Abstract
Aptamers are nucleic acid-based reagents that bind to target molecules with high affinity and specificity. However, methods for generating aptamers from random combinatorial libraries (e.g., SELEX) are often labor-intensive and time-consuming. Recent studies suggest that microfluidic SELEX (M-SELEX) technology can accelerate aptamer isolation by enabling highly stringent selection conditions through the use of very small amounts of target molecules. We present here an alternative M-SELEX method, which employs a disposable microfluidic chip to rapidly generate aptamers with high affinity and specificity. The Micro-Magnetic Separation (MMS) chip integrates microfabricated ferromagnetic structures to reproducibly generate large magnetic field gradients within its microchannel that efficiently trap magnetic bead-bound aptamers. Operation of the MMS device is facile, robust and demonstrates high recovery of the beads (99.5%), such that picomolar amounts of target molecule can be used. Importantly, the device demonstrates exceptional separation efficiency in removing weakly-bound and unbound ssDNA to rapidly enrich target-specific aptamers. As a model, we demonstrate here the generation of DNA aptamers against streptavidin in three rounds of positive selection. We further enhanced the specificity of the selected aptamers via a round of negative selection in the same device against bovine serum albumin (BSA). The resulting aptamers displayed dissociation constants ranging from 25 to 65 nM for streptavidin but negligible affinity for BSA. Since a wide spectrum of molecular targets can be readily conjugated on magnetic beads, MMS-based SELEX should provide a general platform for rapid generation of specific aptamers.
Aptamers are nucleic acid molecules1,2 that have been selected in vitro to bind to molecular targets—including small molecules,3,4 proteins,5–7 tissues,8,9 organisms10 and inorganic materials11,12—with high affinity and specificity. Compared to traditional affinity reagents (e.g., monoclonal antibodies), aptamers can be advantageous because they can be chemically synthesized, are thermo-stable, undergo reversible denaturation and the selection process is not limited by immunogenicity of the target molecule within a given host organism.13–15 The general method of isolating aptamers from a library of random nucleic acid molecules, Systematic Evolution of Ligands by EXponential Enrichment (SELEX), involves an iterative process of nucleic acid binding, separation and amplification. Usually, multiple rounds of selection (typically 8–15 rounds) are necessary in order to isolate aptamers with sufficient affinity and specificity, requiring significant time and labor.16,17
A variety of advanced separation techniques have been employed to increase the efficiency and throughput of aptamer isolation, including flow cytometry,18,19 surface plasmon resonance (SPR)20 and capillary electrophoresis (CE).21–23 In particular, CE-based separation methods have shown remarkable selection efficiencies for protein targets; aptamers with Kd in the low nM range have been isolated after a few selection rounds (~1–4).21–23 However, these methods require sufficient change in the electrophoretic mobility (μe) of proteins upon aptamer binding, and thus they are less effective for classes of targets that do not produce such a μe shift (e.g., small molecules and cell surface marks). Recently, our group described the theoretical advantages of utilizing microfluidics technology for SELEX, demonstrating single-round isolation of DNA aptamers against the light chain of recombinant Botulinum Neurotoxin type A via magnetic separation in the Continuous-flow Magnetic Activated Chip-based Separation (CMACS) device.24 This microfluidic SELEX (M-SELEX) method provided a universal and automatable approach to rapidly generate affinity reagents, and exploited a number of unique phenomena that occur at the micro-scale to achieve exceptional selection efficiencies. First, the embedded ferromagnetic structures within the microchannel enabled precise and reproducible generation of local magnetic field gradients which allowed the manipulation of a small number of magnetic beads. Second, the implementation of a multi-stream, laminar-flow fluidic architecture enabled high purity separation by ensuring that the unbound DNA molecules do not diffuse into the collection port. However, the use of CMACS device suffered from a number of practical disadvantages. For example, the presence of micro-bubbles distorted the flow streams, and any blockage in the microchannel caused bead aggregations that had a severe impact on the aptamer purity and recovery of collected beads. Thus, very careful tuning of the device under microscopy was necessary to reach a high partition efficiency and recovery of aptamer-bound beads.
