SUMMARY
Piwi proteins play essential roles in germline development, stem cell self-renewal, epigenetic regulation, and transposon silencing, and are implicated in oncogenesis [1-7]. Piwi proteins bind to a complex class of small non-coding RNAs called Piwi-Interacting RNAs (piRNAs) [8]. Mammalian Piwi proteins such as Mili are localized in the cytoplasm of spermatogenic cells, where they are associated with a germline-specific organelle called the nuage or its derivative, the chromatoid body, as well as with polysomes [9]. To investigate the molecular mechanisms mediated by Mili, we searched for Mili interacting proteins using co-immunoprecipitation and mass spectrometry. Here we report that Mili specifically interacts with Tudor Domain Containing Protein 1 (Tdrd1; a.k.a. Mouse Tudor Repeat 1, Mtr-1), a germline protein that contains multiple Tudor domains [10, 11]. This RNA-independent interaction is mediated through the N-terminal domain of Mili and the N-terminal region of Tdrd1 containing the Myeloid Nervy DEAF-1 (MYND) domain and first two Tudor domains. In addition, Mili positively regulates Tdrd1 expression at the mRNA level. Furthermore, Mili and Tdrd1 mutants share similar spermatogenic defects. However, Tdrd1, unlike Mili, is not required for piRNA biogenesis. Our results suggest that Mili interacts with Tdrd1 in the nuage and chromatoid body. This interaction does not contribute to piRNA biogenesis, but represents a regulatory mechanism critical for spermatogenesis.
Keywords: Mili, Piwi, spermatogenesis, piRNA, Tdrd1
RESULTS AND DISCUSSION
Mili is expressed in round spermatids in addition to its known expression in spermatogonia and spermatocytes
It was previously reported that Mili is specifically expressed in spermatogonia and primary spermatocytes [9, 12]. However, in the immunostaining of wildtype mouse testicular sections prepared with a shorter fixation time, we observed Mili staining in round spermatids (Figure S1A). To test whether this truly reflects the expression of Mili in these cells, we isolated round spermatids using fluorescence-activated cell sorting (FACS) based on cell size (forward scattering) and cell density (side scattering). 1.2 million spermatids were isolated from 10 million total cells (Figure S1B & C, spermatid population is labeled as P1 in B). The identity of the collected spermatids was verified by DAPI staining and examination under a confocal microscope; this analysis showed that 100% of the isolated cells were round spermatids (Figure S1D). We then conducted Western blot analysis on this isolated population to test for Mili expression. We found that Mili is present in isolated round spermatids and 13 dpp mili+/+ testicular extracts.
Mili interacts with Tdrd1 in an RNA-independent manner
To investigate the molecular mechanisms through which Mili acts in spermatogenic cells, we immunoprecipitated endogenous Mili using a peptide-affinity-purified rabbit anti-Mili antibody that specifically recognizes Mili in mouse testes. This specific recognition allowed us to conduct immunoprecipitation using blocking peptide as a valid negative control. Co-immunoprecipitated samples were analyzed on silver-stained SDS-PAGE gels. The two most abundant specific bands (~100 and 140kDa) were identified as Mili and Tdrd1 by mass spectrometry (Figure 1A). We verified the Mili-Tdrd1 interaction by reciprocal co-immunoprecipitation using total cell lysate from 2-month-old adult mouse testes. Tdrd1 was present in Mili-immunoprecipitates, but not in peptide-blocked negative controls (Figure 1B). Furthermore, immunoprecipitation with an anti-Tdrd1 antibody [10] pulled down endogenous Mili (Figure 1C). This interaction was not affected by RNase A treatment of testicular lysate, suggesting that it is RNA independent (Figure 1B and C).
Figure 1. Mili specifically interacts with Tdrd1 in an RNA-independent manner.
