Abstract
Epithelial–mesenchymal transition (EMT) confers destabilization of cell–cell adhesion and cell motility required for morphogenesis or cancer metastasis. Here we report that zyxin, a focal adhesion-associated LIM protein, is essential for actin reorganization for cell migration in TGF-β1–induced EMT in normal murine mammary gland (NMuMG) cells. TGF-β1 induced the relocation of zyxin from focal adhesions to actin fibers. In addition, TGF-β1 up-regulated zyxin via a transcription factor, Twist1. Depletion of either zyxin or Twist1 abrogated the TGF-β1–dependent EMT, including enhanced cell motility and actin reorganization, indicating the TGF-β1-Twist1-zyxin signal for EMT. Both zyxin and Twist1 were predominantly expressed in the cardiac atrioventricular canal (AVC) that undergoes EMT during heart development. We further performed ex vivo AVC explant assay and revealed that zyxin was required for the reorganization of actin fibers and migration of the endocardial cells. Thus, zyxin reorganizes actin fibers and enhances cell motility in response to TGF-β1, thereby regulating EMT.
INTRODUCTION
Epithelial–mesenchymal transition (EMT) is an essential event during embryogenesis for the formation of many tissues including the mesoderm, for migration of neural crest cells, and for development of the heart valves and septa. The picture emerging from diverse EMT-related studies suggests that precise molecular and cellular control of EMT is complex and context-dependent (Nakaya et al., 2008). An example of developmentally regulated EMT occurs during the initial stages of cardiac morphogenesis. At embryonic day 9.5 (E9.5), endocardial cells undergo EMT (“endocardial EMT”); delaminating from the endothelial sheet, invading the matrix tissue called cardiac jelly, and engaging in endocardial cushion cellularization required for valve and septum formation (Chang et al., 2004). Because a number of congenital heart diseases are caused by abnormal atrioventricular canal (AVC) development (Bruneau, 2008), understanding of the molecular basis of AVC morphogenesis has been long sought, but still not fully achieved. Several EMT-inducing transcription factors such as Snail (Cano et al., 2000) and Twist1 (Ma et al., 2005) are expressed in endocardium during development. Although among them, Twist1 is well analyzed as a potent EMT inducer during cancer metastasis (Yang et al., 2004); however, the role for Twist1 in AVC development is not fully understood.
An important hallmark of EMT is an increase in cell motility with actin cytoskeletal rearrangement. On EMT induction, the structure of actin cytoskeleton changes dynamically from cortical actin network to stress fiber (Zavadil and Bottinger, 2005). Cell motility also depends on the adhesion of the cell to the substratum where the integrin family molecules bind to the extracellular matrix to form focal adhesion complexes. Stress fibers link the focal adhesion to retract the cells. Furthermore, the nascent focal complexes at the leading edge of the migrating cells are needed for cell crawling by connecting the actin fibers between the front and rear focal adhesions. Thus, coordinated control of the various focal adhesion proteins is important for actin fiber reorganization. Among focal adhesion molecules, paxillin, focal adhesion kinase (FAK) and Hic-5 are reported to be involved in EMT (Hetey et al., 2005; Tumbarello et al., 2005). EMT induction in normal murine mammary gland (NMuMG) cells by TGF-β1 induces the phosphorylation and recruitment of paxillin to focal adhesions and the expression of Hic-5 (Tumbarello et al., 2005).
Transforming growth factor β (TGF-β) is a potent EMT inducer and is implicated in a broad kind of EMT in cancer metastasis, tissue fibrosis, or tissue morphogenesis (Zavadil and Bottinger, 2005). TGF-β was first described as an inducer of EMT in normal mammary epithelial cells (Miettinen et al., 1994), and murine mammary epithelial cells treated with TGF-β undergo a complete EMT (Thiery, 2003). Accumulating evidences revealed the involvement of TGF-β in vital steps during development (Sanford et al., 1997; Bartram et al., 2001; Sridurongrit et al., 2008).
Zyxin belongs to a LIM protein family and harbors distinct actin polymerization activity independent of the Arp2/3 complex (Fradelizi et al., 2001). Zyxin has three tandem LIM domains in its C terminus that support specific protein interactions (Schmeichel and Beckerle, 1994) and targeting to focal adhesions (Nix et al., 2001). The proline-rich N-terminal domain called ActA mediates actin polymerization (Fradelizi et al., 2001). Zyxin locates primarily at focal adhesions and regulates actin cytoskeleton dynamics, cell movement, and signal transduction (Beckerle, 1998; Kadrmas and Beckerle, 2004). Despite the understanding of zyxin, its role in the cellular actin rearrangement and its function in vivo is still unknown. A role for zyxin during development also remains to be elucidated.
In this study, we investigated the function of zyxin in TGF-β1–induced EMT and the EMT during heart morphogenesis and explored the requirement of zyxin in TGF-β1–dependent EMT and the contribution of zyxin to EMT for valvulogenesis in the heart.
Here, we show that zyxin regulates EMT by accelerating cell motility through controlling actin fiber reorganization in cultured epithelial cells and the endocardial cells of the embryonic heart.
MATERIALS AND METHODS
Cell Culture
NMuMG cells were obtained from ATCC (Manassas, VA) and maintained in DMEM supplemented with 10% FCS and 10 μg/ml insulin. We cloned the cells by limiting dilution and obtained 13 different clones. Among them, we used a cell line designated C7 that showed typical epithelial morphology and robust response to TGF-β1. The cells were maintained in DMEM (Nacalai Tesque, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS). TGF-β1–stimulation experiments were performed with recombinant human TGF-β1 (2 ng/ml; R&D Systems, Minneapolis, MN).
Deletion Mutants and Retroviral Vector Construction
Zyxin deletion mutants were generated by PCR amplifying the amino acid 1-375 (ΔLIM) or aa 376-564 (LIM only) region and inserting into pCAGIP-EGFP (enhanced green fluorescent protein) vector.
pCX4 puro vector was kindly provided by Tsuyoshi Akagi (Akagi et al., 2003). pCX4 puro-EGFP-Snail, pCX4 puro-EGFP-Twist1, or pCX4 puro-HMGA2 were generated by inserting the full-length Snail cDNA from pCAGGS-EGFP-Snail, Twist1 cDNA from pCAGIP-Twist1 (Hayashi et al., 2007), or HMGA2 cDNA from pENTR-HMGA2 into the pCX4 puro vector, respectively. Each plasmid above was cotransfected with pGP and pE-eco (Takara Bio, Tokyo, Japan) into BOSC to produce ecotropic retrovirus vector.