To address this problem, we have developed an improved microfluidic separation device: the Micro-Magnetic Separation (MMS) chip, which allows reproducible molecular separation with very high recovery of beads (i.e., ~ 99.5%), and enables exceptionally high partition efficiencies (PE ~ 106) without any tuning or optimization. Practically, it is important to note that the MMS offers significantly more robust and facile operation compared to the CMACS device; this device is relatively immune to micro-bubbles and allows a wide range of flow rates without the requirement of microscopy. Using the MMS device, we report here the first work to demonstrate both positive and negative selection in a microfluidic device for rapid isolation of aptamers with high affinity and specificity. As a model, we used streptavidin, a widely used protein for bio-conjugation, as the target. In comparison to conventional magnetic column separation methods, we show that aptamers with higher affinities can be isolated after significantly fewer rounds of selection using the MMS system.
EXPERIMENTAL SECTION
Reagents and Instruments
Streptavidin (SA), bovine serum albumin (BSA), N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) were purchased from Sigma-Aldrich, Inc. (Saint Louis, MO) and used without further purification. The DNA library, unlabeled and labeled PCR primers (see supporting information Table S1) were synthesized and purified by Integrated DNA Technologies (Coralville, IA). Each DNA library component consists of a central random region of 60 bases flanked by two specific 20-base sequences that function as primer-binding sites for PCR. HotStar™ Master Mix and water for PCR were purchased from Qiagen Inc. (Hilden, Germany). The magnetic beads, including Dynabeads® MyOne™ C1 SA-coated beads (for Streptavidin positive selection) and M-270 carboxylic acid-coated beads (for BSA negative selection), were purchased from Invitrogen (Carlsbad, CA). Fluorescence measurements were performed in black 96-well microplates (Microfluor 2, Thermo Scientific, Waltham, MA) using a microplate reader (Tecan, San Jose, CA), and surface plasmon resonance (SPR) measurements were performed on a BIAcore 3000 instrument (GE Healthcare, Waukesha, WI). Real-time PCR equipment (IQ5, Bio-Rad, Hercules, CA) was used to measure the single-stranded (ss) DNA concentrations. The HBS-EP buffer (10 mM HEPES, pH 7.4, including 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) P20 surfactant) was used for the immobilization of the protein onto the SPR sensor chip. The binding buffer (20 mM Tris-HCl, pH 7.6, including 150 mM NaCl, 2 mM MgCl2, 5mM KCl, 1 mM CaCl2 and 0.02% Tween 20) was used for SA aptamer selection.
MMS Chip Fabrication
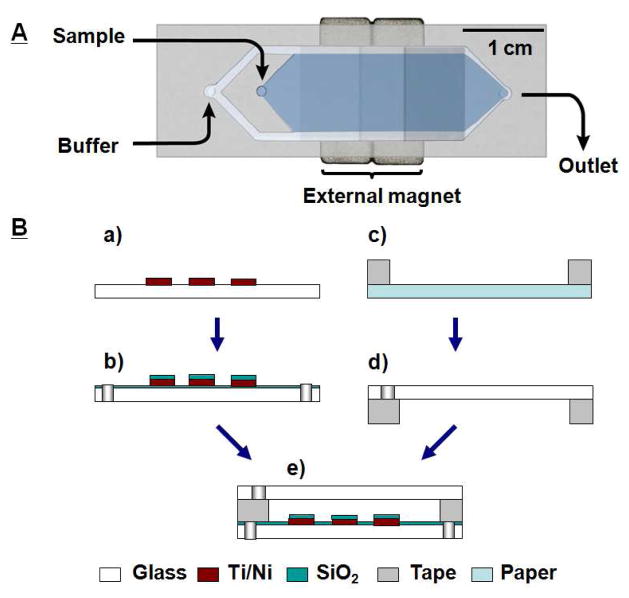
The MMS device (Figure 1A) was fabricated on borosilicate glass substrates with 25-μm-thick double-coated tape (9019, 3M, St. Paul, MN). The micropattern was defined on the bottom glass substrate with 20-nm-thick titanium and 200-nm-thick nickel films using standard photolithography methods (Figure 1B, step a). Pitch distances of 200, 100, and 50 μm were used in the nickel grid pattern to provide increasing grid density. Fluidic vias were drilled through the glass substrates using a computer-controlled CNC mill (Flashcut CNC, San Carlos, CA) and diamond bit (Triple Ripple, Abrasive Technology, Lewis Center, OH) (Figure 1B, step b). After cleaning with acetone, the Ti/Ni layer was passivated with a 100-nm-thick layer of SiO2 by plasma-enhanced chemical vapor deposition (Plasma-Therm, St. Petersburg, FL). Microfluidic channels were cut out of the 25-μm-thick double-sided tape using a plotting cutter (CE5000-60, Graphtec, Santa Ana, CA) (Figure 1B, step c). The patterned tape was overlaid onto the top glass substrate manually (Figure 1B, step d). The channel and micropattern were then manually aligned, and the device was cured in an oven at 65 °C for one hour with light clamping pressure (Figure 1B, step e). A brass eyelet was used as the buffer inlet, and Tygon tubing (S-54-HL, Saint Gobain, Valley Forge, PA) was used for the sample inlet and outlet fluidic connections. All connections were glued in place using 5-minute epoxy (Devcon, Danvers, MA). The external magnets consisted of two 1/2″ × 1/4″ × 1/16″ thick neodymium magnets (grade N42, K&J Magnetics, Jamison, PA). A 1/16″ cubic permanent magnet was placed on top of the chip to fix the function magnets.