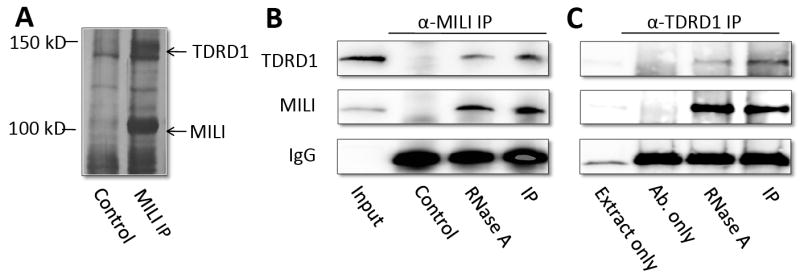
A, Silver-stained SDS-PAGE gels revealing the products of Mili antibody immunoprecipitation of testicular extract. The immunoprecipitation in the presence of blocking peptide was used as negative controls. Two specific bands (~100 and 140kDa) were identified as Mili and Tdrd1 by mass spectrometry. B, Western blot showing that Tdrd1 was co-immunoprecipitated by Mili antibody. The Mili-Tdrd1 interaction is RNA-independent. Peptide-blocked Mili antibody was used as a negative control. IgG band was used as loading control. C, Western blot showing that Mili was co-immunoprecipitated by Tdrd1 antibody. Extract plus beads, and antibody plus beads were used as negative control for IP. The Mili-Tdrd1 interaction is RNA-independent. IgG band was used as loading control.
Mili and Tdrd1 preferentially interact with each other versus with the interactor’s respective family members
We tested the specificity of the Mili-Tdrd1 interaction by expressing tagged versions of Tdrd proteins and Argonaute/Piwi family proteins in 293T cells, followed by co-immunoprecipitation. We found that myc-Tdrd1 preferentially interacts with FLAG-tagged Mili, although association with several other Ago and Piwi family members was observed (Figure 2A). Conversely, we found that Mili specifically interacts with Tdrd1. When tested against a panel of five Tudor domain-containing proteins (Tdrd1, 2, 5, 7, and 9), Mili strongly interacted only with Tdrd1, and a 20-fold weaker interaction with Tdrd2 was also observed (Figure 2B).
Figure 2. The specificity and domains of Mili-Tdrd1 interaction.
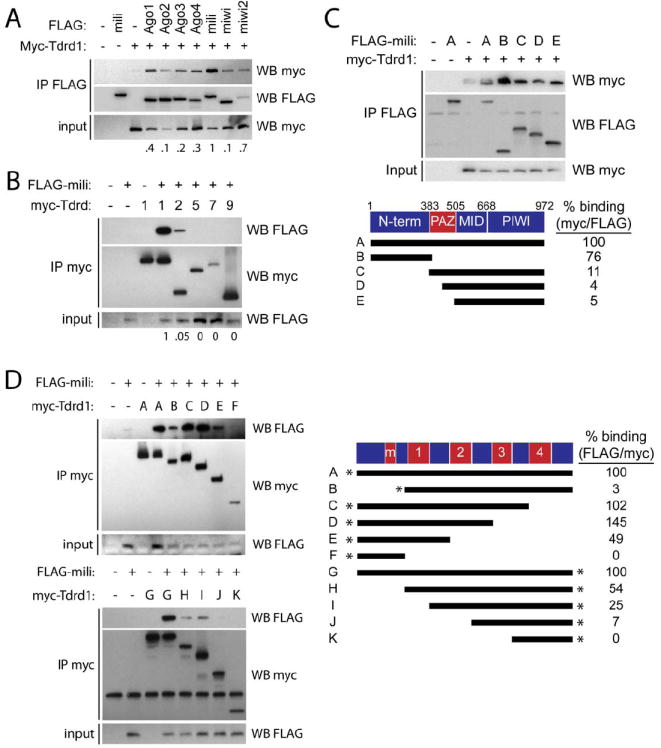
A, Mili preferentially interacts with Tdrd1. 293T cells were transfected with indicated FLAG plasmids and myc-tagged Tdrd1, followed by immunoprecipitation with FLAG beads and subjected to western blotting. The numbers under the blots represent the myc/FLAG signal in the IP FLAG blots. B, Tdrd1 preferentially interacts with Mili. 293T cells were transfected with FLAG-tagged Mili and constructs encoding the indicated myc-taged Tdrd protein, followed by immunoprecipitation with myc antibodies and subjected to western blotting. C, The N-terminal portion of Mili mediates its interaction with Tdrd1. Indicated FLAG-tagged Mili deletion constructs and myc-tagged Tdrd1 were transfected into 293T cells and analyzed as described in (A). D, Multiple domains of Tdrd1 are required for interaction with Mili. Indicated myc-tagged Tdrd1 deletion constructs were transfected into 293T cells with FLAG-tagged Mili and analyzed as described in (B). * indicates either N-terminal or C-terminal myc-tag, m=zinc finger MynD domain.
