Abstract
Cell polarization is a key prerequisite for directed migration during development, tissue regeneration, and metastasis. Integrin-linked kinase (ILK) is a scaffold protein essential for cell polarization, but very little is known about the precise mechanisms whereby ILK modulates polarization in normal epithelia. Elucidating these mechanisms is essential to understand tissue morphogenesis, transformation, and repair. Here we identify a novel ILK protein complex that includes Engulfment and Cell Motility 2 (ELMO2). We also demonstrate the presence of RhoG in ILK–ELMO2 complexes, and the localization of this multiprotein species specifically to the leading lamellipodia of polarized cells. Significantly, the ability of RhoG to bind ELMO is crucial for ILK induction of cell polarization, and the joint expression of ILK and ELMO2 synergistically promotes the induction of front-rear polarity and haptotactic migration. This places RhoG–ELMO2–ILK complexes in a key position for the development of cell polarity and forward movement. Although ILK is a component of many diverse multiprotein species that may contribute to cell polarization, expression of dominant-negative ELMO2 mutants is sufficient to abolish the ability of ILK to promote cell polarization. Thus, its interaction with ELMO2 and RhoG is essential for the ability of ILK to induce front-rear cell polarity.
INTRODUCTION
Directed cell migration is a critical component of multiple biological and pathological processes, including embryonic development, tissue regeneration after injury, and metastasis of transformed cells. Forward cell movement occurs in integrated cycles that involve cell polarization, extension of lamellipodia and formation of stable focal adhesions, followed by disassembly of focal contacts and retraction of the trailing tail (Broussard et al., 2008). Although it is well established that these processes require proteins that link and transduce signals from the extracellular matrix to the actin cytoskeleton, such as integrins and their associated molecules, detailed understanding of how cell polarization and migration occur continues to be limited by the complexity of the pathways involved.
A number of studies have identified integrin-linked kinase (ILK) as a key scaffold protein involved in cell polarization, migration, survival, and differentiation (Bock-Marquettee et al., 2004; Legate et al., 2006; McDonald et al., 2008a; Nakrieko et al., 2008b). ILK contributes to focal adhesions through its association with integrins, paxillin, PINCH, and parvins (Legate et al., 2006), but it also interacts with a variety of other proteins, including the phosphatase ILKAP (Leung-Hagesteijn et al., 2005), tubulin and other centrosomal proteins (Dobreva et al., 2008), rictor (McDonald et al., 2008b), and a renal chloride/bicarbonate exchanger (Keskanokwong et al., 2007). The association of ILK with integrins may play a role in the induction of cell polarity and directional movement via focal adhesion complexes. Although the numerous interactions that ILK exhibits with multiple types of proteins suggests that ILK may also modulate establishment of cell polarity by additional mechanisms independent of focal adhesions, this possibility has remained unexplored.
Polarization involves the heterogeneous distribution of cellular components and is a key event for migration of adherent cells, neuronal and lymphocytic function, maintenance of epithelial apico-basal surfaces, and asymmetric cell division (Iden and Collard, 2008). In spite of the fact that establishment of cell polarity is regulated through numerous pathways, a common element in all polarized systems examined to date involves the Rho family of small GTPases, including RhoG and Rac1. For example, forward cell movement requires localization and activation of Rac1 at the cell front, triggering the formation of lamellipodial extensions. The latter process occurs in synchrony with integrin-mediated transduction of adhesion signals required for migration at the leading edge. During this process, Rac1 activation results from the joint contribution of several signaling modules, including ELMO, Engulfment and Cell Motility (ELMO)–Dock complexes.
ELMO proteins are scaffolds that bind to members of the Dock family, leading to the activation of Rac1 at the cell front (Katoh et al., 2005; Santy et al., 2005). Analysis of the broad tissue distribution of the three known mammalian ELMO homologues has shown extensive overlap, and their biochemical and biological characterization has yet to identify major differences in their functions (Gumienny et al., 2001). In addition to migration, ELMO proteins are important mediators of engulfment of apoptotic cells, bacterial invasion, and neurite outgrowth (Gumienny et al., 2001; Katoh and Negishi, 2003; Handa et al., 2007). ELMO–Dock complexes are recruited to cell protrusions by various mechanisms, including interaction with RhoG, Arf6 or the phosphatidylserine receptor (Katoh and Negishi, 2003; Wang et al., 2003; Santy et al., 2005). Significantly, notwithstanding the recognized scaffolding properties of ELMO, few ELMO-binding proteins have been identified.
In this study, we characterize a novel, specific interaction between ILK and Engulfment and Cell Motility 2 (ELMO2), which occurs in a variety of primary nontransformed cell types. We map the ILK-binding domain of ELMO2 to the N-terminus and demonstrate that ELMO2 serves as a bridge that links active RhoG to ILK. Notably, ILK colocalizes with ELMO2 or with RhoG to leading lamellipodia. The biological role of ILK–ELMO2 complexes includes promotion of cell polarity and directional migration, as evidenced by the ability of ELMO2 to cooperate with ILK in formation of lamellipodia and acquisition of polarized morphology in response to laminin stimulation. Our results show crucial roles for ILK–ELMO2 complexes in epithelial cell polarization and motion.
MATERIALS AND METHODS
Cell Culture and Transfections
Primary mouse keratinocytes isolated from 1–2-d-old CD-1 mice were cultured in Ca2+-free Eagle's minimum essential medium (EMEM; 06–174G, Lonza, Rockland, ME) with supplements (low-Ca2+ medium) as described (Ivanova et al., 2007). Keratinocytes were induced to differentiate by increasing the Ca2+ concentration in the growth medium to 1 mM (high-Ca2+ medium). Primary human dermal microvascular endothelial cells were purchased from and cultured as recommended by Lonza. IMDF cells (Apostolova et al., 2002) were cultured in HyQ DMEM-RS (SH30565.01, HyClone, Logan, UT) containing 4% fetal bovine serum (FBS). Cells were transfected as described (Hildinger et al., 2007), with some modifications. A 60-μl aliquot of polyethyleneimine (PEI) stock solution (1 mg/ml, pH 7.0, 25-kDa linear PEI, cat. no. 23966, Polysciences, Warrington, PA) was mixed by vortexing with 8 μg vector DNA dissolved in 440 μl 150 mM NaCl. Complex formation was allowed to proceed for 10 min, and the DNA mix was added to the culture medium of cells plated in 100-mm dishes. The DNA-containing medium was replaced with normal culture medium 4 h later. Cells were cultured 24–48 h before processing.
Antibodies, Reagents, and Plasmids
Antibodies and their sources are as follows: β-tubulin (E7, Developmental Studies Hybridoma Bank, University of Iowa), green fluorescent protein (GFP; sc8334, Santa Cruz Biotechnology, Santa Cruz, CA), FLAG-M2 (Sigma, St. Louis, MO), V5 (Invitrogen, Carlsbad, CA), glutathione S-transferase (GST; 91G1, Cell Signaling, Beverly, MA), ELMO2 (sc21655, Santa Cruz Biotechnology), ILK (mouse monoclonal, 611802, Transduction Laboratories, Lexington, KY, and rabbit polyclonal, 1979–1, Epitomics, Burlingame, CA). Horseradish peroxidase–conjugated goat anti-mouse and goat anti-rabbit IgG were from Jackson ImmunoResearch Laboratories (West Grove, PA). AlexaFluor-conjugated goat anti-mouse IgG were purchased from Molecular Probes/Invitrogen (Eugene, OR). Glutathione Sepharose 4B and protein A/G UltraLink resin were, respectively, from Amersham Biosciences (Piscataway, NJ) and Pierce Chemical (Rockford, IL). FLAG peptide and all other chemicals were from Sigma.
The vectors encoding V5-tagged wild-type and mutant ILK, as well as thioredoxin (TRX)-ILK have been described (Vespa et al., 2005; Nakrieko et al., 2008a). mCherry-tagged ILK was generated by PCR amplification of a V5-tagged human ILK cDNA and cloning into mCherry-C1 (Clontech, Palo Alto, CA). A cDNA encoding FLAG-tagged human RhoG was generated by PCR and cloned into pET-28a (Novagen, Madison, WI), which provides N- and C-terminus His tags. GFP-tagged ILK and dsRed-tagged paxillin were generously provided by Dr. C. Turner (State University of New York, Upstate Medical University, New York) and Dr. A. F. Horwitz (University of Virginia, VA), respectively. The vectors encoding GFP-tagged wild-type and mutant RhoG have been described (Wennerberg et al., 2002). The vectors encoding FLAG- and GFP-tagged ELMO proteins have been described (Gumienny et al., 2001).
