Abstract
The non-steroidal anti-inflammatory drug tolfenamic acid (TA) inhibits proliferation of SEG-1 and BIC-1 esophageal cancer cells with half-maximal growth inhibitory concentration values of 36 and 48 μM, respectively. TA also increased Annexin V staining in both cell lines, indicative of proapoptotic activity. Treatment of SEG-1 and BIC-1 cells with TA for up to 72 h decreased expression of specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 and this was accompanied by decreased expression of the well-characterized Sp-regulated genes cyclin D1, vascular endothelial growth factor and survivin. TA also decreased hepatocyte growth factor receptor, (c-Met), a receptor tyrosine kinase that is overexpressed in esophageal cancer cells and tumors and is an important drug target. Knockdown of Sp1, Sp3 and Sp4 by RNA interference in SEG-1 and BIC-1 cells also decreased c-Met expression, demonstrating that c-Met is an Sp-regulated gene in esophageal cancer cells. Sp1 was overexpressed in esophageal cancer cells and tumors and increased Sp1 staining was observed in esophageal tumors from patients. TA (20 mg/kg/day) also decreased tumor growth and weight in athymic nude mice bearing SEG-1 cells as xenografts and this was accompanied by increased apoptosis and decreased Sp1 and c-Met staining in tumors from treated mice. Thus, TA-dependent downregulation of Sp transcription factors and c-Met defines a novel chemotherapeutic approach for treatment of esophageal cancer.
Introduction
Esophageal cancer is a highly aggressive disease, and adenocarcinoma has now replaced squamous cell cancer of the esophagus as the most common tumor, particularly in Caucasian males (1,2). Esophageal cancer has been increasing over the past 10–20 years and it is estimated that in 2008, over 16 470 new cases of esophageal cancer will be diagnosed in the U.S. and 14 280 patients will die from this disease (3). Poor survival rates for esophageal cancer are primarily due to patient delays in seeking diagnosis or treatment of tumors that are already advanced and/or metastasized (4). Five year survival rates from esophageal cancer are generally <15% (5,6). Risk factors for esophageal cancer include alcohol, smoking and achalasia, and individuals with Barrett's esophagus, which results from chronic bile and acid reflux into the esophagus, are also at higher risk for this disease (7–10).
Conventional treatment for esophageal cancer is dependent on the type and stage of the tumor and this may include immediate surgery, neoadjuvant radiotherapy and chemotherapy, followed by surgery and adjuvant chemotherapy (11–14). Several different cytotoxic drugs alone or in combination are used to treat esophageal cancer (EC) and these include 5-fluorouracil, platinum derivatives, paclitaxel and related taxanes. There is also considerable interest on the development of targeted therapies for esophageal cancer and these include various inhibitors of growth factor signaling and blockade of angiogenic pathways (13,14). For example, antibodies such as bevacizumab (Avastin), directed against vascular endothelial growth factor (VEGF), are being used in combination chemotherapy in several Phase II clinical trials for esophageal cancer (13) and preliminary results appear to be promising (15).
Several reports show that Sp1 protein is overexpressed in different tumor types including gastric, colorectal, pancreatic, epidermal, thyroid and breast cancers and recent studies in this laboratory clearly show overexpression of the Sp1, Sp3 and Sp4 proteins in cancer versus non-cancer cells (16–23). Although the mechanism of specificity protein (Sp) overexpression has not been determined, Lou et al. (24) have shown that malignant transformation of human fibroblasts resulted in an 8- to 18-fold increase in Sp1 expression and the transformed cells formed tumors in athymic nude mouse xenografts. In contrast, Sp1 knockdown gave cells that were non-tumorigenic in the same mouse xenograft model. RNA interference studies in which Sp1, Sp3 and Sp4 expression is abrogated in various cancer cell lines show that genes involved in cell proliferation (cyclin D1), survival (bcl2, survivin) and angiogenesis [VEGF, VEGF receptor (VEGFR1) and VEGFR2] are Sp-dependent genes (25–32). This suggests that Sp transcription factors may contribute to cancer cell proliferation, survival and angiogenesis and studies in this laboratory have demonstrated that an underlying mechanism of action of anticancer agents such as tolfenamic acid (TA), betulinic acid and curcumin is the targeted repression of Sps in cancer cells (29–32). Studies with TA and structurally related analogs showed that TA was the most potent of these non-steroidal anti-inflammatory drugs as inhibitors of Sp expression (32). In this study, we show that Sps are also overexpressed in esophageal cancer cells and human tumors and TA decreases esophageal cancer cell survival, represses Sps and Sp-dependent genes/proteins including hepatocyte growth factor receptor (c-Met) and also inhibits esophageal tumor growth in a mouse xenograft model. Moreover, in RNA interference studies, we demonstrate that c-Met is an Sp-dependent gene in these cell lines.