Mili and Tdrd1 interact via their N-terminal domains
We then mapped the domains of Mili and Tdrd1 that mediate their interaction. The N-terminal domain of Mili alone was sufficient to interact with Tdrd1 almost as efficiently as full-length Mili (76%; see Figure 2C). In contrast, the remaining Mili region, containing the PAZ, Mid, and Piwi domains, was severely abrogated in its ability to co-IP Tdrd1. Further mapping showed that the PAZ domain may play a minor role in the interaction, as removal of the N-terminal and either half or all of the PAZ domain further reduced binding from 11% of the wildtype level to only 4-5% of the wild-type level (Figure 2C). In comparison, the Mili-interacting region of Tdrd1 was found to be more complex: the N-terminal MYND domain of Tdrd1 was necessary but not sufficient for full interaction potential (Figure 2D), as a naturally occurring splice variant lacking this region displayed greatly reduced binding. An additional interaction surface likely resides between the end of the first Tudor and the beginning of the third Tudor domain, whereas the C-terminal fragment containing the fourth Tudor domain is not involved in binding (Figure 2D).
Mili co-localizes with Tdrd1 in cytoplasm of spermatogonia and spermatocytes
Because Mili is mostly localized in the cytoplasm [9, 12], yet has been implicated in transposon function [7, 13], we further examined whether Mili and Tdrd1 interact in the cytoplasm or in the nucleus. We first modified a cytoplasm-nuclear fractionation method [14] to achieve highly effective separation of these compartments and their contents (see Experimental Procedures). Under our conditions, the nuclear fraction contains approximately 4% cytoplasmic proteins, as judged by using GAPDH as a cytoplasmic marker. Likewise, the cytoplasmic fraction consists of approximately 5% nucleoplasm, as judged by western blotting for the nuclear marker RNA polymerase II. Western blotting of fractionated mouse testicular extracts revealed that approximately 89% of Mili is in the cytoplasm, with approximately 11% present in the nucleus (Figure 3A). Likewise, Tdrd1 is mostly localized in the cytoplasm; only approximately 19% of Tdrd1 is in the nucleus.
Figure 3. Mili colocalizes with Tdrd1 in cytoplasm and in the nuage and chromatoid body.
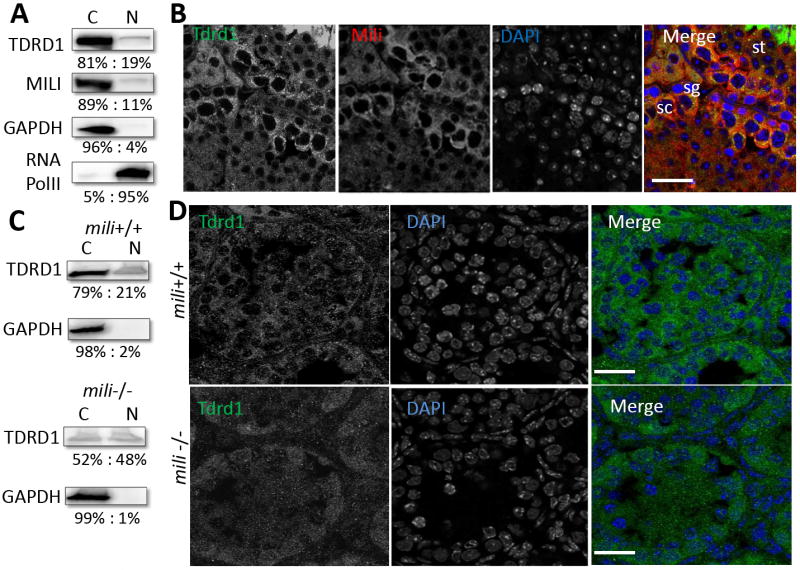
A, Western blots showing that Tdrd1 and Mili are primarily expressed in cytoplasm. Purity of the fractionations was confirmed by western blotting for GAPDH as a cytoplasmic marker and RNA PolII as a nuclear marker. B, Tdrd1 (green) was localized in the cytoplasm and enriched in the dense granule/nuage in spermatogonia and spermatocytes and chromatoid body of spermatids. Mili (red) co-localized with Tdrd1 in the cytoplasm, especially in these granules. DNA was stained with DAPI (as shown in blue). C, Western blotting of Tdrd1 in 13 dpp mili mutant showing that cytoplasmic Tdrd1 is drastically reduced in the mili mutant. D, In 13 dpp mili+/+ testes, Tdrd1 is primarily localized to the cytoplasm of spermatogonia and spermatocytes. While in 13 dpp mili-/- testes show drastic loss of cytoplasmic Tdrd1 in spermatogonia and spermatocytes. The bars are 25μm.