Immunocytochemistry
NMuMG-C7 cells were plated onto collagen-coated glass-bottom dishes (MatTek, Ashland, MA). The cells were fixed in 4% paraformaldehyde (PFA), permeabilized in 0.1% Triton X-100 for 5 min, and probed with mouse anti-zyxin antibody (1:40, 164D4, Synaptic Systems; 1:100, clone ZOL301, Sigma-Aldrich, St. Louis, MO), mouse anti-E-cadherin antibody (1:50, clone 36, BD Biosciences, San Jose, CA), mouse anti-N-cadherin antibody (1:50, clone 32, BD Biosciences), and rabbit anti-ZO-1-antibody (1:100, 61-7300, Invitrogen, Carlsbad, CA). The primary antibody was detected by goat anti-mouse-Alexa 488, goat anti-mouse-Alexa 546, or goat anti-rabbit-Alexa 546 (1:1000, Invitrogen). Nuclei were stained with 4′-6-diamidino-2-phenylindole (DAPI, Invitrogen). F-actin was stained with Alexa Fluor 546-phalloidin (Invitrogen).
Fluorescence Imaging
NMuMG-C7 cells were stably transfected with pCAGIP-EGFP-zyxin plasmid. Fluorescence images were recorded with a Nikon eclipse TE300 inverted microscope (Melville, NY) or with a confocal microscope with a Bio-Rad laser scanning system (Radiance 2100; Richmond, CA) coupled to a Nikon eclipse TE2000-U inverted microscope with a 60× oil immersion objective lens. For time-lapse recording of zyxin images, the cells were grown in 35-mm glass-bottom dishes (MatTek) and placed in a CO2 chamber (Model CZI-3; Zeiss) at 37°C, attached to the stage of an inverted microscope (Axiovert 200; Zeiss). Images were acquired every 3 min using a 40 × 1.3 NA objective lens (Plan-NEOFLUAR, Zeiss) and a CCD camera (model HRm; Zeiss), and analyzed using Axiovision software (version 4.6, Zeiss).
SDS-PAGE and Western Blot Analysis
Cells were harvested in a RIPA lysis buffer. Protein concentration was determined by the method of Lowry (Lowry et al., 1951). Protein samples (10 μg per lane) were separated by SDS-PAGE and transferred to a Hybond-P PVDF membrane (GE Healthcare, Little Chalfont, Buckinghamshire, England). Western blot was performed with the following antibodies: anti-zyxin (1:1000, ZOL301, Sigma-Aldrich), anti-vinculin (1:1000, V9264), anti-β-actin (1:5000, AC-15), anti-E-cadherin (1:5000, clone 36, BD Biosciences), anti-N-cadherin (1:5000, clone 32), anti-paxillin (1:1000, clone 165), anti-FAK (1:200, clone 77), anti-p130Cas (1:1000, clone 21), anti-Hic-5 (1:200, clone 34), anti-GFP (1:1000, 598, MBL, Nagoya, Japan), anti-HMGA2 (1:200, ab41878, Abcam, Cambridge, MA) and anti-GAPDH (1:5000, 6C5). All secondary antibodies were horseradish peroxidase conjugated (GE Healthcare) and were used at 1:1000 dilution. Immunoreactive bands were detected with Chemi-LumiOne L (Nacalai Tesque). Quantification was done by densitometry using ImageJ software (NIH; http://rsb.info.nih.gov/ij/).
Modified Boyden's Chamber Assay
Modified Boyden's chamber assay was carried out as previously described (Saito et al., 2006). Briefly, 27 μl DMEM with 10% FBS was added to the lower chamber, and 50 μl of cell suspension (5 × 105 cells/ml) in DMEM with 10% FBS was added to the upper chamber. After 4 h of incubation, the cells on the lower side of the membrane were stained with Diff-Quick (Sysmex, Hyogo, Japan) to facilitate visualization and counting of cells. The count was done in eight randomly chosen fields.
RNA Interference Experiments
Zyxin or Twist1 Stealth small interfering RNA (siRNA) oligonucleotides were purchased from Invitrogen. The double-strand RNA (dsRNA) oligonucleotides against zyxin were as follows: (sense) 5′-AACAAAUGGAGUGGCAACUGGUGGG-3′ and (antisense) 5′-CCCACCAGUUGCCACUCCAUUUGUU-3′. The dsRNA oligonucleotides against Twist1 were as follows: (sense) 5′-UUGAGGGUCUGAAUCUUGCUCAGCU-3′ and (antisense) 5′-AGCUGAGCAAGAUUCAGACCCUCAA-3′. The scrambled siRNA oligonucleotides were as follows: (sense) 5′-UUCCUCAAUAGAUGCGUGUTT-3′ and (antisense) 5′-ACACGCAUCUAUUGAGGAATT-3′. These sequences did not correspond to any sequence in the mouse genome when subjected to a BLAST search. Transfections were carried out using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
RT-PCR Analysis and Real-Time PCR Quantification
Total RNA was isolated using an RNeasy RNA isolation kit (QIAGEN, Valencia, CA). cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). The sequences of PCR primers are shown in the Supplementary Table SI. The reactions were performed as follows: 98°C for 30 s; 25 cycles of 98°C for 10 s, 52°C for 30 s, and 72°C for 60 s. PCR products were separated in a 1.5% agarose gel. Real-time PCR was performed using an Applied Biosystems 7900 HT Fast Real-Time PCR system with TaqMan Probe for zyxin (Mm00496120_m1) and GAPDH (Mm99999915_g1) and TaqMan Universal PCR Master Mix (Applied Biosystems). The reactions were incubated at 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The relative expression level was calculated from a standard curve obtained by using log dilutions of cDNA containing the gene of interest. The average of two independent analyses for each gene and sample was calculated and was normalized to the endogenous reference control gene GAPDH.
In Situ Hybridization
In situ hybridization was performed as previously described (Koibuchi and Chin, 2007). The cDNAs used for generation of Dig-labeled mouse riboprobes were zyxin (nucleotides 2002-2404; GenBank no. NM_011777) and Twist1 (nucleotides 644-1524; GenBank no. NM_011658).
Lentivirus Vector
pcDNA6.2-shZyxin and pcDNA6.2-shLacZ were generated by inserting the oligonucleotide containing the specific siRNA target sequence into pcDNA6.2 vector (Invitrogen). pLenti6-shZyxin and pLenti6-shLacZ were generated from pcDNA6.2 constructs. Lentivirus was generated by cotransfection of the above construct with packaging plasmids into 293FT cells according to the manufacturer's instruction (Invitrogen). The sequences of shZyxin DNA oligo and shLacZ control oligo are available in the Supplementary Table SII.