Figure 1.
Micro-Magnetic Separation (MMS) device and the fabrication process. A) Photograph of the MMS device with the external rare earth magnet. The MMS device is 64 mm × 15.7 mm × 1 mm (L × W × H) and contains two inlets (sample and buffer) and one outlet. The microchannel inside the device is 25 μm in height and 12 mm in width. The microchannel contains 10-μm-wide microfabricated nickel strips, arrayed in grids at pitches of 200 (near the inlets), 100, and 50 μm (near the outlet). During operation, the sample stream (dark) is flanked by two buffer streams (light, false colors for clarification). B) The MMS fabrication process. a) Ni strips are patterned on the bottom glass substrate by electron-beam evaporation of 20 nm titanium and 200 nm nickel thin-films after standard lithography and lift-off process. b) Microfluidic vias are drilled into the glass substrate with a computer-controlled milling machine. A 100-nm-thick layer of SiO2 is then deposited on the surface for passivation. c) The 25-μm-thick polymer tape is patterned into the geometry of the microchannel with a cutting plotter. d) The patterned tape is overlaid onto the top glass substrate. e) The two substrates are manually aligned and bonded at 65 °C for 1 h, and microfluidic connectors to the inlets and outlet are manually attached with epoxy.
Characterization of MMS Separation
The recovery of magnetic beads in the MMS chip was measured by the fluorescence-labeled magnetic beads. 10 μL of stock MyOne C1 SA-coated beads (108 beads) with 2.5 μL biotin-phycoerythrin (PE) conjugate (4 mg/ml as stock concentration, Invitrogen) were incubated at room temperature for 30 min. After incubation, the biotin-PE conjugated beads were washed 3 times with 1× PBS (11.9 mM sodium phosphates, pH 7.5, including 137 mM NaCl, 2.7 mM KCl) with 0.1% Tween 20 and the unbound DNA supernatant was removed in a Magnetic Particle Concentrator separator (MPC, Invitrogen). The 108 beads were then diluted 102-(107 final), 103-(106 final) or 2×103-fold (5 ×105 final) for recovery experiments. 100 μL of these diluted magnetic beads were loaded into the MMS chip and separated under standard operating conditions (i.e., sample flow rate = 10.8 mL/hr, buffer flow rate = 3 mL/hr). The number of magnetic beads before and after MMS separation was determined using a fluorescence activated cell sorter (FACSAria, BD Biosciences, San Jose, CA), and each concentration was independently measured three times.
To measure the effectiveness of microfluidic washing in the MMS chip, a mixture containing a random DNA library and MyOne C1 SA-coated magnetic beads was loaded into the MMS chip under the same conditions described above. The wash buffer was then continuously flowed over the trapped beads in the MMS chip. Consecutive 500 μL fractions (6 samples total) were collected at the outlet and the initial copy number of DNA molecules in each sample was determined using real-time PCR. The real-time PCR reactions were conducted in 20 μL volume with three replicates for each sample using iQ SYBR green supermix (Bio-Rad) and PCR primers at 500 nM.