We then further determined the co-localization of Mili and Tdrd1 in spermatogenic cells in two month-old wild-type mouse testes by immunofluorescence microscopy. Tdrd1 is localized in the cytoplasm, where it is enriched in the nuage of spermatogonia and spermatocytes, as well as in the chromatoid body of spermatids (Figure 3B). Mili co-localizes with Tdrd1 in the cytoplasm of spermatogonia and primary spermatocytes; this co-localization is especially pronounced in the nuage and the chromatoid body (Figure 3B).
Tdrd1 expression is down-regulated in the mili mutant during spermatogenesis
To examine whether the interaction between Mili and Tdrd1 implicates a regulatory relationship of one towards the other, we first examined whether the level and/or localization pattern of Tdrd1 in spermatogonia and primary spermatocytes is dependent on Mili. The level of cytoplasmic Tdrd1 protein is drastically reduced in 13 dpp mili-/- testes (which contain only spermatogonia and primary spermatocytes in the germline), whereas the levels of Tdrd1 in the nucleus are not drastically affected (Fig. 3C). This indicates that Mili is required for maintaining the level of Tdrd1 in spermatogonia and spermatocytes, either by up-regulating its expression or controlling its stability. Because Tdrd1 and Mili co-localize in both spermatogonia and spermatocytes, we further examined whether the localization pattern of Tdrd1 in these cells is disrupted in 13dpp mili mutant mice. In wild-type testes, Tdrd1 is primarily localized to the cytoplasm of spermatogonia and spermatocytes (Figure 3D). In contrast, levels of Tdrd1 protein in mili-/- testes are dramatically reduced in cytoplasm of both cell populations, and no longer show any obvious localization pattern (Figure 3D). This provides further evidence that Mili is likely required for the expression and/or stability of Tdrd1 in these spermatogenic cells.
To examine whether Mili regulates Tdrd1 expression or stability at the mRNA or protein level, we determined the levels of Tdrd1 mRNA in mili mutant at 7 dpp, before the mili phenotype first manifests at 8 dpp [9], and at 13 dpp, before final spermatogenic arrest in the mili mutant at 14.5 dpp [12]. Real-time RT-PCR revealed a 2- and 5-fold reduction in tdrd1 mRNA levels in 7 dpp mili-/- testes as compared to mili+/- and mili+/+ testes, respectively. At 13 dpp, tdrd1 mRNA in mili-/- mice is reduced to ~24% of mili+/+ and ~53% of mili+/- testes levels, respectively (Figure S2A). These results indicate that Mili promotes the expression of tdrd1 at the mRNA level.