Ex Vivo EMT Assay
The experiments were approved by the Ethics Committee for Animal Experiments of Osaka University Graduate School of Medicine. C57BL/6 mice were purchased from Charles River Breeding Laboratories. The ex vivo EMT assay was performed as described (Camenisch et al., 2002) with some modifications. After putting E9.5 AVC explants on collagen gels, lentivirus-containing medium was added and incubated for 24 h. Then the explants were incubated with M199 media (Invitrogen/BRL, Rockville, MD) supplemented with 0.01% insulin, transferrin and selenium (ITS, Invitrogen/BRL) for 48 h, before determining the extent of outgrowth and matrix invasion. The criteria for “Migratory” were the appearance of the stellate cells migrating outward, frequently invading the collagen gel matrix. The criteria for “Nonmigratory” were the appearance of polygonal cells that form cobblestone-like colonies on the gel surface (Camenisch et al., 2002). XZ confocal images were acquired to visualize collagen gel invasion. Cells that migrated into the collagen gel were stained with DAPI and counted in randomly collected areas. Phalloidin staining of the explants was done after the fixation with 4% PFA for 20 min on the collagen gel.
Statistics
Statistical analysis was performed with StatView 5.0 software (SAS Institute, Cary, NC). All results were expressed as mean ± SEM. Data were compared using ANOVA, followed by the Dunnett test for pairwise comparisons against control and by the Tukey test for multiple comparisons. Significance was defined as p < 0.05.
RESULTS
Zyxin Is Up-Regulated in TGF-β1–induced EMT and Mobilizes to Actin Fibers
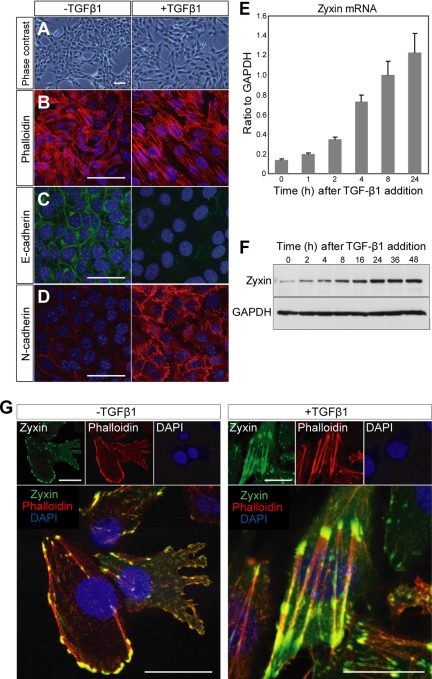
Normal murine mammary gland (NMuMG) cells undergo EMT upon treatment with TGF-β1 (Tumbarello et al., 2005). As reported, original NMuMG cells have heterogeneous morphology and responsiveness to TGF-β1. Thus, we cloned the cells by limiting dilution and established a cell line of NMuMG cells named NMuMG-C7, which altered the morphology in response to TGF-β1. On TGF-β1 treatment, NMuMG-C7 cells exhibited a series of EMT hallmarks such as cell scattering (Figure 1A), actin stress fiber formation (Figure 1B), down-regulation of E-cadherin (Figure 1C), and up-regulation of N-cadherin (Figure 1D). We observed that TGF-β1 up-regulated zyxin mRNA (Figure 1E) and induced expression of zyxin (Figure 1F) in a time-dependent manner, suggesting the possible participation of zyxin in TGF-β1–dependent EMT. An increase of zyxin protein was seen as early as 2 h after treatment and was dramatically augmented later on.
Figure 1.
Zyxin is up-regulated and relocates to actin stress fibers in TGF-β1–induced EMT. (A–D) TGF-β1–induced in vitro EMT in NMuMG-C7 cells. NMuMG-C7 cells treated with TGF-β1 for 24 h exhibit the cell scattering (A), actin stress fiber formation (B), down-regulation of E-cadherin (C), and up-regulation of N-cadherin (D). Scale bars, 100 μm. (E) Real-time PCR analysis of zyxin mRNA in NMuMG-C7 cells treated with TGF-β1. Results are shown as the mean ± SEM (n = 3). (F) Immunoblot analysis of zyxin in NMuMG-C7 cells treated with TGF-β1. GAPDH was used as a loading control. (G) TGF-β1 induced zyxin relocation from focal adhesions to actin fibers. Confocal microscopy of EGFP-tagged zyxin (green) in NMuMG-C7 cells treated or not with TGF-β1 for 24 h. Zyxin locates at focal adhesions before TGF-β1 treatment and mobilizes to actin fibers after the treatment (Also see Supplementary Video 1). The cells were counterstained with phalloidin (red) and DAPI (blue). Scale bars, 50 μm.
To study how zyxin participates in TGF-β1–dependent EMT, we examined the localization of zyxin using EGFP-tagged zyxin before and after the TGF-β1 treatment. Previous reports confirmed the localization of EGFP-tagged zyxin corresponded to the endogenous zyxin (Nix et al., 2001; Hotulainen and Lappalainen, 2006). Without stimulation, consistent with a previous report (Yoshigi et al., 2005), zyxin localized to focal adhesions and focal complexes in the lamellipodia (Figure 1G, −TGF-β1). Of note, upon TGF-β1 stimulation, zyxin mobilized from focal adhesions to actin fibers (Figure 1G, +TGF-β1). The localization of zyxin in confluent cells was shown in Supplementary Figure S1. Time-lapse imaging revealed the translocation of zyxin to actin fibers in NMuMG-C7 cells treated with TGF-β1 (Supplementary Video 1). These findings lead us to assume the switch of the role for zyxin from the regulation of cell-extracellular matrix adhesion to that of actin fibers and suggest the involvement of zyxin in EMT.