Affinity Measurements of Selected Aptamers
The average dissociation constant (Kd) of the selected aptamer pool was measured via a fluorescence binding assay.25 Briefly, the Alexa488-labeled aptamers were diluted to several different concentrations (from 0 to 200 nM) in 80 μL of the binding buffer; these dilutions were heated to 95 °C for 10 min and rapidly cooled down to 0 °C in an ice bath, and then incubated for another 10 min at room temperature. Subsequently, the heat-treated aptamer solutions were incubated with 1.5×108 SA-coated beads at room temperature for 1 h in a total volume of 80 μL. After this incubation, these magnetic beads were washed 5 times with the binding buffer and the unbound aptamers were removed with an MPC separator. The bound aptamers were then released into 160 μL of the binding buffer by heating the aptamer-bound beads at 95 °C for 10 min with shaking. The amount of the released aptamers was determined by fluorescence measurement (λEx = 488 nm, λEm = 519 nm) and the dissociation constants (Kd) were calculated from the calibrated curve fitting.
Cloning and Sequencing of Selected Aptamers
PCR products were amplified with unlabeled primers at the optimized cycle number, purified with the MiniElute® PCR Purification Kit (Qiagen), and cloned using the TOPO TA Cloning Kit (Invitrogen). Fifty colonies were randomly picked and sequenced at Genewiz Inc. (South Plainfield, NJ). The sequences were then analyzed and aligned with Vector NTI Explorer software (Invitrogen). Six representative aptamer sequences from corresponding colonies were amplified with biotinylated reverse primers and Alexa488-labeled forward primers for their fluorescence affinity measurements.
RESULTS AND DISCUSSION
MMS-based Microfluidic Selection
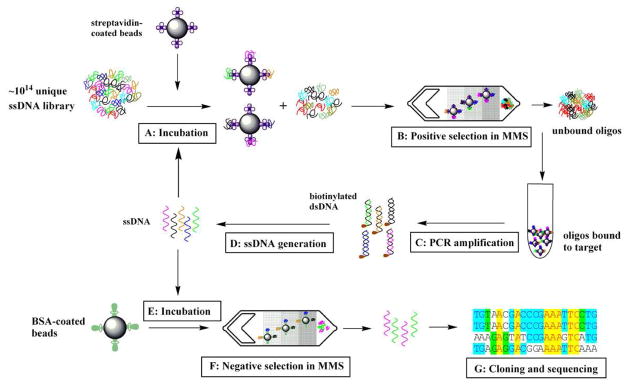
The microfluidic selection process begins with the incubation of 1 nanomole of the random ssDNA library (~1014 molecules) with target proteins conjugated to magnetic beads (Figure 2A). To ensure highly stringent selection conditions, the molar ratio between the ssDNA and target protein is maintained at ~100:1 by using a small number of magnetic beads (106 beads total) coated with a controlled amount of SA (~106 molecules/bead), as described in our previous work.24 After incubation, the partitioning step to separate the target-bound aptamers from the unbound oligos is performed in the MMS chip (Figure 2B). The separation is performed at a flow rate of 10.8 mL/hr (sample) and 3 mL/hr (buffer). During this step, the microfabricated nickel grids are magnetized with an external magnet, efficiently trapping aptamer-bound beads. Stringent washing conditions then are imposed in the microchannel by applying a higher buffer flow rate (20 mL/hr) to continuously elute weakly- and unbound oligos from the device. After the separation, the external magnets are removed and the beads carrying the selected aptamers are released from the device. The entire separation process (trapping, washing and bead release) only requires ~5 minutes. Finally, the selected aptamers are amplified via PCR using a biotinylated reverse primer and an Alexa 488-labeled forward primer (Figure 2C).
Figure 2. Microfluidic SELEX process using the MMS device.
A)Incubation of the bead-immobilized target protein with ssDNA library in binding buffer. To ensure highly stringent binding conditions, the ratio between ssDNA and the protein ratio was kept at 100:1. B) Positive selection of target-bound aptamers. The magnetic beads carrying the target protein and target-bound aptamers are trapped at the nickel patterns within the microchannel. After highly stringent washing, the external magnets are removed, and the beads are eluted into a collection vessel. C) PCR amplification of selected aptamers. The forward primer is Alexa 488-labeled for fluorescence measurements, and the reverse primer is biotinylated for ssDNA isolation. D) Single stranded DNA generation. The biotin-labeled double-stranded PCR product was incubated with Dynabeads® MyOne™ Streptavidin C1 beads for 2 hr at room temperature to separate the ds-DNA from the PCR reaction mixture. Single-stranded DNA was then generated in solution by incubating the beads with 15 mM NaOH for 4 min at room temperature. The supernatant was collected and neutralized with 1 M HCl. E) The positively selected aptamer pool is incubated with BSA-coated magnetic beads. The [BSA]/[ssDNA] ratio is 1:1. F) Negative selection in the MMS device. The unbound ssDNA fraction is collected. G) Cloning and sequencing of the isolated aptamers.