To investigate whether Mili also promotes Tdrd1 expression at the protein level, we examined Tdrd1 levels in 7dpp and 13 dpp mili+/+, mili+/-, and mili-/- mouse testes by Western blotting. At 7 dpp mili-/- mouse testis, Tdrd1 protein levels are reduced to ~15% and ~17% of mili+/+ and mili+/- testes levels, respectively (Figure S2B and C). At 13 dpp mutant testis, the Tdrd1 levels are reduced to ~18% of mili+/+ and ~28% of mili+/- testes levels, respectively (Figure S2B and C). These reductions are similar to the decrease seen for tdrd1 mRNA levels at the corresponding time points, indicating that Mili is mostly, if not exclusively, required for the expression or stability of Tdrd1 at the mRNA level.
Tdrd1 deficiency does not appear to affect expression of Mili
Because Tdrd1 expression was down-regulated in the mili mutant, we investigated whether the level of Mili is also regulated by Tdrd1. We performed Mili immunostaining on sections of 18 dpp tdrd1+/+ and tdrd1-/- mouse testis. At this time point, tdrd1-/- testes are phenotypically normal [11]. Mili is still clearly detected in the tdrd1-/- testes, present in the cytoplasm in spermatogonia and primary spermatocytes, as well as in the smaller chromatoid body in the mutant spermatids (Figure S3). These results suggest that Tdrd1 is not required for Mili expression in the cytoplasm or localization to the chromatoid body.
The Mili-Tdrd1 interaction occurs independently of polysomes
We previously showed that a fraction of Miwi and Mili molecules associate with translational machinery [9, 15]. To test whether the Mili-Tdrd1-containing complex is associated with the translation machinery, we performed a 15-50% (W/W) sucrose gradient polysome fractionation using two-month old mouse testes to resolve the distribution of Tdrd1 in RNP, ribosomal, and polysome fractions (Figure 4A). As previously reported [9], Mili is found mainly in the ribonucleoprotein (RNP) and polysome fractions. However, Tdrd1 was primarily found in the non-RNP fractions, containing ribosomal subunits, monosomes, and polysomes (Figure 4A). To determine whether the co-fractionation of Tdrd1 with various ribosomal fractions reflects association of this protein with ribosomes, we dissociated polysomes and monosomes to their large and small subunits by EDTA treatment (Figure 4B). We also treated the lysates with micrococcal nuclease to degrade polysomes into monosomes (Figure 4C). Both treatments eliminated Mili in the polysome fractions, as would be expected for a ribosome-associated protein. However, neither treatment affected the sedimentation of Tdrd1. To further confirm this polysome-independent interaction, we determined whether the distribution of Mili in polysome fractions is affected in tdrd1-/- mouse testes. As shown in Figure 4D, Mili distribution in polysome fractions did not depend on Tdrd1 (Figure 4D). These results suggest that Tdrd1 does not interact with the pool of Mili that is associated with the cytoplasmic translational machinery, but rather most likely with Mili molecules in the nuage and chromatoid body.
Figure 4. The interaction between Mili and Tdrd1 is not related to translation and piRNA biogenesis.
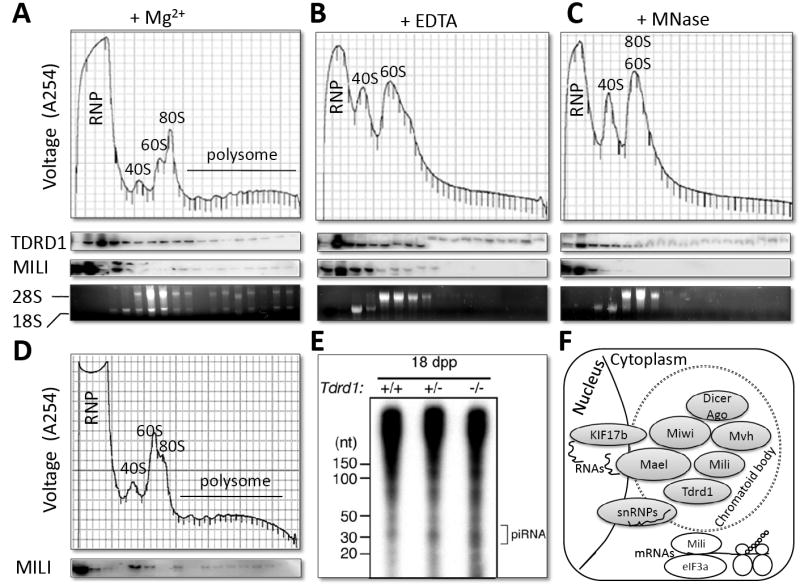
A, Tdrd1 and Mili fraction profile of two-month old wildtype testis in a 15-50% (W/W) sucrose gradient. B and C, fractionation profiles of wildtype testis following treatment with EDTA and micrococcal nuclease respectively. D, Mili distribution on polysome fractions in tdrd-/- testes. E, a polyacrylamide gel showing piRNAs are present in similar abundance in Tdrd1+/+. Tdrd1+/-, and Tdrd1-/- testes. F, A working model on the interaction of Mili, Tdrd1, and other proteins in the nuage and chromatoid body, which is distinct from the Mili-eIF3a interactions in the translational machinery in the cytosol.