In-Gel Tryptic Digestion and Mass Spectrometry Analysis
Primary human keratinocytes were isolated from neonatal foreskins and cultured as described (Chang et al., 2006). The cells were infected with a recombinant adenovirus encoding V5-tagged ILK (Vespa et al., 2003) and cultured for 96 h in growth medium containing 1.0 mM Ca2+. To isolate ILK-interacting proteins, V5-tagged ILK was immunoprecipitated from keratinocyte extracts, prepared by harvesting and lysing the cells in coimmunoprecipitation (CoIP) buffer (1% Triton X-100, 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 5 mM NaF, 2 mM Na3VO4, 1 mM PMSF, 2 μg/ml aprotinin, 2 μg/ml leupeptin, and 2 μg/ml pepstatin; 45 min, 4°C). Cell debris in the lysates were removed by centrifugation, and samples containing each 10 mg protein were precleared with 100 μl protein A/G azlactone beads (53132; Pierce Chemical) for 2 h at 4°C. Precleared lysates were incubated with mouse IgG or with mouse monoclonal anti-V5 antibodies (4 h, 4°C). Immunocomplexes were isolated with protein A/G azlactone beads (30 min, 4°C) and resolved by denaturing gel electrophoresis (SDS-PAGE). Bands were excised from silver-stained gels, cut into 1-mm cubes, and washed in distilled water. Proteins in the gel were digested by incubation with nonself cleaving trypsin (Promega, Madison, WI) for 16 h at 37°C, as described (Shevchenko et al., 1996; Erdjument-Bromage et al., 1998; Gobom et al., 1999). Peptides were extracted with 30 μl acetonitrile and 30 μl 5% formic acid, with vortexing and sonication. The extracted peptides were desalted on columns consisting of Eppendorf gel-loader tips packed with 1 μl each of POROS R2 and R3 resins (Applied Biosystems, Foster City, CA). Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis was conducted on a Micromass Reflectron spectrometer (Manchester, United Kingdom) in positive ion reflector mode. Spectra were collected within a m/z range of 900–4000, with calibration of the MALDI-TOF analyzer using a 1:2:3 ratio of angiotensin I, renin, and adenocorticotrophic hormone clip 18-39. Spectra were analyzed with MassLynx 3.5 software. Peptide mapping and database analyses were conducted using ProFound (http://prowl.rockefeller.edu/cgbin/ProFound) and Mascot (www.matrixscience.com) software. This experiment was repeated four times.
Immunoblot Analysis and IP
Protein lysates were prepared and analyzed as described (Ivanova and Dagnino, 2007). For IP experiments, harvested cells were lysed in CoIP buffer (1% Triton X-100, 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 5 mM NaF, 2 mM Na3VO4, 1 mM PMSF, 2 μg/ml aprotinin, 2 μg/ml leupeptin, and 2 μg/ml pepstatin) for 45 min at 4°C. Cell debris in the lysates were removed by centrifugation, and 1–2 mg protein was precleared with 10 μl protein A/G UltraLink resin (53132; Pierce Chemical) for 2 h at 4°C. Precleared lysates were incubated with antibodies indicated in individual experiments (16 h, 4°C). Immunocomplexes were isolated with protein A/G resin (30 min, 4°C), resolved by SDS-PAGE, and analyzed by immunoblot. For tandem IPs, immunocomplexes first isolated using anti-FLAG antibodies were eluted from the protein A/G resin with 20 μg FLAG peptide (100 μg/ml). The eluate was subjected to a second IP step using anti-V5 antibodies, and the resulting complexes were analyzed as above. Results shown are representative of at least three experiments.
Recombinant Protein Purification and In Vitro Binding Assays
Bacterially produced GST fusion proteins were obtained and purified as described (Ivanova et al., 2007). Trx, Trx-ILK, and RhoG recombinant proteins were isolated from bacterial lysates prepared in buffer A (10 mM imidazole, pH 8.0, 300 mM NaCl, 50 mM NaH2PO4, 1 mM DTT, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM PMSF) and purified using NiNTA agarose (Qiagen, Chatsworth, CA). Proteins were recovered by sequential elutions with buffer A supplemented with 250 or 500 mM imidazole. Eluted proteins were desalted using Amicon Ultra centrifugal filter devices (Millipore, Bedford, MA) and stored in 10% glycerol at −80°C, in single-use aliquots. Purified RhoG proteins (2 μg) were allowed to bind guanine nucleotides by incubation in buffer B (20 mM Tris-HCl, pH 7.5, 0.1 mM DTT, and 5 mM EDTA) containing 0.1 mM GDPβS or GTPγS for 15 min at 22°C immediately before use. Protein-binding assays were conducted by incubation of 2 μg of each purified recombinant protein in buffer C (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 0.5 mg/ml bovine serum albumin (BSA), 1 mM DTT, and 1 μg/ml each aprotinin, leupeptin, and pepstatin, 1 mM PMSF, 10% glycerol) for 16 h at 4°C, followed by IP as described above, with antibodies indicated in individual experiments. Results shown are representative of at least three experiments.
Fluorescence and Confocal Microscopy
Forty-eight hours after transfection, keratinocytes were briefly trypsinized, resuspended in serum- and Ca+-free EMEM supplemented with 2.5% BSA, and plated onto laminin 332 matrix-coated coverslips (Nakrieko et al., 2008b). Cells were cultured for 1.5–2 h to allow attachment and spreading and processed for direct or immunofluorescence microscopy, as described (Ivanova and Dagnino, 2007). In experiments assessing attachment and polarization of cells exogenously expressing ILK and ELMO2 proteins, 1000 cells were examined in each experiment, and statistical significance was set at p < 0.05 (ANOVA). Photomicrographs were obtained with a Leica DMIRBE fluorescence microscope (Deerfield, IL) equipped with an Orca-ER digital camera (Hamamatsu Photonics, Hamamatsu City, Japan), using Volocity 4.3.2 software (Improvision, Coventry, United Kingdom). Confocal images were captured with a Zeiss LSM5 DUO scanning laser confocal microscope (Jena, Germany), using ZEN 2007 SP1 software (Zeiss). Results shown are representative of at least three experiments using triplicate samples.
Migration Assays
Primary keratinocytes were transfected with plasmids encoding a GFP- and an mCherry-tagged proteins, as indicated in the experiment of Figure 7E, and cultured in growth medium for 48 h. The cells were briefly trypsinized and resuspended in warm serum- and Ca2+-free EMEM supplemented with 2.5% BSA at a density of 2.5 × 105 cells/ml, and a 400-μl portion of this cell suspension was added a tissue culture insert (Transwell, 8-μm pore size; 353097, BD Falcon, Bedford, MA) that had been precoated with laminin 332 matrix on the surface facing the lower chamber. Culture medium on both upper and lower chambers consisted of serum- and Ca2+-free EMEM supplemented with 2.5% BSA. The cells were cultured at 37°C for 2 h, and those cells that migrated through the membrane were fixed with 4% freshly diluted paraformaldehyde and stained with Hoescht 33528 (10 μg/ml, Sigma). The number of GFP- and mCherry-double–positive cells that migrated was determined by microscopic examination of the lower surface of the insert and normalized to the total number of GFP- and mCherry-double–positive cells that had been originally added to the upper chamber of the culture insert. Each experiment was conducted with duplicate samples.
Figure 7.