Zyxin Is Required for Cell Migration and Actin Fiber Reorganization in EMT
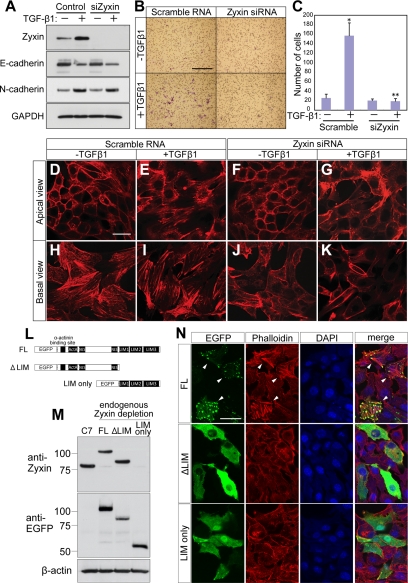
To assess the requirement of zyxin in TGF-β1–induced EMT, we performed knockdown experiments of zyxin. The effectiveness of zyxin siRNA was confirmed in NMuMG-C7 cells (Figure 2A). TGF-β1 induced the expression of N-cadherin and reduced the expression of E-cadherin (Figure 2A). Depletion of zyxin affected the expression of neither E-cadherin nor N-cadherin and altered the localization of neither E-cadherin nor N-cadherin (Figure 2A and Supplementary Figure S2). We further assessed the effect of depletion of zyxin on TGF-β1–dependent cell motility. Knockdown of zyxin almost completely abolished TGF-β1–induced migration (Figure 2, B and C). These results indicate that zyxin is essential for TGF-β1–induced migration, a hallmark of EMT. Next, we examined the effect of zyxin depletion on actin cytoskeleton by phalloidin staining. Polarized epithelial cells including NMuMG cells have distinct actin networks in the apical and the basal side of the cells (Supplementary Figure S3). The apical actin fibers adherent to adherens junctions (Yonemura et al., 1995; Vasioukhin et al., 2000; Perez-Moreno et al., 2003) and the basal actin stress fibers adherent to focal adhesions. We found that TGF-β1 induced both apical and basal actin fiber formation (Figure 2, E and I). Knockdown of zyxin resulted in inhibition of actin fiber formation at the basal level and, to a lesser extent, at the apical level. (Figure 2, G and K). To examine how essential zyxin was in EMT, we carried out rescue experiments using EGFP-tagged zyxin mutants (Figure 2L). The endogenous zyxin was depleted with short hairpin RNA (shRNA) that targets 3′ untranslated region (UTR) of zyxin mRNA (Figure 2M). The expression of exogenous zyxin mutants were confirmed (Figure 2M). The formation of actin stress fibers induced by TGF-β1 was rescued by the expression of EGFP-tagged full-length (FL) zyxin construct, supporting the specificity of zyxin siRNA and shRNA against zyxin. To the contrary, ΔLIM mutant dispersed in cytosol and failed to rescue. LIM only mutant exhibited localization to cytosol, focal adhesions, and nuclei and failed to rescue. Zyxin has ActA sequence in its N terminus, which has a distinct actin fiber polymerization activity (Fradelizi et al., 2001), and in LIM domains, which is necessary for targeting to focal adhesions (Nix et al., 2001). These experiments suggested that in the induction of EMT, both the localization and the activity were essential for zyxin. These findings suggest that zyxin is involved in TGF-β1–dependent EMT by regulating cell migration via controlling actin fiber formation. Considering that one of the most important features of EMT is the acquisition of motility (Thiery, 2003; Sung et al., 2007), Zyxin appears to be essential for the progression of EMT.
Figure 2.
Zyxin is essential for cell migration and actin fiber reorganization in TGF-β1–induced EMT. (A) Immunoblot analysis of zyxin and E-and N-cadherin in NMuMG-C7 cells transfected with zyxin siRNA or scramble RNA. After treatment with TGF-β1 for 24 h, total lysates were subjected to Western blot. GAPDH was used as a loading control. (B) Zyxin depletion abolished cell motility in TGF-β1-EMT. Cell motility was assessed by modified Boyden's chamber assay. NMuMG-C7 cells were transfected with zyxin siRNA or scramble RNA and treated or not with TGF-β1 for 24 h. The cells that migrated to the lower surface of the membrane were stained after 4 h of incubation. Scale bar, 250 μm. (C) The number of migrated cells in B. *p = 0.0344 versus Scramble RNA, TGF-β1 (−); **p = 0.0159 versus Scramble RNA, TGF-β1 (+). Results are shown as the mean ± SEM of four separate migration assays from two independent experiments. (D–K) Zyxin is essential for actin fiber formation in TGF-β1-EMT. Phalloidin staining was performed with NMuMG-C7 cells transfected with zyxin siRNA or scramble RNA and treated or not with TGF-β1 for 24 h. Apical and basal actin network were observed separately with a confocal microscope. Note that both the apical and basal actin fiber formation were hampered by zyxin depletion. Scale bar, 50 μm. (L) The scheme of EGFP-tagged zyxin deletion constructs. EGFP–zyxin constructs were introduced into NMuMG-C7 cells by transient transfection. FL: full length. (M) NMuMG-C7 cells were transduced with lentivirus vector that expresses shZyxin that targets 3′UTR. The resultant cells were then transfected with the EGFP–zyxin constructs that lack 3′UTR. The suppression of endogenous zyxin and the expression of transfected genes were confirmed by blotting with anti-zyxin and ant-EGFP antibodies. LIM only construct lacks the epitope recognized by zyxin antibody. (N) Neither ΔLIM nor LIM only constructs rescued the endogenous zyxin depletion. Endogenous zyxin-depleted cells were transfected with EGFP–zyxin constructs and treated with TGF-β1 for 24 h. Actin fiber formation was observed in the cells transfected with FL (arrowhead), but not with ΔLIM or LIM only constructs. Scale bar, 50 μm.
Zyxin Is Expressed in AVC of Developing Heart and Valves in Mice
We sought the physiological role for zyxin in EMT. During development, EMT occurs during AVC morphogenesis that gives rise to valves and septa. We therefore investigated the expression of zyxin during development by in situ hybridization in mouse fetus. We found that zyxin was strongly expressed in AVC at E13.5 (Figure 3A, arrowhead). We examined the expression of zyxin in more detail during development and revealed that its expression at AVC was induced during development, peaked in AVC at E13.5 and waned in valves and septa at E14.5 (Figure 3B, arrowhead). These expression patterns of zyxin prompted us to assume that zyxin was involved in the endocardial EMT and that the cells expressing zyxin contributed to valvulogenesis. We further examined the expression of Twist1 and found the concordance of the expression between Twist1 and zyxin in AVC (Figure 3B). These findings are in agreement with the in vitro results of zyxin regulation by Twist1 (see below). Furthermore, analyses in adults revealed the expression of zyxin in valves (Figure 3C, arrowhead). These findings imply that zyxin-expressing cells that undergo endocardial EMT participate in valvulogenesis.
Figure 3.
Zyxin is expressed in AVC during cardiac morphogenesis and valves in adult. (A) In situ hybridization for zyxin in E13.5 mouse embryo. Arrowheads indicate the strong expression of zyxin in AVC. RA, LA, RV, and LV stand for right atrium, left atrium, right ventricle, and left ventricle, respectively. Scale bar, 250 μm. (B) In situ hybridization for zyxin or Twist1 from E9.5 to E14.5. Arrowheads indicate the expression of each gene. Note that the expression pattern of zyxin and Twist1 are highly correlated with each other. (C) In situ hybridization for zyxin in adult heart. The black square indicates the region of higher magnification shown to the right. Arrowhead indicates the zyxin expression in the mitral valve.
Next, we examined whether the involvement of zyxin in TGF-β1-EMT was similarly seen in vascular endothelial cells (Supplementary Figure S4) in vitro. TGF-β1 induced cell scattering and actin fiber formation in murine vascular endothelial cells (Supplementary Figure S4A). Knockdown of zyxin (Supplementary Figure S4B) repressed the actin fiber formation induced by TGF-β1 (Supplementary Figure S4C). Moreover, in bovine arterial endothelial cells, TGF-β1 treatment induced the cell shape change into fusiform (Supplementary Figure S4D) and up-regulation of zyxin (Supplementary Figure S4E).