Tdrd1 does not contribute to piRNA biogenesis
As Mili is involved in the biogenesis of piRNAs [8, 16], we investigated whether the interaction of Tdrd1 with Mili is required for piRNA biogenesis. Total RNAs were isolated from 18 dpp tdrd1+/+, tdrd1+/-, and tdrd1-/- mouse testes, labeled at the 5’-end with 32P, and fractionated by electrophoresis. A similarly abundant population of piRNA is detected in tdrd1+/+, tdrd1+/-, and tdrd1-/- mouse testes (Figure 4E). This result suggests that the Mili-Tdrd1 interaction does not contribute to piRNA biogenesis.
Conclusions
We have identified Tdrd1 as an interactor of Mili. This interaction is not associated with polysomes, but likely takes place within the nuage and chromatoid body where Tdrd1 and Mili colocalize (Figure 4F). The nuage and chromatoid body contain abundant RNA binding proteins and have been implicated in the storage, metabolism, and cytoplasm-to-nuclear transport of mRNA, small RNA, and related proteins such as KIF17b, a testis-enriched kinesin [17]. Because tdrd1-/- and mili-/- mutants share similar spermatogenic defects, and because Tdrd1-/- mice exhibit defects in the integrity of the nuage and chromatoid body, Mili might act together with Tdrd1 to recruit nuage and chromatoid body component proteins for proper formation of these structures and their function in the metabolism and transport of mRNAs and small RNAs (Figure 4F). This function is essential for spermatogenesis and is distinct from the interaction of Mili with translational initiation factors (eIF3a and eIF4G) in translational regulation outside the nuage/chromatoid body [9]. Furthermore, since Tdrd1 is not involved in piRNA biogenesis, our data further indicates that the Mili-Tdrd1 interaction represents a novel regulatory mechanism, either downstream or independent of piRNA biogenesis, that is crucial for spermatogenesis.
EXPERIMENTAL PROCEDURES
Isolation of round spermatids
Round spermatids were isolated using flow cytometry as described in [18]. Briefly, testes from one mouse were freshly dissected and washed twice with 1xPBS. The tunica albuginea and the remaining blood vessels were then removed under a dissecting microscope. The tubules were rinsed twice with 1xPBS, minced forceps, and placed into 1 ml PBS. Spermatogenic cells were dissociated with repeated pipetting. The resulting suspension was stored for 15 min at room temperature. The resulting supernatant was centrifuged at 600 g for 5 min. The pellet was resuspended in 1 ml PBS.
To enrich spermatogenic cells, the resulting suspension was applied to the top of a discontinued Percoll gradient (45, 30, 22, and 15% Percoll in 1xPBS) and centrifuged at 600 g for 25 min. Cells in 22% Percoll were recovered and applied to BD FACSAria Flow Cytometer for cell sorting. The cell size (forward angle light scattering, FSC) and cell density (90° light scattering, SSC) were measured simultaneously, and P1 area was collected into collecting tubes coated with 4% BSA in PBS overnight at 4°C. Cells were collected at 1500g for 5 min for further analysis.