ILK and ELMO2 cooperate to promote cell polarization and migration. (A) Primary mouse keratinocytes were cotransfected with mCherry-tagged ILK and GFP-tagged ELMO2. Forty-eight hours after transfection, the cells were briefly trypsinized and replated onto a laminin 332 matrix, and 2 h later they were processed for fluorescence microscopy. Bar, 20 μm. (B) Relative abundance of attached, nonpolarized, and polarized cells expressing mCherry-tagged ILK and/or GFP-tagged ELMO2, as indicated. Results are expressed as the means; error bars, SEM (n = 3). The asterisks indicate p < 0.05 (ANOVA) relative to cells expressing mCherry and GFP. (C) Relative abundance of attached, nonpolarized and polarized cells expressing mCherry-tagged ILK in the presence or absence of either GFP or the indicated GFP-tagged ELMO2 proteins. The abbreviations used for the ELMO2 proteins are as follows: wt, wild type; N, ELMO2 1–481; C, ELMO2 482–718. ILK (−) and ELMO2 (−) indicate, respectively, that the cells were transfected with a vector encoding mCherry or GFP. Results are expressed as the means; error bars, SEM (n = 3). *p < 0.05 (ANOVA) relative to cells expressing mCherry and GFP. (D) Relative abundance of attached, nonpolarized, and polarized cells expressing mCherry-tagged ILK in the presence or absence of GFP or the indicated GFP-tagged wild-type (wt) or F37A mutant RhoG. ILK (−) and RhoG (−) indicate, respectively, that the cells were transfected with a vector encoding mCherry or GFP. Results are expressed as the means; error bars, SEM (n = 3). *p < 0.05 (ANOVA) relative to cells expressing mCherry and GFP. (E) Primary cultured keratinocytes were transfected and briefly trypsinized, as described for C. The cells were resuspended in serum- and Ca2+-free medium containing 2.5% BSA, and 1 × 105 cells were added to the upper chamber of a Transwell insert in which the lower side had been coated with laminin 332 matrix. The cells were cultured at 37°C for 2 h to allow migration through the insert pores, and cells that were positive for both GFP and mCherry fluorescence and that had migrated to the lower side of the insert were quantified. An aliquot of the cell suspension was analyzed to determine the total number of GFP- and mCherry double–positive cells initially dispensed into the cell insert. The fraction of cells expressing GFP and mCherry that had migrated is set to 1 and represents the control population. The results show the migration of cells expressing mCherry-ILK and/or GFP-ELMO2 proteins relative to the control. The results are expressed as the means; error bars, SEM (n = 4). *p < 0.05 (ANOVA) relative to cells expressing mCherry and GFP.
RESULTS
Interaction of ILK with ELMO2
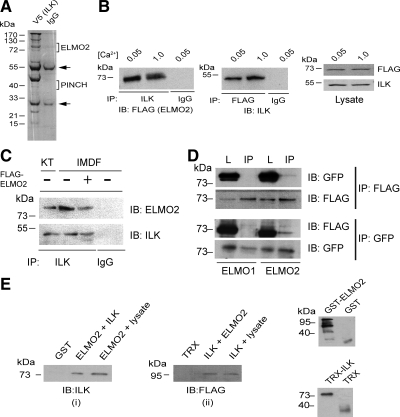
To identify candidate proteins that interact with ILK in the epidermis, we conducted a proteomic analysis of V5-tagged ILK immunoprecipitates isolated from lysates prepared from primary human epidermal keratinocytes induced to differentiate by culture in medium containing 1.0 mM Ca2+. This analysis identified ELMO2 as a novel ILK-interacting protein, as well as other well-established ILK-binding proteins, such as PINCH (Legate et al., 2006; Figure 1A). To confirm this interaction and to determine whether ILK–ELMO2 complex formation depends on differentiation status, we transiently transfected primary murine epidermal keratinocytes with FLAG-tagged ELMO2 and isolated endogenous ILK immunoprecipitates from undifferentiated cells cultured in low-Ca+2 medium (≤0.05 mM Ca2) or from keratinocytes induced to terminally differentiate by culture in high-Ca2 medium (1.0 mM Ca2). We found the presence of exogenously expressed ELMO2 in all endogenous ILK immunocomplexes, irrespective of the differentiation status of the cells (Figure 1B). Reciprocally, we found endogenous ILK in FLAG-ELMO2 immunoprecipitates isolated from undifferentiated or differentiated keratinocytes (Figure 1B). Given that ELMO2 was isolated from lysates of cells overexpressing ILK, we next investigated the association of endogenous ILK with endogenous ELMO2 in various tissues and cell types. To this end, we isolated cellular ILK immunoprecipitates from the epidermis, from primary cultured human microvascular endothelial cells and from cultured IMDF dermal fibroblasts. We detected cellular ELMO2 in ILK immunocomplexes in those three cell types (Figure 1C and data not shown), indicating that ILK-ELMO2 complexes naturally occur in a wide variety of cell types. To further investigate the ability of ILK to interact with ELMO proteins, we expressed FLAG-tagged ELMO1 or ELMO2 in IMDF cells, which exhibit nearly 100% transfection efficiency and, consequently, allow excellent detection of exogenously expressed proteins. In spite of the high degree of similarity between ELMO1 and ELMO2 (Gumienny et al., 2001), we did not detect any complexes containing ILK and ELMO1, consistent with the concept that ILK selectively interacts with ELMO2 (Figure 1D).
Figure 1.
Interaction of ILK with ELMO2. (A) Lysates prepared from primary cultured human keratinocytes infected with a V5-tagged ILK encoding recombinant adenovirus 96 h before harvest were incubated with anti-V5 antibodies or mouse IgG, as indicated. Immunocomplexes were resolved by denaturing gel electrophoresis. The gel was silver-stained, and 1-cm bands in each lane were excised and processed for mass spectrometry analysis. Brackets indicate the excised bands in which ELMO2 and PINCH were identified. Arrows indicate the position of IgG heavy and light chains. (B) Primary undifferentiated mouse keratinocytes were cultured in low-Ca2 medium (0.05 mM) and were transfected with a vector encoding FLAG-tagged ELMO2. After transfection, the cells were cultured in medium containing 0.5 mM Ca2, or were induced to differentiate by culture in medium with 1.0 mM Ca2 for 24 h. Cell lysates were prepared and endogenous ILK or FLAG-ELMO2 was immunoprecipitated. A control sample was immunoprecipitated with an unrelated antibody (IgG). The immunocomplexes were analyzed by immunoblot with antibodies against FLAG, to visualize exogenous ELMO2, or ILK, as indicated. Portions of the cell lysates (50 μg) before IP were resolved by SDS-PAGE and analyzed by immunoblot with antibodies against FLAG (to visualize ELMO2) or ILK. (C) Endogenous ILK–ELMO2 complexes in epidermis and IMDF cells. Lysates from keratinocytes freshly isolated from epidermis (KT) or from IMDF cells were immunoprecipitated with a rabbit anti-ILK antibody or with an irrelevant antibody (IgG). Immunocomplexes were analyzed by immunoblot with an antibody against ELMO2 or a mouse anti-ILK antibody, as indicated. The third lane represents a lysate from IMDF cells transfected with a FLAG-tagged ELMO2-encoding vector, used as additional control. (D) IMDF cells were transfected with FLAG-tagged ELMO1 or ELMO2, together with GFP-tagged ILK. Twenty-four hours after transfection, cell lysates were prepared and exogenous ILK or ELMO proteins were immunoprecipitated (IP). Immunocomplexes were analyzed by immunoblot with antibodies against FLAG (to visualize ELMO proteins) or GFP (to visualize ILK). A 25-μg aliquot of total cell lysate before IP (L) was included in the blots. (E) Bacterially produced GST-ELMO2 was allowed to interact in vitro with TRX-ILK or with keratinocyte lysates, and the coprecipitation of ILK was assessed by immunoblotting with an anti-ILK antibody (i). Similarly, bacterially produced TRX-ILK was allowed to interact in vitro with FLAG-tagged GST-ELMO2 or with lysates from keratinocytes transfected with FLAG-tagged ELMO2, and the coprecipitation of ELMO2 was assessed by immunoblotting with antibodies against FLAG (ii). Bacterially produced proteins were verified by immunoblot, using anti-GST or anti-TRX antibodies (iii).