Zyxin Is Required for Morphogenetic Movement of the Endocardial Cells
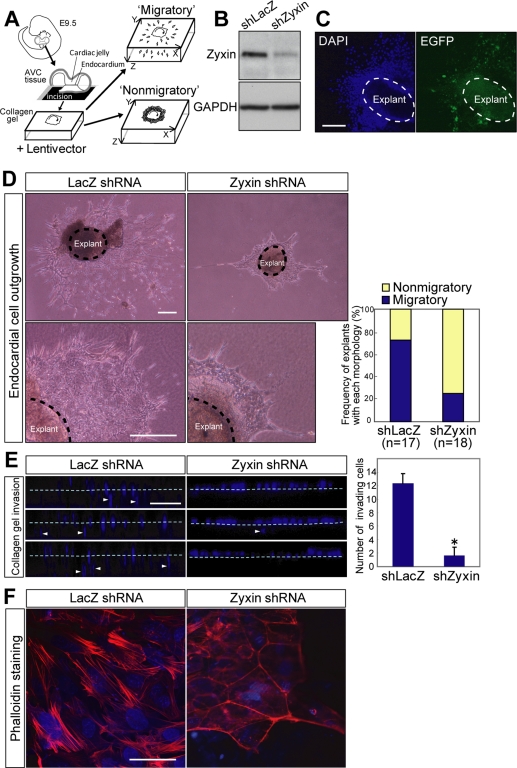
To understand how zyxin regulates EMT in the endocardial cells, we performed an ex vivo EMT assay (Figure 4A). The AVC explants isolated from E9.5 embryos recapitulate the process of EMT extracorporeally (Runyan and Markwald, 1983). We generated lentivirus vector expressing either zyxin shRNA or control LacZ shRNA to evaluate the effect of loss of zyxin expression in endocardial EMT. The efficacy of knockdown was confirmed after zyxin shRNA transduction (Figure 4B). The transduction efficiency was more than 85% by the estimation on the basis of EGFP fluorescence coexpressed on a single transcript (Figure 4C). The explants transduced with control shLacZ showed extensive radial migration of stellate cells with abundant protrusions (Figure 4D, LacZ shRNA). In contrast, the explants transduced with shZyxin showed significantly reduced cell migration and exhibited cobblestone-like appearance composed of polygonal cells (Figure 4D, zyxin shRNA). Although 76% of the explants transduced with shLacZ showed robust outgrowth of cells (Figure 4D, Migratory), only 22% of the shZyxin-transduced explants exhibited endocardial cell migration and the majority remained cobblestone-like appearance (Figure 4D, Nonmigratory).
Figure 4.
Zyxin is required for morphogenetic cell movement of the endocardial cells. (A) Scheme of ex vivo EMT assay. The AVC explants from E9.5 embryos were incised and put the endocardial surface down on collagen gels. After 24 h of incubation with lentivirus expressing shZyxin or shLacZ, the explants were cultured for another 48 h before the subsequent analyses. (B) Immunoblot analysis of zyxin after transduction with zyxin shRNA or LacZ shRNA. GAPDH was used as a loading control. (C) Transduction efficiency was assessed by the expression of EGFP coexpressed with shRNA. After 48 h of incubation after transduction, the explants were stained with DAPI and subjected to fluorescent microscopy. Scale bar, 100 μm. (D) Depletion of zyxin abolished the outgrowth of the endocardial cells. Phase-contrast images of AVC explants after 48 h of incubation after transduction. Scale bars, 250 μm. Right graph shows the proportion of colonies with the morphology defined by criteria described in Materials and Methods. (E) Cell invasiveness was suppressed by zyxin depletion. Collagen gel invasiveness assessed by XZ confocal microscopy. After 48 h of incubation after transduction, the explants were fixed and stained with DAPI. Broken lines indicate the surface of the gel. Scale bar, 100 μm. Right panel shows the number of the cells invading into the gel per unit area presented as mean ± SEM (n = 6). *p = 0.0048 versus shLacZ. (F) Zyxin is required for actin fiber reorganization in the endocardial cells. After 48 h of incubation after transduction, the explants were fixed and stained with phalloidin and DAPI and subjected to confocal microscopy. Note the decreased actin stress fibers in the endocardial cells that underwent EMT upon the collagen gel. Scale bar, 50 μm.
Invasiveness into extracellular matrices is an important feature for the endocardial cells that undergo EMT, because it is required for the endocardial cushion cellularization (Niessen et al., 2008). Therefore, we examined the invasiveness of the explanted endocardial cells into collagen gel by confocal microscopy. Consequently, invasion of the zyxin-depleted cells was significantly reduced compared with the control cells (Figure 4E). Finally, the effect of zyxin depletion on actin fiber formation was addressed in the explanted cells. Although the prominent actin stress fiber formation was observed in shLacZ-transduced explants (Figure 4F, LacZ shRNA), only scarce actin network formation was seen in the shZyxin-transduced explants (Figure 4F, zyxin shRNA). These findings suggest that zyxin contributes to the endocardial EMT by regulating cell motility though the reorganization of actin fibers.
Twist1 Regulates Zyxin in TGF-β1-EMT
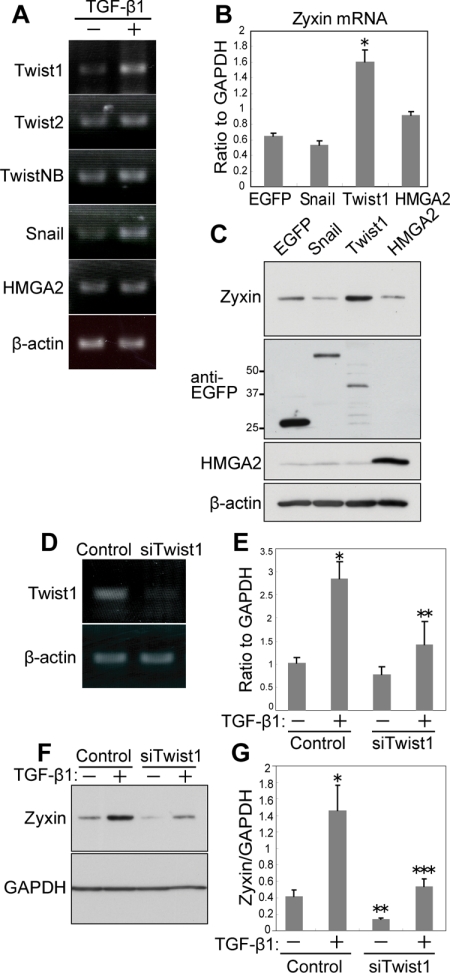
To clarify how zyxin expression is regulated during EMT, we tested the potential transcription factors that are reported to promote EMT (Thuault et al., 2006). NMuMG-C7 cells expressed Twist1, Twist2, TwistNB, Snail, and HMGA2 (Figure 5A). Among them, the expression of Twist1 and Snail was increased by TGF-β1 treatment (Figure 5A). Subsequently, we transduced NMuMG-C7 cells with retrovirus vectors expressing Snail, Twist1, HMGA2 (Thuault et al., 2006), or control EGFP. Among them, Twist1 up-regulated zyxin mRNA (Figure 5B) and induced zyxin expression (Figure 5C). These results suggest that Twist1 can promote zyxin expression. We then studied the requirement of Twist1 for zyxin up-regulation by TGF-β1. On knockdown of Twist1 (Figure 5D), TGF-β1 failed to up-regulate zyxin mRNA (Figure 5E) and to induce the expression of zyxin (Figure 5, F and G). Twist1 knockdown also decreased the basal expression of zyxin, suggesting the contribution of Twist1 to the basal expression of zyxin (Figure 5, F and G). We then studied whether focal adhesion-related proteins were generally regulated by TGF-β1 or Twist1. We investigated the expression of paxillin, FAK, vinculin, and p130Cas. Among them, paxillin was up-regulated by TGF-β1 (Supplementary Figure S5A). Twist1 depletion by siTwist1, however, did not abrogate the up-regulation of paxillin by TGF-β1 (Supplementary Figure S5B). These results suggest that focal adhesion proteins are not generally regulated by a single transcription factor in TGF-β1–induced EMT. Next, we investigated the possibility that paxillin was regulated by Snail, which was up-regulated by TGF-β1 (Figure 5A). We found paxillin was not up-regulated by Snail, either (Supplementary Figure S5C). These results delineate the signaling pathway, TGF-β1-Twist1-zyxin in the EMT and raise the possibility that different sets of focal adhesion proteins are regulated by distinct transcription factors.