Materials and methods
Cell lines
SEG-1 and BIC-1 esophageal cancer cell lines were obtained from the Cancer Research Institute at MD Anderson Cancer Center Orlando, Orlando Regional Healthcare, Orlando, Florida. Fetal bovine serum (FBS) was obtained from JRH Biosciences (Lenexa, KS). SEG-1 cells were maintained in RPMI 1640 medium (Sigma–Aldrich, St. Louis, MO) supplemented with 0.22% sodium bicarbonate, 0.011% sodium pyruvate, 0.45% glucose, 0.24% 4-(2-hydroxyethyl)-piperazineethane-sulfonic acid, 10% FBS and 10 ml/l 100X antibiotic/antimycotic solution (Sigma-Aldrich). BIC-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM)/Ham's F-12 (Sigma–Aldrich) supplemented with 0.22% sodium bicarbonate, 0.011% sodium pyruvate, 10% FBS and 10 ml/l of 100X antibiotic/antimycotic solution (Sigma–Aldrich). Cells were maintained at 37°C in the presence of 5% CO2.
Antibodies, chemicals and reagents
The Sp1 antibody was purchased from Upstate Biotechnology (Lake Placid, NY). Antibodies against Sp3, Sp4, VEGF, survivin and CD1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for c-Met were purchased from Cell Signaling Technology (Danvers, MA). Reporter lysis buffer and luciferase reagent for luciferase studies were purchased from Promega (Madison, WI). β-Galactosidase (β-gal) reagent was obtained from Tropix (Bedford, MA). LipofectAMINE reagent was supplied by Invitrogen (Carlsbad, CA). TA was purchased from LKT Laboratories (St. Paul, MN).
Cell proliferation assay
Esophageal cancer cells (2 × 104 per well) were plated in 12-well plates and allowed to attach for 24 h. The medium was then changed to DMEM/Ham's F-12 medium containing 2.5% charcoal-stripped FBS, and either vehicle (dimethyl sulfoxide) or different concentrations of the compound were added. Fresh medium and compounds were added every 48 h, and cells were then trypsinized and counted after 48 and 96 h using a Coulter Z1 cell counter. Each experiment was carried out in triplicate, and results are expressed as means ± SEs for each set of experiments.
Immunoblotting
Cell lysates were prepared using lysis buffer [20 mmol/l Tris–HCl (pH 7.4), 150 mmol/l sodium chloride, 1 mmol/l ethylenediaminetetraacetic acid, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 1 mmol/l sodium orthovanadate, 1 mmol/l phenylmethylsulfonyl fluoride, 1 μmol/l leupeptin and 1 μg/ml aprotinin]. After centrifugation of the lysate at 15 000g for 20 min, the supernatants were recovered, and protein was quantified by the Bradford protein assay using a reagent kit from Bio-Rad Laboratories (Hercules, CA). Protein samples (20–60 μg) were size separated by electrophoresis on sodium dodecyl sulfate–polyacrylamide gels under non-reducing conditions. Separated proteins were electroblotted onto nitrocellulose membranes. The blot was blocked by incubating in blocking buffer [5% skim milk, 10 mmol/l Tris (pH 7.5), 10 mmol/l sodium chloride and 0.1% Tween 20] for 1 h at 20°C and then incubated with the primary antibody overnight at 4°C. Incubation with a horseradish peroxidase-conjugated antimouse or rabbit secondary antibody was then carried out at 20°C for 4 h. Antibody-bound proteins were detected by the enhanced chemiluminescence western blotting analysis system (PerkinElmer Life and Analytical Sciences, Boston, MA).