For each round of selection, we chose the optimal PCR cycle number via a “pilot PCR” process 24,26 to eliminate any undesired PCR products. We then generated Alexa 488-labeled ssDNA from the amplified doubled-stranded PCR product (Figure 2D), and measured the DNA concentration via UV-vis absorption. We performed three rounds of positive selection (Figure 2, A to D) to increase the affinity of the selected aptamer pool to SA. In order to raise the specificity of selected aptamers, we also performed an additional round of negative selection in the MMS device against BSA, the most abundant blood protein. We coupled BSA to carboxylic-coated magnetic beads through a modified EDC-NHC coupling process,27 and the aptamer pool was incubated with the BSA-coated beads at a 1:1 molar ratio ([BSA]/[DNA]) (Figure 2, E to F). Finally, the selected aptamer pool was cloned into E. coli and sequenced (Figure 2G).
Micromagnetic Separation
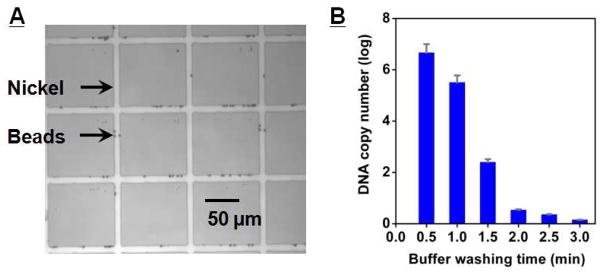
The MMS device offers significant advantages over conventional magnetic separation apparatus (e.g. magnetic columns) because it enables precise control over the hydrodynamic and magnetophoretic forces within the microchannel. The lithographically-patterned nickel microstructures on the bottom glass layer are a key component of the device; due to the large difference in relative magnetic permeabilities between nickel and the buffer medium (μr,nickel = 200, μr,buffer ~ 1), the placement of an external magnet on the chip results in the automatic and reproducible generation of large magnetic field gradients(∇B),28 which cause the magnetic beads to be efficiently trapped at the edges of the pattern, where the gradient is the largest (Figure 3A). We calculated the magnitude of ∇B to be ~104 T/m, and the resulting magnetophoretic force (F⃗m) is on the order of tens of nano-Newtons, as approximated by F⃗m = m∇B, where m is the bead magnetization.29
Figure 3.
MMS device performance. A) Optical micrograph of the trapped magnetic beads. The beads are preferentially captured at the edge of the nickel patterns, where the magnetic field gradients are the highest. B) Real-time PCR measurement of eluted DNA as a function of wash duration. After 3 minutes of washing at 20 mL/hr in the microchannel, all weakly- and unbound DNA were eluted from the MMS device. The measurements were taken in triplicate and real-time PCR was performed in 20 μL volume with forward and reverse primers at 500 nM.
After trapping the beads, we impose highly stringent washing conditions in situ by injecting the washing buffer at a rate of 20 mL/hr. During this step, we ensure that the fluidic drag force (F⃗d) is smaller than F⃗m such that the beads carrying the bound aptamers do not escape from the magnetic trap, and only unbound and weakly-bound oligos are washed away. Since the magnetic beads are held at the bottom of the microchannel and laminar-flow conditions are upheld during the washing step (Reynolds number ~ 1), the calculated fluidic drag force exerted on the beads (F⃗d = 6πηav⃗f, where η is the fluid viscosity, a is the diameter of the bead, and v⃗f the velocity of the wash buffer) is less than a pico-Newton—significantly smaller than F⃗m, which is at the nano-Newton scale.
The partition efficiency of the MMS chip was quantified by measuring the initial copy number of eluted ssDNA as a function of washing time. We found that virtually all weakly- and unbound ssDNAs were eluted within three minutes (Figure 3B), and the resulting partition efficiency is ~106. This is comparable to that of the previously reported CMACS device;24 however, we note that the operation of the MMS device is significantly more reproducible and robust and does not require tuning. Furthermore, we found that the MMS device allows efficient manipulation of very small number of beads (< 106) without loss; upon measuring the number of beads before and after MMS separation via flow cytometry, we found that almost all of the beads loaded in the device were successfully recovered (recovery = 99.5 %). In contrast, the recovery in CMACS is typically ~50 % (see supporting information Figure S1). Finally, the MMS device allows molecular separation at a significantly higher volumetric throughput. Typical sample flow rate in the MMS chip is 10.8 ml/hr, approximately an order of magnitude higher than that of the CMACS device, significantly decreasing the separation time (see supporting information Figure S2).