Figure 5.
Twist1 regulates zyxin in TGF-β1-EMT. (A) Expression analysis of EMT inducers in NMuMG-C7 cells. RT-PCR performed with total RNA isolated from NMuMG-C7 cells treated with or without TGF-β1. β-Actin was used as a control. (B) Twist1 up-regulates zyxin. Real-time PCR performed with total RNA from NMuMG-C7 cells transduced with EGFP, Snail, Twist1, or HMGA2. *p = 0.0122 versus EGFP. Error bars, the SEM of three technical replicates. (C) Twist1 induced the expression of zyxin. Immunoblot analysis of zyxin in NMuMG-C7 cells transduced with EGFP, EGFP-Snail, EGFP-Twist1, or HMGA2. The expression of transduced genes was confirmed by blotting with anti-EGFP and anti-HMGA2 antibodies. (D) Efficacy of Twist1 RNAi assessed by RT-PCR. Total RNA from NMuMG-C7 cells transfected with either Twist1 siRNA or scramble RNA were subjected to RT-PCR. (E) Twist1 depletion abrogated up-regulation of zyxin by TGF-β1. Real-time PCR analysis of zyxin mRNA in NMuMG-C7 cells transfected with Twist1 siRNA or scramble RNA and treated or not with TGF-β1. Results are shown as the mean ± SEM (n = 3). *p = 0.011 when compared with control, TGF-β1(−); **p = 0.0236 when compared with control, TGF-β1(+). (F) Twist1 depletion suppressed the expression of zyxin. Immunoblot analysis of zyxin in NMuMG-C7 cells transfected with Twist1 siRNA or scramble RNA and treated or not with TGF-β1. GAPDH was used as a loading control. (G) Results of F were quantitated by densitometry using NIH ImageJ. Error bars, SD from triplicate experiments. *p = 0.0084 when compared with control, TGF-β1(−); **p = 0.035 when compared with control, TGF-β1(−); ***p = 0.012 when compared with control, TGF-β1(+).
Twist1 Uses Zyxin to Execute EMT
Twist1 is a potent EMT inducer, the function of which is well-characterized in cancer metastasis (Ansieau et al., 2008). We explored whether zyxin is required for Twist1-induced EMT. NMuMG-C7 cells transduced with Twist1 (NMuMG-Twist1) underwent EMT and exhibited cell scattering, numerous protrusions (Figure 6A, arrowhead) and stress fiber formation (Figure 6C). In contrast, NMuMG-Twist1 cells transfected with zyxin siRNA exhibited fewer protrusions and rectangular cell morphology (Figure 6B). Actin stress fiber formation was also attenuated (Figure 6D). These findings suggest that zyxin is required for the cell shape change and actin fiber formation induced by Twist1. In addition, although NMuMG-Twist1 cells displayed enhanced migratory activity, depletion of zyxin strongly suppressed the migration of NMuMG-Twist1 cells (Figure 6, E and F). These results suggest that Twist1 induces EMT by up-regulating zyxin and by potential zyxin-dependent effects on actin reorganization to promote cell migration.
Figure 6.
Twist1 uses zyxin to execute EMT. (A and B) Twist1-induced cell morphological change was abrogated by zyxin depletion. NMuMG-C7 cells transduced with Twist1 were transfected with zyxin siRNA or scramble RNA. Phase-contrast microscopy was done 48 h after transfection. (A) Arrowheads indicate the cell protrusions induced by Twist1 transduction. Scale bar, 100 μm. (C and D) Twist1-induced actin fiber reorganization was abrogated by zyxin depletion. Phalloidin staining in NMuMG-Twist1 cells transfected with zyxin siRNA or scramble RNA. Scale bar, 100 μm. (E) Modified Boyden's chamber assay performed with NMuMG-Twist1 cells transfected with zyxin siRNA or scramble RNA. After incubation for 4 h, the cells that migrated to the lower surface of the membrane were stained. Scale bar, 250 μm. (F) The number of migrated cells in E. *p = 0.0013 versus NMuMG-EGFP, Control; **p = 0.0144 versus NMuMG-Twist1, Control; ***p = 0.0184 versus NMuMG-EGFP, Control. Results are shown as the mean ± SEM of four separate migration assays from two independent experiments.
Thus, zyxin plays a significant role in motility acquisition in EMT by promoting actin fiber formation and confers the endocardial cells the motility required for cardiac morphogenesis.
DISCUSSION
We delineate the signaling pathway, TGF-β1-Twist1-zyxin, in the EMT in cultured cells and demonstrate the involvement of this signaling in valvulogenesis in the development of the heart in vivo. Depletion of zyxin abrogated either Twist1-dependent or TGF-β1–dependent EMT in NMuMG cells, confirming this signaling.