Plasmids
The pSVV-259-luc (Survivin) construct was kindly provided by Dr M.Zhou (Emory University, Atlanta, GA). The Sp1 and Sp3 promoter constructs (pSp1-FOR4-luc and pSp3-FOR5-luc) were given to us by Drs Carlos Cuidad and Veronique Noe (University of Barcelona). The c-Met construct was provided by Dr M.Ladanyi (Memorial Sloan-Kettering Cancer Center).
Luciferase assay
SEG-1 and BIC-1 cells were plated in 12-well plates at 1 × 105 cells per well in DMEM/Ham's F-12 media supplemented with 2.5% charcoal-stripped FBS. After growth for 16–20 h, various amounts of reporter gene constructs (i.e. 0.4 μg pGL3/pGL2-Luc; 0.04 μg β-gal and 0.4 μg pSp1For4-Luc, pSp3For5-Luc, pVEGF-Luc, pc-Met-Luc and pSurvivin-Luc) were transfected by LipofectAMINE 2000 reagent (Invitrogen) according to the manufacturer's instructions. After 5 h, the media was replaced with complete medium containing either vehicle (dimethyl sulfoxide) or the indicated ligands for 20–22 h. Cells were then lysed with 100 μl of 1X reported lysis buffer (Promega) and 30 μl cell extract was used for luciferase and β-gal assays. Lumicount was used to quantitate luciferase and β-gal activities, and the luciferase activities were normalized to β-gal activity. Results are expressed as mean ± SD of at least three independent determinations for each treatment group.
Small inhibitory RNA studies
Small inhibitory RNAs for Sp1, Sp3 and Sp4 and non-specific oligonucleotide (iScr) were purchased from Dharmacon RNA Technologies (Lafayette, CO). The siRNA complexes used in this study are indicated as follows:
Lamin: 5′-CUG GAC UUC CAG AAG AAC ATT
Sp1: SMARTpool L-026959-00-0005, Human Sp1, NM_138473ss
Sp3: 5′-GCG GCA GGU GGA GCC UUC ACU TT
Sp4: 5′-GCA GUG ACA CAU UAG UGA GCT T
The two esophageal cancer cell lines, SEG-1 and BIC-1, were seeded (1 × 105 cells per well) in 12-well plates in DMEM:Ham's F-12 medium supplemented with 2.5% charcoal-stripped FBS without antibiotic and left to attach for 1 day. The triple Sp siRNA knockdown (iSp1, iSp3 and iSp4 complex) was performed using Lipofectamine 2000 transfection reagent as per the manufacturer's instructions both for protein knockdown and effects on Met promoter activity by transfecting the cells with either pGL3 basic or pc-Met promoter constructs. The knockdown studies use 20 nmol Sp1 and 100 nmol of Sp3 and Sp4 small inhibitory RNAs essentially as described (29).
Annexin V staining
Vybrant apoptosis assay kit was purchased from Molecular Probes (Eugene, OR). BIC-1 and SEG-1 cells were seeded on Lab-Tek four chambered cover glass and allowed to attach overnight. After treatment with the desired compounds, cells were washed with cold phosphate-buffered saline (PBS) twice and incubated with Annexin V conjugate and propidium iodide for 15 min according to the manufacturer's instructions. The cells were then washed with the Annexin-binding buffer twice and detected for fluorescence with Stallion digital imaging confocal microscope.
Mouse xenograft study
Male athymic nude mice (NCI-nu) were purchased from the Animal Production Area of the National Cancer Institute, Frederick Cancer Research and Development Center. The mice were housed and maintained under specific pathogen-free conditions in facilities approved by the American Association for Accreditation of Laboratory Animal Care and in accordance with current regulations and standards of the USA Department of Agriculture, USA Department of Health and Human Services and National Institutes of Health. The mice were used in accordance with institutional guidelines when they were ages 8–12 weeks of age. SEG-1 cells (1 × 106) were implanted with Matrigel (BD Biosciences, San Jose, CA) subcutaneously into the flank of each mouse. Ten days after cell inoculation when palpable tumors are observed, mice were randomly assigned to receive one of the following treatments: (i) thrice weekly oral administrations of vehicle i.e. corn oil (control group) and (ii) thrice weekly oral administrations of TA (20 mg/kg/day of body wt) in corn oil. The mice (six per treatment group) were weighed, and tumor areas were measured throughout the study. Treatments were continued for 4 weeks, and the mice were killed by CO2 asphyxiation, weighed and subjected to necropsy. The volume and weights of xenograft tumors were recorded. Selected tissues were further examined by routine hematoxylin and eosin staining and immunohistochemical analysis.
