dom34Δ Strain in S. cerevisiae
Dom34p and its homologues belong to a highly conserved family of proteins, suggesting that the process of NGD is conserved in other organisms. For example, Pelota is a Dom34p homolog in Drosophila and has been characterized as essential for mitotic steps during the cell cycle and for Bam-independent control of regeneration (Xi et al., 2005). To determine if Pelota also functions in NGD, we tested whether expression of Pelota in S. cerevisiae could complement a dom34Δ for NGD. We expressed Pelota under the control of a GAL promoter in a dom34Δ xrn1Δ strain and examined the production of the PGK1-SL 3′ mRNA fragment by NGD. We observed that expression of Pelota in a dom34Δ xrn1Δ mutant restored production of the mRNA fragment arising by NGD, although at low levels (Figure 5). This correlates with independent observations that Pelota complements the sporulation and growth defects in dom34Δ mutants at low temperatures (Davis and Engebrecht, 1998). This observation suggests that Pelota functions similarly to Dom34 and that NGD is conserved in other eukaryotes.
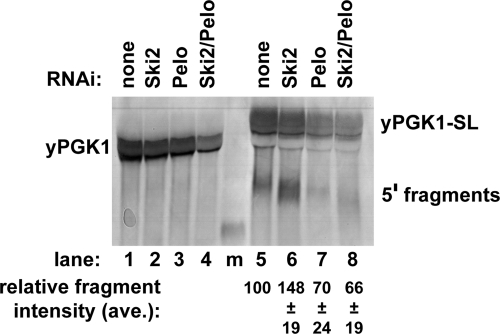
To directly test if Pelota functions in NGD in Drosophila cells, we expressed the yeast PGK1 and PGK1-Sl mRNAs in S2 cells. We observed that expression of the PGK1-SL mRNA, but not the PGK1 mRNA without the elongation stall, led to the accumulation of a mRNA fragment corresponding to the 5′ fragment generated by NGD (Figure 6, lanes 1 and 5). Consistent with this fragment being produced by NGD and then degraded 3′ to 5′, the levels of this fragment were increased by dsRNA knockdowns of Ski2 (lane 6), which is required for 3′ to 5′ exonucleolytic degradation by the exosome (Anderson and Parker, 1998). Importantly, we observed that dsRNA knockdowns of Pelota led to a reduction of the mRNA fragment with or without knockdowns of Ski2 (Figure 6, cf. lanes 5 and 7 with lanes 6 and 8). Taken together, these results indicate that NGD and the function of Dom34 orthologues is conserved between yeast and insect cells and presumably in other eukaryotes as well.
Figure 6.
NGD functions in Drosophila S2 cells and is dependent on Pelota. Northern analysis of yPGK and yPGK-SL reporter mRNA expressed in S2 cells depleted of Ski2, Pelota, or both by RNA interference. The Northern probe is designed to hybridize upstream of the stall site in PGK-SL. Percent fragment intensities were calculated as the intensity of the fragment band divided by the sum of the intensities of the fragment and full-length bands for each lane.
Implications for NGD
Our results indicate that NGD and the role of Dom34 and its orthologues is conserved within eukaryotic cells. We also identify three loops on the Dom34 structure that are required for optimal NGD. Our results also suggest that the proposed nuclease activity of Dom34 is not required for NGD, because mutations inactivating the proposed active site do not affect NGD in vivo, and that mRNA cleavage can be partially restored in dom34Δ strains when Rps30a is overexpressed. In addition, we note that the proposed Dom34 nuclease activity is not of broad enough specificity for NGD because it was specific for double-stranded regions (Lee et al., 2007), whereas NGD can occur in response to translation pauses at rare codons and premature translation termination sites, which would not necessarily be double-stranded (Doma and Parker, 2006). Finally, we have been unable to document nuclease activity of recombinant Dom34 suggesting that any nuclease activity of this protein is highly sensitive to biochemical conditions (Figure 4). It remains possible that the Dom34 nuclease activity contributes to NGD, but that there are redundant nucleases that can cleave the mRNA. Nevertheless, an unresolved issue is the mechanism of endonucleolytic cleavage during NGD and the precise role for Dom34 in that process.