To examine whether ILK and ELMO2 interact directly, we analyzed the ability of bacterially produced GST-ELMO2 and TRX-ILK to associate in vitro. GST-ELMO2, but not GST, was able to precipitate recombinant TRX-ILK, as well as endogenous ILK from cell lysates (Figure 1E). Reciprocally, TRX-ILK, but not TRX, formed complexes with GST-tagged ELMO2 or with FLAG-tagged ELMO2 isolated from cell lysates. Thus, the interaction between ILK and ELMO2 is direct.
Protein Domains that Mediate the Interaction of ILK and ELMO2
We exogenously expressed a series of FLAG-tagged ELMO2 deletion mutants (Figure 2A) together with GFP-tagged ILK to determine the region in ELMO2 that binds ILK. These experiments demonstrated that the presence of a GFP tag on ILK did not interfere with its ability to associate with ELMO2 (Figure 2B). Deletions of the ELMO2 C-terminus had no significant effect on its ability to associate with ILK, and an ELMO2 region comprising the first N-terminal 307 amino acid residues, containing four of the five putative Armadillo repeats, was sufficient for this interaction (Figure 2B). In contrast, an ELMO2 mutant lacking all Armadillo repeats, but containing the pleckstrin homology domain and the regions in ELMO that bind Dock family proteins (ELMO2 482-718) was not detected in ILK immunoprecipitates.
Figure 2.
ELMO2 domains that mediate interaction with ILK. (A) Schematic representation of FLAG-tagged ELMO2 mutants used. The numbers on the left indicate amino acid residues present in the mutants. Domains in ELMO2 shown are the armadillo repeats (A1–A5), the pleckstrin homology domain (PH) and the proline-rich region (PxxP). wt, wild type. (B) IMDF cells were transiently transfected with GFP-tagged ILK and the ELMO2 protein indicated, and 24 h later cell lysates were prepared. Exogenous ILK was immunoprecipitated with anti-GFP antibodies. The presence of ELMO2 proteins (indicated with an asterisk) in immunocomplexes was analyzed by immunoblot with anti-FLAG antibodies. Replicate samples of lysates before IP were analyzed by immunoblot to verify expression of ELMO2 proteins and ILK using, respectively, anti-FLAG and anti-ILK antibodies. Note that we occasionally detect GFP-tagged ILK as a doublet.
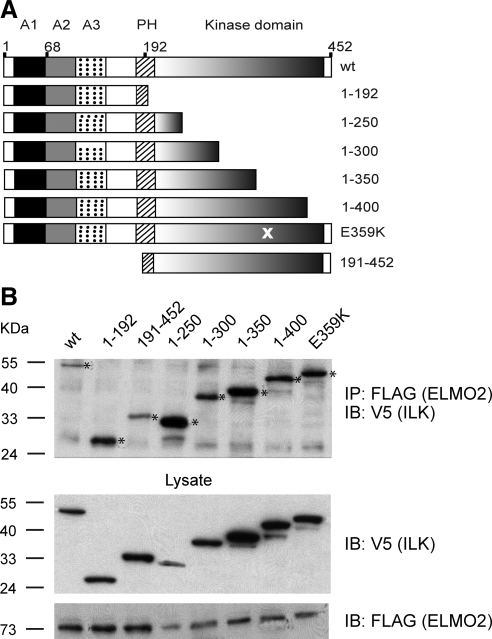
Through a similar approach, we expressed a series of ILK deletion and point mutants to map the region(s) in ILK that interact with ELMO2 (Figure 3A). As shown in Figure 3B, mutants with various deletions in the C-terminal kinase domain were able to associate with ELMO2, as did the point mutant ILK E359K, which exhibits abnormal kinase activity, indicating that the ILK kinase domain is dispensable. Of note, we also found a fragment lacking the N-terminal ankyrin repeats and containing an intact kinase domain (ILK 191-452) in full-length ELMO2 or ELMO2 (1-482) immunoprecipitates (Figure 3B and data not shown). Together, these data suggest that several regions in ILK contribute to optimal binding to the N-terminus of ELMO2.
Figure 3.
ILK domains that mediate interaction with ELMO2. (A) Schematic representation of V5-tagged ILK mutants used. Numbers on the right indicate amino acid residues present in the mutants. Domains in ILK shown are the ankyrin repeats (A1–A3), the pleckstrin homology domain (PH) and the kinase domain. Wild-type ILK is indicated as wt. (B) IMDF cells were transiently transfected with indicated V5-tagged ILK proteins and FLAG-tagged ELMO2. Twenty-four hours after transfection, cell lysates were prepared. Exogenous ELMO2 was immunoprecipitated with anti-FLAG antibodies, and the presence of ILK proteins (indicated with an asterisk) in immunocomplexes was analyzed by immunoblot with anti-V5 antibodies. Replicate samples of lysates before IP were analyzed by immunoblot to verify expression of exogenous ELMO2 and ILK proteins using, respectively, anti-FLAG and anti-V5 antibodies.
Several putative ankyrin repeats have been identified in ILK (amino acid residues 33-65, 66-98, and 99-131). Essential for the structure of these repeats are tetrapeptide motifs (Mosavi et al., 2002), which correspond in ILK to regions 36SPLH39, 69TPLH72, and 102VPLH105. We determined the role of each of these domains for ILK interactions with ELMO2 by ectopically expressing GFP-tagged ILK (1-192) forms in which these tetrapeptide motifs were individually mutated to alanines (Figure 4A). We found that ELMO2 associated with each of these ILK mutants, indicating that, individually, the ankyrin repeats of ILK are dispensable for interactions with ELMO2.
Figure 4.
Role of ILK ankyrin repeats in interaction with ELMO2. (A) Schematic representation of GFP-tagged ILK mutants used. Putative ankyrin repeats in ILK are shown with key amino acid residues in each repeat. The following mutations were introduced in the ankyrin repeat regions: S, residues 36SPLH39→AAAA; T, residues 69TPLH72→AAAA; V, residues 102VPLH105→AAAA. Wild-type ILK is indicated as wt. (B) IMDF cells were transiently transfected with indicated GFP-tagged ILK proteins together with FLAG-tagged ELMO2. Twenty-four hours after transfection, cell lysates were prepared. Exogenous ILK was immunoprecipitated with anti-GFP antibodies. The presence of ELMO2 in immunocomplexes was analyzed by immunoblot with anti-FLAG antibodies. The lane marked IgG denotes IP of lysates containing ELMO2 and wt ILK with an unrelated antibody, whereas the lane marked vector indicates IP with anti-GFP antibodies of lysates containing FLAG-ELMO2 and an empty control vector. Replicate samples of lysates before IP were analyzed by immunoblot to verify expression of exogenous ELMO2 and ILK proteins using, respectively, anti-FLAG and anti-GFP antibodies.
Modulation of ILK Subcellular Localization by ELMO2
To begin to assess the biological role of ILK–ELMO2 complexes, we first investigated the subcellular distribution of these two proteins. Because endogenous ELMO2 could not be detected by immunofluorescence microscopy, we transiently expressed GFP-tagged ELMO2 and mCherry-tagged ILK in primary murine keratinocytes. Transfected cells were trypsinized and replated at low density on a laminin 332 matrix to induce focal adhesion formation, cell spreading, as well as cell polarization and directional migration mediated by stimulation of integrin α3β1 (Choma et al., 2004). We used laminin 332 as it is the major extracellular matrix protein to which keratinocytes attach in normal, undamaged epidermis (Nguyen et al., 2000). Under these conditions, and in agreement with previous reports (Nikolopoulos and Turner, 2002; Vespa et al., 2003; Nakrieko et al., 2008a), mCherry-ILK was detected throughout the cytoplasm and, at low levels, in the nucleus when expressed in the absence of exogenous ELMO2 (Figure 5A). GFP-ELMO2 also distributed throughout the cell and, in cells with lamellipodia, ribbons of GFP-ELMO2 fluorescence were visualized in discrete regions of such extensions (Figure 5A). Significantly, exogenous ELMO2 was capable of recruiting ILK to membrane ruffles and lamellipodia formed in polarized cells, as evidenced by the presence of intense mCherry-ILK and GFP-ELMO2 fluorescence along cell protrusions (Figure 5, A and B). Further, coexpression of ILK and ELMO2 frequently gave rise to formation of extensive lamellipodia generally localized to the cell front, and occasionally localized to two different cell areas (Figure 5, A and B). Similarly, ELMO2 (1-307) was able to recruit ILK to cell extensions, although the cell protrusions observed with this mutant were generally substantially smaller than those generated by wild-type ELMO2 (Figure 5B). In contrast, an ELMO2 mutant lacking the N-terminus (ELMO2 482-718) was neither able to localize to lamellipodia, nor to recruit ILK to the plasma membrane (Figure 5B), in keeping with its inability to interact with ILK, as well as the reported failure of N-terminal truncations of ELMO1 proteins to localize to membrane ruffles (Grimsley et al., 2004).