Human tissue and immunohistochemistry
Patients with a diagnosis of esophageal cancer treated surgically at MD Anderson Cancer Center in Orlando were identified and pathological records were reviewed. Paraffin-embedded tissue blocks from random patients (without preoperative radiation) were then extracted and cut to give representative sections of tumor and surrounding normal tissue. Tissue sections both from mouse xenograft tumors and human esophageal tumors, mounted on poly-L-lysine-coated slide, were deparaffinized by standard methods. Endogenous peroxidase was blocked by the use of 3% hydrogen peroxide in PBS for 10 min. Antigen retrieval was done for 5 min in 10 mmol/l sodium citrate buffer (pH 6) heated at 95°C in a steamer followed by cooling for 15 min. The slides were washed with PBS and incubated for 42 min at room temperature with a protein blocking solution (VECTASTAIN ABC kit, Vector Laboratories, Burlingame, CA). Excess blocking solution was drained, and the samples were incubated overnight at 4°C with one of the following: a 1:1000 dilution of Sp1 antibody or a 1:50 dilution of Met antibody. Sections were then incubated with biotinylated secondary antibody followed by streptavidin (VECTASTAIN ABC kit). The color was developed by exposing the peroxidase to diaminobenzidine reagent (Vector Laboratories), which forms a brown reaction product. The sections were then counterstained with Methyl green (Sigma). The brown staining identified Sp1 and Met expressions.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining was carried out using DeadEnd Colorimetric TUNEL System (Promega). Paraffin-embedded sections (4–6 μmol/l thick) were processed as per the manufacturer's protocol. Briefly, sections were deparaffinized in xylene and then treated with a graded series of alcohol [100, 95, 85, 70 and 50% ethanol (vol/vol) in double-distilled water] and rehydrated in PBS (pH 7.4). Tissues were then treated with proteinase K solution for permeabilization and then refixed with 4% paraformaldehyde solution. Slides were then treated with recombinant terminal deoxynucleotidyl transferase reaction mix and incubated at 37°C for 1 h. Reaction was terminated by immersing the slides in 2× SSC solutions for 15 min at room temperature. After blocking the endogenous peroxidases activity (by 0.3% hydrogen peroxide), slides were washed with PBS and then incubated with streptavidin horseradish peroxidase solution for 30 min at room temperature. After washing, slides were incubated with 3,3′-diaminobenzidine (substrate) solution until a light brown background appears (10 min) and then rinsed several times in deionized water. After mounting, slides were analyzed by light microscope.
Results
TA inhibits proliferation and induces apoptosis in EC cells
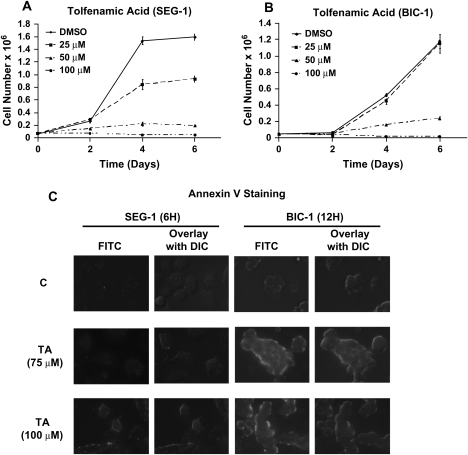
Figure 1A and B summarize the effects of 25, 50 and 100 μM TA on proliferation of SEG-1 and BIC-1 esophageal cancer cells over a 6 day period. Significant growth inhibition was observed at 25 and 50 μM concentrations in SEG-1 and BIC-1 cell lines and half-maximal growth inhibitory concentration values for this response after treatment for 6 days was 36 and 48 μM, respectively. These results are comparable with previous studies on the effects of TA on pancreatic cancer cell survival (32). Figure 1C summarizes the effects of 75 and 100 μM TA on the Annexin V staining percentage distribution of SEG-1 and BIC-1 cells. Annexin V staining, which is an early measure of proapoptotic activity, was induced in both cell lines demonstrating that TA not only inhibits proliferation of EC cells but also induces apoptosis in BIC-1 and SEG-1 cells.