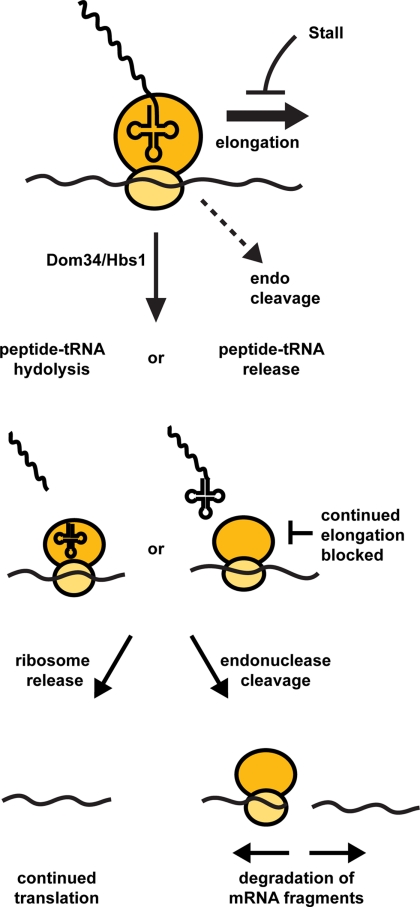
On the basis of these experiments and information in the literature, we propose a model for the process of NGD that is based on the kinetic competition between different molecular events that can occur at a given codon in the mRNA and how those events are influenced by elongation stalls and Dom34/Hbs1 (cartooned in Figure 7). Specifically, we propose that at any given codon three events can occur; continued elongation, endonuclease cleavage of the mRNA, or interaction of Hbs1/Dom34 with the empty A-site, which seems likely given the similarity of these two proteins to eRF1 and eRF3 and the fact that a stalled ribosome might be expected to have an empty A site. At a normal codon, continued elongation would be kinetically favored and any endonuclease cleavage event would be extremely slow in comparison. However, during translation stalls the rate of Hbs1/Dom34 interaction would become competitive with elongation. By analogy to eRF1 and eRF3 function, interaction of Hbs1/Dom34 with the ribosome is most likely to lead to peptide-tRNA hydrolysis and release of the peptide (reviewed in Petry et al., 2008; Jackson, 2007), or alternatively could lead to release of a tRNA-peptide conjugate. The consequence of such an action would be to prevent any further elongation and commit the translation complex to alternative fates. We consider it likely that the next step would be a competition between ribosome disassembly and release from the mRNA with endonuclease cleavage, which is based on the observation that NGD is generally not extremely fast (t1/2 = 8 min for PGK1-SL), unlike NMD (t1/2 for PGK1 early nonsense codon = 1 min; Cao and Parker, 2003) or NSD (t1/2 for PGK1 with no stop codon <2 min, van Hoof et al., 2002) and therefore is unlikely to be triggered by every stalled ribosome (Doma and Parker, 2006). Note that the endonuclease cleavage could be caused by recruitment of a nuclease by the ribosome or might be an intrinsic activity of the ribosome. Thus, in this model the function of Dom34/Hbs1 in NGD is to terminate translation, thereby preventing continued elongation and committing the mRNA either to degradation or ribosome release. This model predicts that extremely strong translation stall might trigger NGD independent of Dom34/Hbs1, and that Dom34/Hbs1 might function as termination factors for some mRNAs in a stop codon independent manner.
Figure 7.
A model for NGD.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0028) on May 6, 2009.
REFERENCES
- Anderson J. S., Parker R. P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- Carr-Schmid A., Pfund C., Craig E. A., Kinzy T. G. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol. Cell. Biol. 2002;22:2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C., Blanchet C., Geourjon C., Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Davis L., Engebrecht J. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics. 1998;149:45–56. doi: 10.1093/genetics/149.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K., Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Eurwilaichitr L., Graves F. M., Stansfield I., Tuite M. F. Mol. Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P. A., van Hoof A., O'Donnell K., Guerrerio A. L., Parker R., Dietz H. C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Gandhi R., Manzoor M., Hudak K. A. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. The J. Biol. Chem. 2008;283:32218–32228. doi: 10.1074/jbc.M803785200. [DOI] [PubMed] [Google Scholar]
- Graille M., Chaillet M., van Tilbeurgh H. Structure of yeast Dom34, a protein related to translation termination factor Erf1 and involved in No-Go decay. J. Biol. Chem. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- Inagaki Y., Ford Doolittle W. Evolution of the eukaryotic translation termination system: origins of release factors. Mol. Biol. Evol. 2000;17:882–889. doi: 10.1093/oxfordjournals.molbev.a026368. [DOI] [PubMed] [Google Scholar]
- Inge-Vechtomov S., Zhouravleva G., Philippe M. Eukaryotic release factors (eRFs) history. Biol. Cell (European Cell Biology Organization) 2003;95:195–209. doi: 10.1016/s0248-4900(03)00035-2. [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L. E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Jackson R. J. The missing link in the eukaryotic ribosome cycle. Mol. Cell. 2007;28:356–358. doi: 10.1016/j.molcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Kong C., Ito K., Walsh M. A., Wada M., Liu Y., Kumar S., Barford D., Nakamura Y., Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- Lee H. H., et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol. Cell. 2007;27:938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Maquat L. E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Merkulova T. I., Frolova L. Y., Lazar M., Camonis J., Kisselev L. L. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- Noble C. G., Song H. Structural studies of elongation and release factors. Cell Mol. Life Sci. 2008;65:1335–1346. doi: 10.1007/s00018-008-7495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., Weixlbaumer A., Ramakrishnan V. The termination of translation. Curr. Opin. Struct. Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Song H., Mugnier P., Das A. K., Webb H. M., Evans D. R., Tuite M. F., Hemmings B. A., Barford D. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. 2000. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P. A., Dietz H. C., Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Xi R., Doan C., Liu D., Xie T. Pelota controls self-renewal of germline stem cells by repressing a Bam-independent differentiation pathway. Development. 2005;132:5365–5374. doi: 10.1242/dev.02151. [DOI] [PubMed] [Google Scholar]