Mili immunoprecipitation, protein identification, and Tdrd1 immunoprecipitation
Mili antibody was purified by protein A sepharose beads (GE Healthcare Life Science) and Affi-Gel 15 (Bio-Rad) per manufacture protocol. 10 mg testis lysate from 2 month old CD1 wildtype mouse was used for Mili immunoprecipitation with protein A sepharose beads. Peptide-blocked anti-Mili immunoprecipitation was used as a negative control. Proteins were resolved by 6% SDS-PAGE and stained with silver staining method. The bands present in Mili immunoprecipitation lane were excised and were sent to Prottech, Inc. (Fairview Village, PA) for identification using NanoLC-MS/MS peptide sequencing technology. Briefly, each band was destained and digested with sequencing grade modified trypsin (Promega, Madison, WI). The peptide mixture was analyzed by a NanoLC-MS/MS system (Thermo, Palo Alto, CA). The resulted spectrometric data were searched against the most recent non-redundant protein database (downloaded from NCBI’s GenBank) with ProtTech’s proprietary software suite.
Tdrd1 immunoprecipitation was carried out with rabbit anti-Tdrd1 with two negative controls: lysate plus beads only, and anti-Tdrd1 plus beads only. Briefly, anti-Tdrd1 was incubated with protein A sepharose for 2 hrs at 4°C. The beads were washed with MCB buffer and splitted into two fractions. One was saved as an antibody control. The testicular lysate was added to the other fraction and incubated 2 hrs at 4°C. Meanwhile, the same amount of lysate was added to bare beads to be used as a lysate plus beads control.
RNase A (Sigma) was used to treat one set of testis extract for Mili immunoprecipitation to see whether the interaction is RNA-dependent or not. RNase A was added to lysate at the final concentration of 20 U/ml and incubated for two hrs at 4°C, then 30 mins at room temperature before being used for immunoprecipitation.
Nuclear/cytoplasmic fractionation
Testis tissue were homogenized in MCB buffer (100 mM KoAc, 0.1% Triton, 50mM HEPES pH7.4, 2 mM Mg(OAc)2, 10% glycerol, 1 mM DTT, 20U/mL RNase out (invitrogen), 1X complete mini EDTA-free protease inhibitor cocktail (Roche)). The crude nuclear pellet was collected at the speed of 1300 g for 10 min at 4°C. Supernatant was collected as cytoplasmic fraction. The above crude pellet was homogenized again, centrifuged at 1300 g for 10 min to collect the pellet as the nuclear pellet. Nuclear pellet was resuspended in the same volume of buffer used for the cytoplasmic fraction. Nuclear membranes were broken using sonication. The un-broken nuclei were pelleted at 1300 g for 10 min. The resulting supernatant is the nuclear fraction.
Western blot and immunostaining of Mili and Tdrd1
Polyclonal guinea pig anti-Mili generated against amino acids 483-644 in our laboratory and polyclonal rabbit anti-Tdrd1[10] were used at 1:100 dilution for immunostaining on 8 μm-thick cryosections which either was fixed overnight with 4% paraformaldehyde (PFA) [19] or was fixed 3 hrs with 2% PFA [10]. For Western blot, all the antibodies were used at 1:1500 dilution. Rabbit anti-GAPDH was purchase from Sigma and rabbit anti-RNA polymerase II was bought from Santa Cruz. All secondary antibodies were purchased from Jackson ImmunoResearch Laboratory. 4’-6-Diamidino-2-phenylindole (DAPI) or Hoechst was used to label nuclei in immunostaining. Images were taken with Leica TCS SP5 Spectral Confocal Microscope in the sequential scanning mode.
Western blot quantification
All western blot images were taken with Kodak Gel Logic 2200 Imaging System. The control images (GAPDH, RNA PolII, Actin) were acquired and analyzed from the same corresponding blots. All bands shown in quantification were exposed within the linear range of the instrument. The band density was quantified with Kodak Molecular Imaging (MI) Software.