We show that zyxin is essential for cell migration and actin fiber reorganization in TGF-β1-EMT. Zyxin and paxillin are the prototypes of two related subfamilies of LIM domain proteins that are localized primarily at focal adhesion plaques (Wang and Gilmore, 2003). Although paxillin and Hic-5 are required for the progression of EMT (Tumbarello et al., 2005), the involvement of zyxin in EMT has not been reported. On EMT induction, zyxin mobilized from focal adhesions to actin stress fibers. Although our findings are based on EGFP-fusion protein study, EGFP-tagged zyxin was reported to display a distribution pattern that is indistinguishable from that of the endogenous zyxin (Nix et al., 2001; Hotulainen and Lappalainen, 2006). Mechanostress-induced relocation of zyxin has been reported (Yoshigi et al., 2005). Yoshigi et al. showed that cyclic stretch or sheer stress to cells in vitro resulted in mobilization of zyxin from focal adhesions to actin filaments. Lele et al. (2006) measured the molecular binding kinetics of focal adhesion proteins in living cells using modified fluorescence recovery after photobleaching technique and showed mechanical forces specifically altered the molecular binding kinetics of zyxin, but not of vinculin. What role does the relocation of zyxin play, if any, in endocardial morphogenesis? Yashiro et al. (2007) has shown that left-right asymmetry of aortic arch was determined by asymmetric blood flow during the development of great arteries. Similarly, sheer stress induced by blood flow or mechanostress altered through cardiac morphogenesis might induce the relocation of proteins that sense mechanical forces, thereby contributing to endocardial morphogenesis. The significance of relocation requires further analysis, although it is speculated that zyxin interacts with a set of proteins such as VASP (Yoshigi et al., 2005) as an adaptor protein on the actin stress fibers. The cells transfected with zyxin siRNA failed to show TGF-β1–enhanced cell motility and fiber formation, indicating that zyxin is essential for motility acquisition and actin fiber reorganization induced by TGF-β1 in NMuMG cells.
We delineated the signaling pathway, TGF-β1-Twist1-zyxin in EMT. TGF-β1 induced the expression of Twist1 and Snail. Twist1, but not Snail, was required for the up-regulation of zyxin by TGF-β1. Numerous focal adhesion proteins are regulated in EMT (Zavadil and Bottinger, 2005). In NMuMG-C7 cells, paxillin was up-regulated by TGF-β1 (Supplementary Figure S5A), but neither Twist1 nor Snail induced paxillin expression, suggesting the other transcription factor was responsible for it. It is of note that Twist1 or Snail targets different genes despite that both are potent EMT inducers. These results implied the multiple layers of transcriptional regulation in EMT. Coordinated expression of transcription factors might be important for the efficient progression of EMT. Niessen et al. showed that Snail and Slug, which belong to the same protein family, are regulated differentially and function in combination during AVC morphogenesis (Niessen et al., 2008). Thuault et al. (2006) showed that HMGA2 induces the expression of Snail/Slug and weakly of Twist1 in TGF-β1–induced EMT. These reports support that EMT is more efficiently executed by coordination of multiple transcriptional regulations. Further investigation is necessary to address how the cellular phenomena are regulated by multiple layers of transcriptional regulation.
Twist1 is an EMT regulator well characterized in cancer metastasis (Yang et al., 2004). It is of note that Twist1 uses zyxin to induce the cell morphological change and migration. Therefore, zyxin might be a target of treatment or prevention for cancer metastasis. We confirmed the presence of several E-box sites, which basic helix-loop-helix transcription factors including Twist1 preferentially bind to, in the promoter region of zyxin gene. Further analysis is needed to reveal how Twist1 promotes transcription of zyxin.
We revealed that zyxin was predominantly expressed in AVC in the embryonic heart undergoing EMT and that zyxin was required for migration and actin fiber reorganization in the endocardial cells. The expression of zyxin mRNA began at E10.5 that corresponds to the onset of endocardial EMT and became predominant in AVC of developing heart and in adult valves. The expression of Twist1 in AVC was highly correlated with that of zyxin. TGF-β1 is expressed in AVC and contributes to valvulogenesis (Nakajima et al., 2000; Molin et al., 2003), suggesting functional coordination of TGF-β1-Twist1-zyxin pathway in vivo. Most recently, the role for Twist1 in AVC development was reported (Shelton and Yutzey, 2008), and further molecular dissection would lead to better understanding of cardiac morphogenesis.
We further examined the importance of zyxin in EMT by ex vivo assay using E9.5 AVC tissue explants. The endocardial cells transduced with lentivirus vector expressing zyxin shRNA failed to exhibit outgrowth and invasiveness into collagen gels and to execute actin fiber reorganization. Together, zyxin is essential for the endocardial cells to undergo EMT in which zyxin promotes cell migration through actin fiber reorganization. Although zyxin-null mice are viable and fertile (Hoffman et al., 2003), the hearts of the zyxin-null mice have not been inspected in detail. We considered two possibilities for no phenotype in zyxin-null mice. First, the abnormalities might be minor ones requiring careful observation, such as bicuspid aortic valves (Garg et al., 2005) or malformation of supporting apparatus such as chordae tendineae (Norris et al., 2008). Second, there might be genetic redundancy and chronic loss of zyxin might be overcome. To support this, although single knockout mice of VASP, with which zyxin directly interacts, exhibit no cardiac phenotype, but triple knockout mice of VASP, Ena and Ena-VASP-like show abnormally thin endocardium (Furman et al., 2007). It might be necessary to ablate plural genes including LIM-family proteins to observe abnormal phenotypes due to the absence of zyxin. In Xenopus laevis, Martynova et al. (2008) revealed that zyxin expression increased along gastrulation that is another example of developmental EMT, and administration of zyxin morpholino resulted in disruption of embryos during gastrulation. It implied the essential role for zyxin in EMT during gastrulation. Thus, considering the particularly strong expression of zyxin in AVC undergoing EMT and the result from ex vivo AVC explant assay, we conclude that zyxin is required for migration and actin fiber reorganization of the endocardial cells.
In this study, we delineate the essential signaling, TGF-β1-Twist1-zyxin pathway, in in vitro EMT using cultured cells and in vivo EMT using the developing heart.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tsuyoshi Akagi for providing pCX4 vectors. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan Heart Association, and Miyata Heart Foundation.
Abbreviations used:
- AVC
atrioventricular canal
- EMT
epithelial–mesenchymal transition
- NMuMG
normal murine mammary gland.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0046) on May 13, 2009.