Figure 5.
Colocalization of ILK with ELMO2 and RhoG. (A) Primary mouse keratinocytes were cotransfected with the indicated fluorescent proteins. Forty-eight hours after transfection, the cells were briefly trypsinized and replated onto a laminin 332 matrix. Keratinocytes were allowed to adhere and spread, and after 2 h they were processed for confocal microscopy. (B) Primary mouse keratinocytes were cotransfected with plasmids encoding mCherry-tagged ILK and the indicated GFP-tagged ELMO2 proteins. Forty-eight hours after transfection, the cells were briefly trypsinized and replated onto a laminin 332 matrix. Keratinocytes were allowed to adhere and spread, and after 2 h they were processed for confocal microscopy. Insets represent magnification of boxed areas. (C) Primary mouse keratinocytes were cotransfected with mCherry-tagged ILK and the indicated GFP-tagged RhoG proteins. Forty-eight hours after transfection, the cells were trypsinized and replated onto a laminin 332 matrix. Keratinocytes were allowed to adhere and spread, and after 2 h they were processed for confocal microscopy. Insets represent magnification of boxed areas. Bar, 20 μm.
Formation of Ternary Complexes Containing ILK, ELMO2, and RhoG
ILK and ELMO proteins are linked to signaling pathways involved in Rac1 activation in various cell types (Brugnera et al., 2002; Liu et al., 2005; Boulter et al., 2006; Grimsley et al., 2006; Nakrieko et al., 2008b). Specifically, ILK is essential for normal Rac1 activation triggered by integrin α3β1 stimulation, whereas ELMO cassettes activate Rac1 after their interaction with RhoG. Given that ILK binds to ELMO2, and ELMO2 binds to RhoG, we next investigated the possibility of association between ILK and RhoG. To this end, we first exogenously expressed GFP-tagged wild-type RhoG and mCherry-tagged ILK in epidermal keratinocytes. We briefly trypsinized the cells and replated them on a laminin 332 matrix to induce attachment, spreading and forward movement mediated by integrin α3β1. We first determined in these cells if ILK and RhoG are found at similar subcellular locations. In agreement with previous reports (Hiramoto et al., 2006; Nakrieko et al., 2008b), exogenous expression of RhoG induced formation of cell protrusions, and intense RhoG fluorescence was present at sites of membrane ruffling (Figure 5C). Remarkably, ILK colocalized at the membrane in those same areas enriched for RhoG (Figure 5C), suggesting that these two proteins may be elements of a common protein complex. We next investigated whether ILK was enriched in regions in which exogenously expressed RhoG mutant proteins could be found. We observed that ILK also colocalized with the constitutively active mutant RhoG Q61L, which induced formation of cell protrusions in a very similar manner to that observed with exogenous, wild-type RhoG (Figure 5C). In stark contrast, expression of RhoG T17N, an inactive mutant that binds to and presumably sequesters RhoG activators, such as Trio (Blangy et al., 2000), abolished the ability of the cells to spread on the laminin 332 matrix, irrespective of whether or not exogenous ILK was present, as evidenced by the spherical morphology of the cells expressing this RhoG mutant (Figure 5C). These observations suggested that pathways sensitive to inhibition by RhoG T17N are essential for keratinocyte spreading on laminin 332, but precluded the identification of the subcellular sites of potential colocalization of the two proteins, if any.
To further investigate the potential interaction between ILK, ELMO2, and RhoG, we determined the ability of RhoG to bind ELMO2 or ILK. We transiently transfected IMDF cells with RhoG-encoding vectors, together with plasmids encoding either ELMO2 or ILK, and were able to detect the presence of wild-type RhoG in ELMO2 immunocomplexes (Figure 6A). As previously reported (Katoh and Negishi, 2003), ELMO2 associated also with RhoG Q61L, but not with the T17N mutant (Figure 6A). Remarkably, we also detected ILK in immunocomplexes containing wild-type or Q61L RhoG, but not RhoG T17N (Figure 6B). These observations identify a novel physical interaction of ILK with Rho family GTPases.
Figure 6.
ILK, ELMO2, and active RhoG form trimeric complexes. (A) IMDF cells were cotransfected with vectors encoding FLAG-tagged ELMO2 and GFP or the indicated GFP-tagged RhoG protein. Cell lysates were prepared 24 h later, and exogenous ELMO2 was immunoprecipitated with anti-FLAG antibodies. The presence of GFP or RhoG proteins in immunocomplexes was analyzed by immunoblotting with anti-GFP antibodies. Replicate samples of lysates before IP were analyzed by immunoblot to verify expression of GFP GFP-RhoG proteins and of ELMO2 using, respectively, anti-GFP and anti-FLAG antibodies. The blots were also probed for β-tubulin, as a loading control. (B) IMDF cells were cotransfected with vectors encoding V5- and mCherry-doubly tagged ILK and GFP or the indicated GFP-tagged RhoG protein. Cell lysates were prepared 24 h later, and exogenous ILK was immunoprecipitated with anti-V5 antibodies. The presence of GFP or RhoG proteins in the ILK immunocomplexes was analyzed by immunoblotting with anti-GFP antibodies. Replicate samples of lysates before IP were analyzed by immunoblot to verify expression of GFP or GFP-RhoG proteins using anti-GFP antibodies and of V5-mCherry-ILK using anti-V5 antibodies. The blots were also probed for β-tubulin, as a loading control. (C) IMDF cells were cotransfected with vectors encoding V5- and mCherry-doubly tagged ILK, FLAG-tagged ELMO2, and the indicated GFP-tagged RhoG protein. Cell lysates were prepared 24 h later and subjected to two sequential IPs. The lysates were incubated first with anti-FLAG antibodies, to isolate ELMO2-containing immunocomplexes, which were eluted with excess FLAG peptide, and subjected to a second IP using anti-V5 antibodies, to isolate complexes containing both ELMO2 and ILK. ILK–ELMO2 immunocomplexes were analyzed by immunoblot with antibodies against GFP, to visualize RhoG proteins; V5, to visualize ILK; or FLAG, to detect ELMO2, as indicated. Portions of the cell lysates (50 μg) before IP were resolved by SDS-PAGE and analyzed by immunoblot with antibodies against V5, FLAG, or GFP to visualize, respectively, ILK, ELMO2, or RhoG proteins. The blots were also probed for β-tubulin, as a loading control. (D) In vitro binding assays were conducted with the bacterially produced proteins indicated on the left. FLAG-tagged RhoG was incubated with the indicated GTP or GDP analogue for 15 min at 22°C before binding assays. For binding, proteins were incubated for 2 h at 22°C. Complexes were immunoprecipitated, using antibodies against FLAG or GST, as indicated. Immunocomplexes were resolved on SDS-PAGE and analyzed by immunoblot, using the following antibodies: anti-GST, to detect GST or GST-ELMO2; anti-TRX, to detect TRX or TRX-ILK; and anti-FLAG, to detect FLAG-RhoG. (E) Immunoblots showing expression of the indicated bacterially produced proteins used for in vitro binding experiments.
The experiments described above do not distinguish between the possibility that ILK forms different binary complexes with either ELMO2 or RhoG, or that it participates in the formation of a ternary complex containing both ELMO2 and RhoG. To address this issue, we conducted sequential IPs in lysates from IMDF cells exogenously expressing V5- and mCherry-doubly tagged ILK, FLAG-tagged ELMO2 and one of various GFP-tagged RhoG proteins. ELMO2 immunocomplexes were first obtained from these lysates using anti-FLAG antibodies. After elution in the presence of excess FLAG peptide, the ELMO2-containing immunocomplexes were subjected to a second round of IP using anti-V5 antibodies. This approach allowed us to isolate species that contained both ILK and ELMO2, which were then analyzed by immunoblot for the presence of exogenous RhoG proteins. As shown in Figure 6C, both wild-type and Q61L RhoG were present in ELMO2–ILK immunoprecipitates, whereas T17N RhoG was not detected, indicating the existence in cells of complexes composed of ILK, ELMO2, and active RhoG.