Quantitative real-time PCR
In order to obtain statistically significant data, 3 individual mice were collected for each genotype at 7 dpp and 13 dpp. One testes of each mouse was used for Western blot. The other one was used for isolation of RNA using RNeasy Mini Kit (Qiagen) following manufacture protocol. Reverse transcription was done using SuperScript™ III RT (Invitrogen). Quantitative real-time PCR was carried out on a Roche LightCycler 2.0 system with the Roche SYBR Green Master mix using the following primer sets bought from Integrated DNA Technologies: GAPDH (forward, 5’-CCAGGAGCGAGACCCCACTAACA-3’; reverse, 5’-TTCACACCCATCACAAACAT-3’), Tdrd1 (forward, 5’-TTCAAAGAATGTCCACGCAG-3’; reverse, 5’-GTGTGGTATCTCTTTAGTGG-3’).
Sucrose gradient polysome fractionation
Testicular extract from 2 month old adult mice was applied to sucrose gradient polysome fractionation as in [15]. One mouse testis was freshly dissected, decapsulated and then homogenized in 0.5 mL of MCB buffer mentioned above. The lysate was centrifuged at 1,300g at 4°C for 10 minutes. The supernatant was applied onto the top of a 15%-50% (W/W) linear sucrose gradient made by Density Gradient Fractionation System (Teledyne ISCO Inc.). The gradient was centrifuged at 150,000g for 3 hours (Beckman, CA). Fractions (0.3 ml) were collected. SDS-PAGE was run after TCA precipitation and the blot was probed with rabbit anti-Mili, and rabbit anti-Tdrd1. RNA was isolated using Trizol (Invitrogen). Cycloheximide (CHX) was added to inhibit translational elongation at a final concentration of 200 μM. For the EDTA treatment, the lysate was treated with 20 mM EDTA before applying to the sucrose gradient (supplemented with 20 mM EDTA instead of Mg(OAc)2). For the micrococcal nuclease (MNase, Sigma) treatment, micrococcal nuclease at a final concentration of 60U/mL was added to the lysate (supplemented with 2mM CaCl2 instead of Mg(OAc)2), and incubated at room temperature for 20 minutes, then stopped by 25mM EGTA and applied to the sucrose gradient (supplemented with 25 mM EGTA).
piRNA detection
Total piRNAs were detected mostly as previously described [16]. Briefly, total RNA extracted from Tdrd1 mutant and control testes at 18 dpp was treated with DNase I (Promega, USA) and end-labeled using T4 polynucleotide kinase (NEB, USA) and [gamma-32P] ATP. One microgram of radio-labeled RNA was separated on a 15% denaturing PAGE, and signals were detected with x-ray film.
Supplementary Material
Acknowledgments
We thank Norio Nakatsuji for his enthusiasm and financial support of the part of this work by S.C. and T.T., who are grateful to Prof. Nakatsuji’s guidance. We also thank Prof. Toru Nakano and Dr. Satomi Kuramochi-Miyagawa for the mili mutant mice. This research was supported by NIH42012 and Connecticut Stem Cell Research Foundation to H.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & development. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development (Cambridge, England) 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development (Cambridge, England) 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 4.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science (New York, N Y) 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 5.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & development. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 7.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science (New York, N Y) 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 8.Lin H. piRNAs in the germ line. Science (New York, N Y) 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- 9.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a piRNA binding protein, is required for germline stem cell self-renewal and appears to positively regulate translation. The Journal of biological chemistry. 2008 doi: 10.1074/jbc M809104200. Published online, December 29, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mechanisms of development. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 11.Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development (Cambridge, England) 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 13.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & development. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SY, Park JH, Kim S, Park EJ, Yun Y, Kwon J. A proteomics approach for the identification of nucleophosmin and heterogeneous nuclear ribonucleoprotein C1/C2 as chromatin-binding proteins in response to DNA double-strand breaks. The Biochemical journal. 2005;388:7–15. doi: 10.1042/BJ20042033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 17.Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. Journal of cell science. 2006;119:2819–2825. doi: 10.1242/jcs.03022. [DOI] [PubMed] [Google Scholar]
- 18.Lassalle B, Ziyyat A, Testart J, Finaz C, Lefevre A. Flow cytometric method to isolate round spermatids from mouse testis. Human reproduction (Oxford, England) 1999;14:388–394. doi: 10.1093/humrep/14.2.388. [DOI] [PubMed] [Google Scholar]
- 19.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Developmental cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