Affinity and Specificity of Selected Aptamers
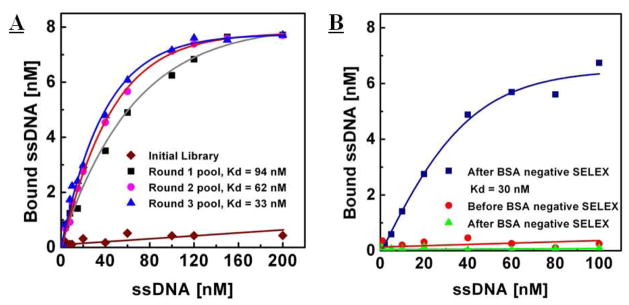
We performed three rounds of positive selection (Figure 2, A–D) to select aptamers with strong SA affinity using the MMS chip with highly stringent selection conditions, and measured the affinity of the enriched aptamer pool from each round via fluorescence assay.25 As expected, the initial random library showed negligible binding affinity to SA target (Figure 4A,  ). For the first round of selection, DNA-target binding was performed at room temperature, and we used the highly competitive binding condition during the incubation step, as described in our previous work.24 More specifically, the molar ratio between the ssDNA and target protein was maintained at ~100:1 by using a small number of magnetic beads (106 beads total) coated with a controlled amount of SA (~106 molecules/bead). The resulting dissociation constant (Kd) of the enriched aptamer pool was 94 ± 10 nM (Figure 4A, ▪). As reported by Romaniuk’s work,30 due to the fact that the entropic contribution is significant in the formation of DNA-protein complexes, selection at elevated temperatures often leads to aptamers with higher affinities. Thus, during the second and third rounds of selection, a higher incubation temperature (37 °C) was used to increase the binding stringency between target protein and aptamers. The resulting aptamer pool yielded Kd of 62 ± 5 nM after the second round (Figure 4A,
). For the first round of selection, DNA-target binding was performed at room temperature, and we used the highly competitive binding condition during the incubation step, as described in our previous work.24 More specifically, the molar ratio between the ssDNA and target protein was maintained at ~100:1 by using a small number of magnetic beads (106 beads total) coated with a controlled amount of SA (~106 molecules/bead). The resulting dissociation constant (Kd) of the enriched aptamer pool was 94 ± 10 nM (Figure 4A, ▪). As reported by Romaniuk’s work,30 due to the fact that the entropic contribution is significant in the formation of DNA-protein complexes, selection at elevated temperatures often leads to aptamers with higher affinities. Thus, during the second and third rounds of selection, a higher incubation temperature (37 °C) was used to increase the binding stringency between target protein and aptamers. The resulting aptamer pool yielded Kd of 62 ± 5 nM after the second round (Figure 4A,  ), and 33 ± 5 nM after the third round of selection (Figure 4A,
), and 33 ± 5 nM after the third round of selection (Figure 4A,  ). Next, in order to further increase the specificity of the selected aptamers, we performed a single round of negative selection against BSA (Figure 2, E to F). In contrast to our positive selection, which used 106 SA-coated beads, we used 108 beads, with 106 BSA molecules per bead, to efficiently deplete the aptamers that bind to BSA from the enriched pool. After a single round of negative selection, the affinity of the enriched aptamer pool was slightly increased for SA (Kd = of 30 ± 5 nM, Figure 4B,
). Next, in order to further increase the specificity of the selected aptamers, we performed a single round of negative selection against BSA (Figure 2, E to F). In contrast to our positive selection, which used 106 SA-coated beads, we used 108 beads, with 106 BSA molecules per bead, to efficiently deplete the aptamers that bind to BSA from the enriched pool. After a single round of negative selection, the affinity of the enriched aptamer pool was slightly increased for SA (Kd = of 30 ± 5 nM, Figure 4B,  ) while the affinity for BSA was decreased by a factor of 5 (Figure 4B, pre-negative selection (
) while the affinity for BSA was decreased by a factor of 5 (Figure 4B, pre-negative selection ( ), post-negative selection (
), post-negative selection ( ). Although we only used BSA in our negative selection, the methodology is general, and virtually any target or combination of targets can be used for the negative selection.