To further investigate the interactions of RhoG with ILK–ELMO2 species, we conducted in vitro binding experiments with bacterially produced FLAG-tagged RhoG, GST-tagged ELMO2, and TRX-tagged ILK (Figure 6E). After incubation of RhoG with GTPγS to activate it, we found that RhoG-GTPγS was able to bind ELMO2 in the presence or absence of ILK (Figure 6D). Similarly, ILK was able to bind ELMO2 irrespective of whether or not RhoG-GTPγS was added to the binding reaction (Figure 6D). In contrast, RhoG-GTPγS associated with ILK only in the presence of ELMO2. Further, neither ILK nor ELMO2 associated with inactive RhoG, present as either RhoG-GDPβS or free bacterial RhoG (Figure 6D). Together, these data are consistent with the concept that ELMO2 acts as a bridge to bring together active RhoG and ILK, to form a ternary complex.
ILK and ELMO2 Cooperate to Modulate Cell Spreading and Polarization
We previously reported that inactivation of the Ilk gene yields ILK-deficient keratinocytes that are incapable of spreading on laminin 332 substrates, of acquiring a polarized phenotype and of sustaining forward movement (Nakrieko et al., 2008b). Similarly, ELMO is involved in normal integrin-mediated cell spreading and migration in various cell types (Grimsley et al., 2004; Katoh et al., 2005). The requirement of lamellipodia formation for cell polarization, together with the ability of ELMO2 and ILK to localize to cell protrusions, prompted us to investigate if ELMO2 and ILK act cooperatively to modulate the establishment of cell polarity. To this end, we seeded keratinocytes exogenously expressing ILK and/or ELMO2 at low density on laminin 332 substrates. Three hours after plating, we determined the fraction of cells that had attached, but not spread (hereafter termed Attached), those that had attached and spread, but did not exhibit a polarized morphology (hereafter termed Nonpolarized), and those that exhibited a polarized morphology (termed Polarized, Figure 7A). Under these conditions, attached and nonpolarized cells that expressed neither exogenous ILK nor ELMO2 constituted, each, ∼40–45% of the population, with the remaining ∼15–18% of the cells showing a polarized morphology (Figure 7B). These proportions were significantly altered in the joint presence of exogenous ILK and ELMO2. Specifically, the fraction of attached and nonpolarized cells decreased, respectively, to ∼20 and 30%, whereas the proportion of polarized cells increased just over threefold to 50–55% relative to control keratinocytes (Figure 7B). Importantly, exogenous expression of ILK alone increased the proportion of polarized cells about twofold, whereas that of ELMO2 increased it just over 1.5-fold (Figure 7C). Together, these observations suggest that ILK and ELMO2 can cooperate to induce establishment of keratinocyte polarity in response to laminin 332.
Given the ability of the N-terminus of ELMO2 to bind ILK and localize to cell protrusions, albeit with lower efficiency than the wild-type protein, we investigated its effect on ILK-induced cell polarization. Strikingly, the presence of ELMO2 (1-481) abolished the ability of ILK to induce cell polarization (Figure 7C), suggesting that full-length ELMO2 may be a required scaffold for the functional interaction of ILK with other downstream effector proteins, necessary for induction of cell polarity. Paradoxically, expression of the N-terminal deletion mutant ELMO2 (482-718), which does not bind ILK, also resulted in loss of ILK induction of cell polarization (Figure 7C). Together, these observations suggest that the association of ILK with the N-terminus of ELMO2 may contribute to localize the former protein to lamellipodia and to transmit signals to downstream effectors likely associated with the C-terminus of ELMO2. These signals induce changes in cell morphology and establishment of front-rear polarity. The ability of the ELMO2 truncation mutants to completely interfere with ILK induction of cell polarization is consistent with the proposal that the ILK–ELMO2 cassette may be an important contributor to the ability of ILK to induce keratinocyte polarization in response to stimulation by laminin 332.
Role of ELMO2–RhoG Interactions in ILK-mediated Cell Polarization
Given that we established the presence of RhoG in ILK–ELMO2 complexes, we next examined whether an ELMO2-mediated link between this small GTPase and ILK is necessary for ILK induction of cell polarization. We reasoned that if ILK induces cell polarization via ILK–ELMO2–RhoG species, exogenous expression of RhoG mutants incapable of binding to ELMO2 would hinder ILK-induced cell polarity. Thus, we scored the ability of cells to attach, spread, and acquire a polarized morphology in response to laminin 332, conducting similar experiments to those described for Figure 7C, but using keratinocytes exogenously expressing ILK and various RhoG proteins. We observed that either exogenous ILK or wild-type RhoG increased the fraction of polarized cells ∼3.5-fold relative to control cultures, mainly at the expense of the Attached (nonspread) cell population (Figure 7D). The joint presence of transfected ILK and RhoG did not significantly increase the proportion of polarized cells relative to ILK or RhoG alone, either because ILK and RhoG also function through other, independent pathways and/or because other elements in the RhoG–ILK complex (such as ELMO2) become limiting under the conditions of these experiments. In contrast, expression of a mutant RhoG F37A incapable of binding to ELMO2 (Katoh and Negishi, 2003 and data not shown) did not increase the fraction of polarized cell (Figure 7D), consistent with the concept that ELMO proteins are important contributors to the effects of RhoG on keratinocytes. Notably, RhoG F37A abolished the ability of ILK to increase the proportion of polarized cells (Figure 7D), indicating that RhoG is a major contributor of ILK-induced keratinocyte polarization in response to laminin 332, likely through mechanisms that involve RhoG interactions with ELMO2–ILK species.
ILK and ELMO2 Cooperate to Induce Directional Migration
The increases in lamellipodia formation and cell polarization induced by the joint expression of ILK and ELMO2 may produce stable leading edges that would result in enhanced directional motility or, alternatively, highly dynamic and/or multidirectional cell protrusions which would in fact hamper migration. To distinguish between these two possibilities, we measured haptotactic migration in keratinocytes exogenously expressing ILK–ELMO2. We transfected cells with GFP-tagged ELMO2 and/or mCherry-tagged ILK and measured the ability of the cells to migrate to the laminin 332–coated underside of Transwell tissue culture inserts in the absence of any chemotactic factors. The presence of exogenous ILK by itself increased 3.8-fold the number of keratinocytes that migrated through the insert, whereas those cells only expressing ELMO2 showed 2.1-fold increased migration that, however, failed to reach statistical significance (Figure 7E). Remarkably, exogenous expression of ILK with ELMO2 gave rise to a 11-fold increase in the number of cells that migrated through the insert (Figure 7E). In contrast, and similar to their effect on ILK-induced cell polarization, ELMO2 mutants 1-481 or 482-718 were incapable of increasing cell motility, and keratinocytes expressing ILK and any one of these mutants did not exhibit migration above levels observed with ILK only (Figure 7E). Together, these studies indicate that ILK and ELMO2 synergistically promote directional migration, possibly via pathways that involve enhanced formation of lamellipodia and extracellular matrix interactions.
DISCUSSION
Complex cellular processes depend on the coordinated actions of many components, which can be achieved through scaffold proteins. Signaling scaffolds are crucial to assemble multiprotein complexes that participate in common and/or interlinked pathways. Integrin-linked kinase is a key scaffold that binds to and coordinates specific interactions among a variety of signaling proteins. In this manner, ILK is a central player in a variety of cell functions related to attachment, spreading, and migration. We now expand this role of ILK to include a novel interaction with ELMO2, another important protein critical for cell polarization and migration, as well as phagocytosis (Cote and Vuori, 2007; Kinchen and Ravichandran, 2007).