REFERENCES
- Akagi T., Sasai K., Hanafusa H. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. USA. 2003;100:13567–13572. doi: 10.1073/pnas.1834876100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansieau S., et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Bartram U., Molin D. G., Wisse L. J., Mohamad A., Sanford L. P., Doetschman T., Speer C. P., Poelmann R. E., Gittenberger-de Groot A. C. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGF-beta(2)-knockout mice. Circulation. 2001;103:2745–2752. doi: 10.1161/01.cir.103.22.2745. [DOI] [PubMed] [Google Scholar]
- Beckerle M. C. Spatial control of actin filament assembly: lessons from Listeria. Cell. 1998;95:741–748. doi: 10.1016/s0092-8674(00)81697-9. [DOI] [PubMed] [Google Scholar]
- Bruneau B. G. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Camenisch T. D., Molin D. G., Person A., Runyan R. B., Gittenberger-de Groot A. C., McDonald J. A., Klewer S. E. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev. Biol. 2002;248:170–181. doi: 10.1006/dbio.2002.0731. [DOI] [PubMed] [Google Scholar]
- Cano A., Perez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., Nieto M. A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Chang C. P., Neilson J. R., Bayle J. H., Gestwicki J. E., Kuo A., Stankunas K., Graef I. A., Crabtree G. R. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118:649–663. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Fradelizi J., Noireaux V., Plastino J., Menichi B., Louvard D., Sykes C., Golsteyn R. M., Friederich E. ActA and human zyxin harbour Arp2/3-independent actin-polymerization activity. Nat. Cell Biol. 2001;3:699–707. doi: 10.1038/35087009. [DOI] [PubMed] [Google Scholar]
- Furman C., Sieminski A. L., Kwiatkowski A. V., Rubinson D. A., Vasile E., Bronson R. T., Fassler R., Gertler F. B. Ena/VASP is required for endothelial barrier function in vivo. J. Cell Biol. 2007;179:761–775. doi: 10.1083/jcb.200705002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V., Muth A. N., Ransom J. F., Schluterman M. K., Barnes R., King I. N., Grossfeld P. D., Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Nimura K., Kashiwagi K., Harada T., Takaoka K., Kato H., Tamai K., Kaneda Y. Comparative roles of Twist-1 and Id1 in transcriptional regulation by BMP signaling. J. Cell Sci. 2007;120:1350–1357. doi: 10.1242/jcs.000067. [DOI] [PubMed] [Google Scholar]
- Hetey S. E., Lalonde D. P., Turner C. E. Tyrosine-phosphorylated Hic-5 inhibits epidermal growth factor-induced lamellipodia formation. Exp. Cell Res. 2005;311:147–156. doi: 10.1016/j.yexcr.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Hoffman L. M., et al. Targeted disruption of the murine zyxin gene. Mol. Cell Biol. 2003;23:70–79. doi: 10.1128/MCB.23.1.70-79.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P., Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas J. L., Beckerle M. C. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Koibuchi N., Chin M. T. CHF1/Hey2 plays a pivotal role in left ventricular maturation through suppression of ectopic atrial gene expression. Circ. Res. 2007;100:850–855. doi: 10.1161/01.RES.0000261693.13269.bf. [DOI] [PubMed] [Google Scholar]
- Lele T. P., Pendse J., Kumar S., Salanga M., Karavitis J., Ingber D. E. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J. Cell Physiol. 2006;207:187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Ma L., Lu M. F., Schwartz R. J., Martin J. F. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Martynova N. Y., Eroshkin F. M., Ermolina L. V., Ermakova G. V., Korotaeva A. L., Smurova K. M., Gyoeva F. K., Zaraisky A. G. The LIM-domain protein zyxin binds the homeodomain factor Xanf1/Hesx1 and modulates its activity in the anterior neural plate of Xenopus laevis embryo. Dev. Dyn. 2008;237:736–749. doi: 10.1002/dvdy.21471. [DOI] [PubMed] [Google Scholar]
- Miettinen P. J., Ebner R., Lopez A. R., Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin D. G., Bartram U., Van der Heiden K., Van Iperen L., Speer C. P., Hierck B. P., Poelmann R. E., Gittenberger-de-Groot A. C. Expression patterns of Tgfbeta1–3 associate with myocardialisation of the outflow tract and the development of the epicardium and the fibrous heart skeleton. Dev. Dyn. 2003;227:431–444. doi: 10.1002/dvdy.10314. [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Yamagishi T., Hokari S., Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP) Anat. Rec. 2000;258:119–127. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Nakaya Y., Sukowati E. W., Wu Y., Sheng G. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat. Cell Biol. 2008;10:765–775. doi: 10.1038/ncb1739. [DOI] [PubMed] [Google Scholar]
- Niessen K., Fu Y., Chang L., Hoodless P. A., McFadden D., Karsan A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008;182:315–325. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix D. A., Fradelizi J., Bockholt S., Menichi B., Louvard D., Friederich E., Beckerle M. C. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- Norris R. A., Moreno-Rodriguez R. A., Sugi Y., Hoffman S., Amos J., Hart M. M., Potts J. D., Goodwin R. L., Markwald R. R. Periostin regulates atrioventricular valve maturation. Dev. Biol. 2008;316:200–213. doi: 10.1016/j.ydbio.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M., Jamora C., Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- Runyan R. B., Markwald R. R. Invasion of mesenchyme into three-dimensional collagen gels: a regional and temporal analysis of interaction in embryonic heart tissue. Dev. Biol. 1983;95:108–114. doi: 10.1016/0012-1606(83)90010-6. [DOI] [PubMed] [Google Scholar]
- Saito Y., et al. Transfection of human hepatocyte growth factor gene ameliorates secondary lymphedema via promotion of lymphangiogenesis. Circulation. 2006;114:1177–1184. doi: 10.1161/CIRCULATIONAHA.105.602953. [DOI] [PubMed] [Google Scholar]
- Sanford L. P., Ormsby I., Gittenberger-de Groot A. C., Sariola H., Friedman R., Boivin G. P., Cardell E. L., Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel K. L., Beckerle M. C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Shelton E. L., Yutzey K. E. Twist1 function in endocardial cushion cell proliferation, migration, and differentiation during heart valve development. Dev. Biol. 2008;317:282–295. doi: 10.1016/j.ydbio.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridurongrit S., Larsson J., Schwartz R., Ruiz-Lozano P., Kaartinen V. Signaling via the Tgf-beta type I receptor Alk5 in heart development. Dev. Biol. 2008;322:208–218. doi: 10.1016/j.ydbio.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S. Y., Hsieh C. L., Wu D., Chung L. W., Johnstone P. A. Tumor microenvironment promotes cancer progression, metastasis, and therapeutic resistance. Curr. Probl. Cancer. 2007;31:36–100. doi: 10.1016/j.currproblcancer.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Thiery J. P. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Thuault S., Valcourt U., Petersen M., Manfioletti G., Heldin C. H., Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello D. A., Brown M. C., Hetey S. E., Turner C. E. Regulation of paxillin family members during epithelial-mesenchymal transformation: a putative role for paxillin delta. J. Cell Sci. 2005;118:4849–4863. doi: 10.1242/jcs.02615. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Yin M., Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Wang Y., Gilmore T. D. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- Yang J., et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yashiro K., Shiratori H., Hamada H. Haemodynamics determined by a genetic programme govern asymmetric development of the aortic arch. Nature. 2007;450:285–288. doi: 10.1038/nature06254. [DOI] [PubMed] [Google Scholar]
- Yonemura S., Itoh M., Nagafuchi A., Tsukita S. Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between non-polarized fibroblasts and polarized epithelial cells. J. Cell Sci. 1995;108(Pt 1):127–142. doi: 10.1242/jcs.108.1.127. [DOI] [PubMed] [Google Scholar]
- Yoshigi M., Hoffman L. M., Jensen C. C., Yost H. J., Beckerle M. C. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 2005;171:209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J., Bottinger E. P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.