). Although we only used BSA in our negative selection, the methodology is general, and virtually any target or combination of targets can be used for the negative selection.
Figure 4.
Fluorescence measurements to determine dissociation constants (Kd) of selected aptamers. A) The initial, naïve library exhibits negligible binding to the streptavidin target  ). After the first round of selection, the average Kd of the enriched pool was 94 ± 10 nM (■). Subsequently, second and third round selections yielded an average Kd of 62 ± 5 nM (
). After the first round of selection, the average Kd of the enriched pool was 94 ± 10 nM (■). Subsequently, second and third round selections yielded an average Kd of 62 ± 5 nM ( ) and 33 ± 5 nM (
) and 33 ± 5 nM ( ), respectively. B) After a single round of negative selection against BSA, the aptamer pool exhibited slightly higher affinity for streptavidin (Kd = of 30 ± 5 nM,
), respectively. B) After a single round of negative selection against BSA, the aptamer pool exhibited slightly higher affinity for streptavidin (Kd = of 30 ± 5 nM,  ). In contrast, the affinity for BSA significantly decreased 5-fold from (
). In contrast, the affinity for BSA significantly decreased 5-fold from ( ) to (
) to ( ).
).
After cloning and sequencing, sequence alignment revealed that the selected aptamers fell into six groups. One representative sequence from each group was synthesized and their Kd were measured with fluorescence. The values of affinities ranged from 25 (clone 10) to 65 nM (clone 24) (Table 1), which is consistent with the value obtained with the enriched aptamer pool. In addition, SPR was used to further verify the specific binding of clone 10 to streptavidin and the negligible binding to BSA (see supporting information Figure S3).
Table 1.
Sequences and dissociation constants of selected aptamers. The enriched aptamer pool was cloned into E. coli and fifty colonies were randomly picked for sequencing. Sequence alignment revealed 6 consensus sequence groups, and one representative sequence from each group was synthesized and its affinity was measured via fluorescence. All sequences exhibited high affinity for streptavidin and their Kd ranged from 25 to 65 nM.
| Clone Name | Sequence | Dissociation Constant (Kd) |
|---|---|---|
| 10 | GTGATCGTCCAGCGACCGAGCAGGAAACTTATGTAACGACCCGAAATTCCTGCTTAGACT | 25 nM |
| 02 | ATTTGCAACACTTACCACTAAAAGGGCCGACGACCGATGATGTTAGGTCCAGTGCTTTCT | 35 nM |
| 50 | CAAGAACTCCTAAGTATAATGTGAGGGATCCGAAATTCTGCTCTTATGTATGGCAAGATT | 35 nM |
| 07 | ATGGCTGTGGTGGAGGGCCGAAGCCTGTACAAAACAGGAATGCGCTTATCTTGGAGTAT | 50 nM |
| 05 | CAGCGGGCATTTCTAGGGCCATCTTTAATCTTCTTTATGGAGAGCCTCGTTCACACGTTG | 60 nM |
| 24 | TCGAATAGCCCCCCGCGTCCGTCTTCCTTTAGCGATCTATGCTCCTTTGTATGGTCGGGC | 65 nM |
CONCLUSIONS
In this work, we demonstrate the first use of microfluidics technology for highly efficient positive and negative selection of aptamers, such that both affinity and specificity can be rapidly matured in a single separation device. The MMS microfluidic device offers significant improvements over the CMACS device, which was previously used for microfluidic aptamer selection24. By integrating ferromagnetic materials within the microchannel, we were able to accurately control the hydrodynamic and magnetophoretic trapping forces, which enabled the use of small numbers of beads and target molecules with minimal loss and resulted in high molecular partition efficiencies. Because the MMS chip allows separations at relatively high flow rates (> 10 mL/hr), the entire process of trapping, washing and release can be performed within 5 minutes. It is noteworthy that our microfluidic selection is significantly more efficient than conventional magnetic separation methods; for example, Stoltenburg and co-workers25 selected DNA aptamers for streptavidin in 13 rounds using a conventional magnetic column, yielding DNA molecules with Kd ~56–86 nM. In contrast, using the MMS device, we isolated aptamers with Kd ranging from 25 to 65 nM in 3 rounds.