The interaction between ILK and ELMO2 occurs in epithelial, endothelial, and mesenchymal cell types, suggesting a general and important role. ILK interacts with the N-terminus of ELMO2, but not with ELMO1. Although ELMO family members are highly homologous, the region in ELMO2 that binds to ILK is relatively less well conserved, potentially explaining the ability of ILK to discriminate between ELMO1 and ELMO2 (Gumienny et al., 2001). Significantly, this novel ILK–ELMO2 interaction has important potential implications vis-à-vis specificity of ELMO1 and ELMO2 functions in mammalian cells, as it constitutes the first major biochemical difference described between the two ELMO homologues.
Another major finding of our study is the identification of ELMO2 as a bridge between ILK and RhoG, directly linking RhoGTPases with ILK, and demonstrating a novel relationship between the ILK and the ELMO signaling platforms. ILK has been linked to Rac1 activation in various cell types (Liu et al., 2005; Boulter et al., 2006; Nakrieko et al., 2008b), although it has yet to be detected in Rac1-containing multiprotein complexes. Together, ILK and ELMO2 appear to fulfill a critical role as scaffolds linking Rac1 GTPase activators and, possibly, effectors.
The interaction of ILK with ELMO2 and RhoG is a potential mechanism that contributes to ILK localization to lamellipodia and Rac1 activation. Unlike integrin-stimulated signaling, which is initiated from focal contacts, Rac1 is activated in membrane-associated regions close to or at lamellipodial projections, thus placing ILK–ELMO2–RhoG species at appropriate intracellular sites to participate in Rac1 activation during cell polarization. Further, the ability of ILK–ELMO2 to induce front-rear polarity is associated with increased directional haptotactic migration, indicating a central role for this complex in forward cell movement. Indeed, we have observed ILK–ELMO2 complexes at the cell front in migrating cells (data not shown). An important future step will be to define the role of ILK–ELMO2 in other types of migration, such as those promoted by chemotactic factors.
ILK is a well-established constituent of focal adhesions, structures composed of integrins, paxillin, focal adhesion kinase, and other proteins. We observed that ELMO2 concentrates at sites of membrane extensions, but does not colocalize with paxillin-containing focal contacts (data not shown). We have also determined that the accumulation of ILK at lamellipodia does not exclude its localization at focal contacts, especially at the rear of the cell, suggesting that distinct ILK pools may be responsible for various cellular processes. ILK localization to focal adhesions participates in cell adhesion and spreading. We now show that ILK induction of cell polarization and migration also occur through its accumulation at lamellipodia, possibly through distinct signaling pathways from those activated at focal contacts. Future experiments are needed to establish the relationship between these two pools of ILK.
ILK deficiency results in impaired spreading, migration, and Rac1 activation in various cell types, likely through the inactivation of multiple pathways. The fractional contribution of ILK–ELMO2–RhoG complexes to cell spreading, polarization, and migration remains to be determined. The increased cell polarization by ILK observed in the presence of exogenous ELMO2, together with the abrogation of this effect by N- or C-terminal truncations of ELMO2 suggests that a major contributing complex to keratinocyte polarization induced by laminin 332 is a species that contains ILK, RhoG and ELMO2. This is one of several pathways functional in these cells, as direct Rac1 activation by Tiam1 in response to laminin 332 has also been implicated in keratinocyte migration and spreading in epidermal keratinocytes (Hamelers et al., 2005). The next key step will be to define the factors and downstream effectors that interact with ILK in the context of ILK–ELMO2–RhoG complexes and to determine how they modulate cell polarity.
Current models propose that active RhoG recruits ELMO bound to a Dock family member to activate Rac1 (Hiramoto et al., 2006). Indeed, although ELMO1 mutants lacking the C-terminal half bind to RhoG, they do not bind Dock proteins and hence cannot promote Rac1 activation at the plasma membrane and cell migration (Grimsley et al., 2004). In the present studies, we have established that exogenous expression of ILK on its own promotes cell polarization in response to laminin 332 and that ELMO2 cooperates with ILK to further increase cell polarization. Together, these observations suggest that ELMO2 may be functioning as a scaffold that allows ILK to interact with and mediate activation of downstream effectors involved in the development of cell polarity. Consistent with this notion, expression of the C-terminal deletion mutants of ELMO2 was sufficient to abrogate the capacity of ILK to promote cell polarization, in spite of the inability of these mutants to bind to ILK. This suggests that a functional ILK–ELMO2 cassette requires the ELMO2 C-terminus, possibly to bridge or facilitate interaction with other proteins, which in turn could be regulated through ILK to induce cell polarization.
Given that exogenous RhoG expression can induce ruffles and lamellipodia and that RhoG can localize to the cell membrane in ILK-deficient keratinocytes (Nakrieko et al., 2008b), it is unlikely that the role of ILK in this complex is to bring RhoG–ELMO2 to the plasma membrane. Rather, ILK may be targeted to the cell front by RhoG–ELMO2. This would allow ILK to serve as a key scaffold to link other proteins to the RhoG–ELMO2 complex. The importance of ELMO2 bridging RhoG and ILK is underlined by the observed interference of RhoG F37A on ILK-induced cell polarization.
Although an intact kinase domain of ILK is not necessary for binding to ELMO2, the possibility that it may be necessary to facilitate interactions with other, still unidentified downstream effectors remains to be tested. Exogenous expression of RhoG can rescue the spreading and protrusion defects of ILK-deficient keratinocytes (Nakrieko et al., 2008b). In some cell types, RhoG is not essential for integrin-mediated cell spreading, but contributes to Rac1-dependent and -independent cell migration (Meller et al., 2008). RhoG may also be involved in multiple Rac1-activation pathways, possibly related and unrelated to ILK. The rescue of ILK-deficient keratinocyte spreading by RhoG may be due to amplification of other, parallel ILK-independent pathways, including those that involve other ELMO family members. Indeed, important implications of our studies are that RhoG may participate in cell spreading and migration through ILK-dependent pathways involving ELMO2 and through ILK-independent pathways potentially involving other ELMO proteins. An additional key aspect of future studies will be to determine whether ELMO2 modules signal exclusively through ILK-containing complexes or through various other pathways.
ACKNOWLEDGMENTS
The authors thank L. Nini and K.A. Nakrieko for helpful comments on the manuscript, as well as A. Pajak and D. Bryce for superb technical assistance. This work was supported by grants to L.D. from the Canadian Cancer Society (Grant 018208) and from the Canadian Institutes of Health Research (CIHR). E.H. was supported with funds from the Children's Health Research Institute “Quality of Life” program. During the development of this work, S.J.A.D. was the recipient of a CIHR New Investigator Award.
Abbreviations used:
- BSA
bovine serum albumin
- EMEM
Eagle's minimum essential medium
- FBS
fetal bovine serum
- ELMO
Engulfment and Cell Motility
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- ILK
integrin-linked kinase
- PEI
polyethyleneimine
- TRX
thioredoxin.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0050) on May 13, 2009.
REFERENCES
- Apostolova M. D., Ivanova I. A., Dagnino C., D'Souza S.J.A., Dagnino L. Active import and export mechanisms regulate E2F-5 subcellular localization. J. Biol. Chem. 2002;277:34471–34479. doi: 10.1074/jbc.M205827200. [DOI] [PubMed] [Google Scholar]
- Blangy A., Vignal E., Schmidt S., Debant A., Gauthier-Rouviere C., Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J. Cell Sci. 2000;113(Pt 4):729–739. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- Bock-Marquettee I., Saxena A., White M. D., DiMaio J. M., Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432:466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- Boulter E., Grall D., Cagnol S., Van Obberghen-Schilling E. Regulation of cell-matrix adhesion dynamics and Rac-1 by integrin linked kinase. FASEB J. 2006;20:1489–1491. doi: 10.1096/fj.05-4579fje. [DOI] [PubMed] [Google Scholar]
- Broussard J. A., Webb D. J., Kaverina I. Asymmetric focal adhesion disassembly in motile cells. Curr. Opin. Cell Biol. 2008;20:85–90. doi: 10.1016/j.ceb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Brugnera E., et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- Chang W. Y., Andrews J., Carter D. E., Dagnino L. Differentiation and injury-repair signals modulate the interaction of E2F and pRB proteins with novel target genes in keratinocytes. Cell Cycle. 2006;5:1872–1879. doi: 10.4161/cc.5.16.3136. [DOI] [PubMed] [Google Scholar]
- Choma D. P., Pumiglia K., DiPersio C. M. Integrin alpha3beta1 directs the stabilization of a polarized lamellipodium in epithelial cells through activation of Rac1. J. Cell Sci. 2004;117:3947–3959. doi: 10.1242/jcs.01251. [DOI] [PubMed] [Google Scholar]
- Cote J. F., Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva I., Fielding A., Foster L. J., Dedhar S. Mapping the integrin-linked kinase interactome using SILAC. J. Proteome Res. 2008;7:1740–1749. doi: 10.1021/pr700852r. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage H., Lui M., Lacomis L., Grewal A., Annan R. S., McNulty D. E., Carr S. A., Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Gobom J., Nordhoff E., Mirgorodskaya E., Ekman R., Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 1999;34:105–116. doi: 10.1002/(SICI)1096-9888(199902)34:2<105::AID-JMS768>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Grimsley C. M., Kinchen J. M., Tosello-Trampont A. C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O., Ravichandran K. S. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J. Biol. Chem. 2004;279:6087–6097. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- Grimsley C. M., Lu M., Haney L. B., Kinchen J. M., Ravichandran K. S. Characterization of a novel interaction between ELMO1 and ERM proteins. J. Biol. Chem. 2006;281:5928–5937. doi: 10.1074/jbc.M510647200. [DOI] [PubMed] [Google Scholar]
- Gumienny T. L., et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Hamelers I.H.L., Olivo C., Mertens A.E.E., Pegtel D. M., van der Kammen R. A., Sonnenberg A., Collard J. H. The Rac activator Tiam1 is required for α3β1-mediated laminin-5 deposition, cell spreading and cell migration. J. Cell Biol. 2005;171:871–881. doi: 10.1083/jcb.200509172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y., Suzuki M., Ohya K., Iwai H., Ishijima N., Koleske A. J., Fukui Y., Sasakawa C. Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat. Cell Biol. 2007;9:121–128. doi: 10.1038/ncb1526. [DOI] [PubMed] [Google Scholar]
- Hildinger M., Baldi L., Stettler M., Wurm F. M. High-titer, serum-free production of adeno-associated virus vectors by polyethyleneimine-mediated plasmid transfection in mammalian suspension cells. Biotechnol. Lett. 2007;29:1713–1721. doi: 10.1007/s10529-007-9441-3. [DOI] [PubMed] [Google Scholar]
- Hiramoto K., Negishi M., Katoh H. Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp. Cell Res. 2006;312:4205–4216. doi: 10.1016/j.yexcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Iden S., Collard J. G. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat. Rev. Mol. Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- Ivanova I. A., Dagnino L. Activation of p38- and CRM1-dependent nuclear export promotes E2F1 degradation during keratinocyte differentiation. Oncogene. 2007;26:1147–1154. doi: 10.1038/sj.onc.1209894. [DOI] [PubMed] [Google Scholar]
- Ivanova I. A., Vespa A., Dagnino L. A novel mechanism of E2F1 regulation via nucleocytoplasmic shuttling: determinants of nuclear import and export. Cell Cycle. 2007;6:2186–2195. doi: 10.4161/cc.6.17.4650. [DOI] [PubMed] [Google Scholar]
- Katoh H., Hiramoto K., Negishi M. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 2005;119:56–65. doi: 10.1242/jcs.02720. [DOI] [PubMed] [Google Scholar]
- Katoh H., Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461–464. doi: 10.1038/nature01817. [DOI] [PubMed] [Google Scholar]
- Keskanokwong T., Shandro H. J., Johnson D. E., Kittanakom S., Vilas G. L., Thorner P., Reithmeier R. A., Akkarapatumwong V., Yenchitsomanus P. T., Casey J. R. Interaction of integrin-linked kinase with the kidney chloride/bicarbonate exchanger, kAE1. J. Biol. Chem. 2007;282:23205–23218. doi: 10.1074/jbc.M702139200. [DOI] [PubMed] [Google Scholar]
- Kinchen J. M., Ravichandran K. S. Journey to the grave: signaling events regulating removal of apoptotic cells. J. Cell Sci. 2007;120:2143–2149. doi: 10.1242/jcs.03463. [DOI] [PubMed] [Google Scholar]
- Legate K. R., Montanez E., Kudlacek O., Fassler R. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- Leung-Hagesteijn C., Hu M. C., Mahendra A. S., Hartwig S., Klamut H. J., Rosenblum N. D., Hannigan G. E. Integrin-linked kinase mediates bone morphogenetic protein 7-dependent renal epithelial cell morphogenesis. Mol. Cell. Biol. 2005;25:3648–3657. doi: 10.1128/MCB.25.9.3648-3657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E., et al. Targeted deletion of integrin-linked kinase reveals a role in T-cell chemotaxis and survival. Mol. Cell. Biol. 2005;25:11145–11155. doi: 10.1128/MCB.25.24.11145-11155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald P. C., Fielding A. B., Dedhar S. Integrin-linked kinase-essential roles in physiology and cancer biology. J. Cell Sci. 2008a;121:3121–3132. doi: 10.1242/jcs.017996. [DOI] [PubMed] [Google Scholar]
- McDonald P. C., Oloumi A., Mills J., Dobreva I., Maidan M., Gray V., Wederell E. D., Bally M. B., Foster L. J., Dedhar S. Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res. 2008b;68:1618–1624. doi: 10.1158/0008-5472.CAN-07-5869. [DOI] [PubMed] [Google Scholar]
- Meller J., Vidali L., Schwartz M. A. Endogenous RhoG is dispensable for integrin-mediated cell spreading, but contributes to Rac-independent migration. J. Cell Sci. 2008;121:1981–1989. doi: 10.1242/jcs.025130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi L. K., Minor D.L.J., Peng Z. -Y. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. USA. 2002;99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakrieko K. A., Vespa A., Mason D., Irvine T., D'Souza S.J.A., Dagnino L. Modulation of integrin-linked kinase cytoplasmic shuttling by ILKAP and CRM1. Cell Cycle. 2008a;7:2157–2166. doi: 10.4161/cc.7.14.6241. [DOI] [PubMed] [Google Scholar]
- Nakrieko K. A., Welch I., Dupuis H., Bryce D. M., Pajak A., St-Arnaud R. S., Dedhar S., D'Souza S.J.A., Dagnino L. Impaired hair follicle morphogenesis and polarized keratinocyte movement upon conditional inactivation of integrin-linked kinase in the epidermis. Mol. Biol. Cell. 2008b;19:1462–1473. doi: 10.1091/mbc.E07-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B. P., Ryan M. C., Gil S. G., Carter W. G. Deposition of laminin 5 in epidermal wounds regulates integrin signaling and adhesion. Curr. Opin. Cell Biol. 2000;12:554–562. doi: 10.1016/s0955-0674(00)00131-9. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos S. N., Turner C. E. Molecular dissection of actopaxin-integrin-linked kinase-paxillin interactions and their role in subcellular localization. J. Biol. Chem. 2002;277:1568–1575. doi: 10.1074/jbc.M108612200. [DOI] [PubMed] [Google Scholar]
- Santy L. C., Ravichandran K. S., Casanova J. E. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr. Biol. 2005;15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Vespa A., D'Souza S.J.A., Dagnino L. A novel role for integrin-linked kinase in epithelial sheet morphogenesis. Mol. Biol. Cell. 2005;16:4084–4095. doi: 10.1091/mbc.E05-02-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa A., Darmon A. J., Turner C. E., D'Souza S.J.A., Dagnino L. Ca2+-dependent localization of integrin-linked kinase to cell-cell junctions in differentiating keratinocytes. J. Biol. Chem. 2003;278:11528–11535. doi: 10.1074/jbc.M208337200. [DOI] [PubMed] [Google Scholar]
- Wang X., et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563–1566. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- Wennerberg K., Ellerbroek S. M., Liu R. Y., Karnoub A. E., Burridge K., Der C. J. RhoG signals in parallel with Rac1 and Cdc42. J. Biol. Chem. 2002;277:47810–47817. doi: 10.1074/jbc.M203816200. [DOI] [PubMed] [Google Scholar]