Importantly, since the magnetophoretic force is unaffected by many useful selection parameters, including buffer composition, salinity, pH, and temperature, we believe our platform can be readily adapted to impose multiple selection pressures such that aptamers can be isolated for specific, desired properties. Finally, given that a wide variety of coupling chemistries are available,29 we believe our method can be extended to many different types of molecular targets (e.g., small molecules, proteins, cell-surface markers and inorganic materials) as well as different classes of molecular libraries.
Supplementary Material
Acknowledgments
We thank ONR, NIH, ARO Institute for Collaborative Biotechnologies, and Armed Forces Institute for Regenerative Medicine (AFIRM) for their financial support. We also thank Prof. Patrick Daugherty for the use of the TECAN microplate reader, BD-FACSAria and SPR BIAcore 3000.
References
- 1.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Mann D, Reinemann C, Stoltenburg R, Strehlitz B. Biochem Biophys Res Commun. 2005;338:1928–1934. doi: 10.1016/j.bbrc.2005.10.172. [DOI] [PubMed] [Google Scholar]
- 4.Sazani PL, Larralde R, Szostak JW. J Am Chem Soc. 2004;126:8370–8371. doi: 10.1021/ja049171k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekiya S, Noda K, Nishikawa F, Yokoyama T, Kumar PKR, Nishikawa S. J Biochemistry. 2006;139:383–390. doi: 10.1093/jb/mvj046. [DOI] [PubMed] [Google Scholar]
- 6.Lee JH, Canny MD, Erkenez AD, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F. Proc Natl Acad Sci USA. 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, Schmidt PG, Warren S. J Biol Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 8.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. Proc Natl Acad Sci USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopinath SCB, Misono TS, Kawasaki K, Mizuno T, Imai M, Odagiri T, Kumar PKR. J Gen Virol. 2006;87:479–487. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami J, Imanaka H, Yokota Y, Sugimoto N. J Inorg Biochem. 2000;82:197–206. doi: 10.1016/s0162-0134(00)00158-6. [DOI] [PubMed] [Google Scholar]
- 12.Wilson C, Szostak JW. Chem Biol. 1998;5:609–617. doi: 10.1016/s1074-5521(98)90289-7. [DOI] [PubMed] [Google Scholar]
- 13.Stoltenburg R, Reinemannn C, Strehlitz B. Biomol Engr. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Tombelli S, Minunni M, Mascini M. Biosens Bioelectron. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Nimjee SM, Rusconi CP, Sullenger BA. Annu Rev Med. 2005;56:555–558. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 16.Bowser MT. Analyst. 2005;130:128–130. doi: 10.1039/b412492h. [DOI] [PubMed] [Google Scholar]
- 17.Bunka DHJ, Stockley PG. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 18.Blank M, Weinschenk T, Priemer M, Schluesener H. J Biol Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Li X, Prow TW, Reece LM, Bassett SE, Luxon BA, Herzog NK, Aronson J, Shope RE, Leary JF, Gorenstein DG. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misono TS, Kumar PKR. Anal Biochem. 2005;342:312–317. doi: 10.1016/j.ab.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Mendonsa SD, Bowser MT. J Am Chem Soc. 2004;126(1):20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 22.Mosing RK, Mendonsa SD, Bowser MT. Anal Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 23.Berezovski M, Drabovich A, Krylova SM, Musheev M, Okhonin V, Petrov A, Krylov SN. J Am Chem Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- 24.Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow Theodore M, Heeger AJ, Soh HT. Proc Natl Acad Sci USA. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoltenburg R, Reinemann C, Strehlitz B. Anal Bioanal Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- 26.Tok JBH, Fischer NO. Chem Commun. 2008;16:1883–1885. doi: 10.1039/b717936g. [DOI] [PubMed] [Google Scholar]
- 27.Hermanson GT. Bioconjugate Techniques. 2. Academic Press; 2008. [Google Scholar]
- 28.Inglis DW, Riehn R, Austin RH, Sturm JC. Appl Phys Lett. 2004;85:5093–5095. [Google Scholar]
- 29.Adams JD, Kim U, Soh HT. Proc Natl Acad Sci USA. 2008;105:18165–18170. doi: 10.1073/pnas.0809795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai G, Iskandar M, Barilla K, Romaniuk PJ. Biochemistry. 2001;40:2032–2040. doi: 10.1021/bi001941r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.