Fig. 1.
TA inhibits cell proliferation and modulates esophageal cancer cell cycle progression. Inhibition of SEG-1 (A) and BIC-1 (B) cell proliferation by TA. Cells were treated with dimethyl sulfoxide (DMSO) or different concentrations of TA for up to 6 days and cells were counted as described in the Materials and Methods. Results are expressed as means ± SEs for three separate determinations for each treatment group. Growth inhibition half-maximal growth inhibitory concentration values in SEG-1 and BIC-1 cells were 36 and 48 μM, respectively. (C) Effects of TA on Annexin V staining in SEG-1 and BIC-1 cells. Cells were treated with dimethyl sulfoxide or TA for 12 h and the Annexin V staining was determined as described in the Materials and Methods.
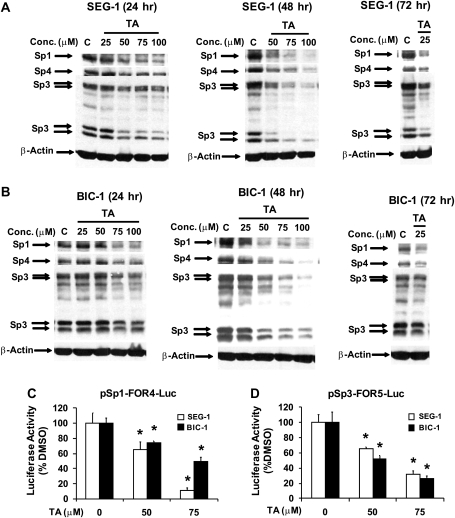
TA represses Sp1, Sp3 and Sp4 transcription factors in EC cells
We also examined the concentration- and time-dependent effects of TA on Sp1, Sp3 and Sp4 protein levels in esophageal cancer cells. Figure 2A shows that TA decreased expression of Sp1, Sp3 and Sp4 proteins in SEG-1 cells. At the earliest time point, 50 μM TA decreased Sp1, Sp4 and the lower molecular weight form of Sp3 and, after 72 h, decreased Sp levels were observed at concentrations at low as 25 μM. In BIC-1 cells, treatment with 75 and 100 μM TA (Figure 2B) decreased Sp1, Sp3 and Sp4 proteins and, after 72 h, 25 μM decreased Sp1 and Sp4 but not Sp3 protein expression, indicating that BIC-1 cells were less sensitive than SEG-1 cells to the effects of TA on cell proliferation and repression of Sps. The effects of TA on transcription of Sp1 and Sp3 were investigated in SEG-1 and BIC-1 cells transfected with the Sp1-FOR4-luc (contains the −751 to −20 region of the Sp1 gene promoter) and pSp3-FOR5-luc (contains the −417 to −38 region of the Sp3 promoter) constructs. TA decreased luciferase activity in both cell lines (Figure 2C and D), demonstrating that TA decreases activity of both the Sp1 and Sp3 promoters and represses transcription of both genes. The Sp4 promoter is not yet available.
Fig. 2.
TA modulates Sp expression in esophageal cancer cells. Effects of TA on Sp1, Sp3 and Sp4 protein expression in SEG-1 (A) and BIC-1 (B) cells. Esophageal cancer cells were treated with dimethyl sulfoxide (DMSO) or different concentrations of TA for 24, 48 or 72 h and whole-cell lysates were analyzed by western blots as described in the Materials and Methods. Similar results were observed in duplicate experiments. TA decreases transactivation in esophageal cancer cells transfected with Sp1 (C) and Sp3 (D) promoter constructs. SEG-1 or BIC-1 cells were transfected with Sp1-FOR4-luc or Sp3-FOR5-luc treated with dimethyl sulfoxide or different concentrations of TA and luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SEs for three replicate determinations for each treatment group and significantly (*P < 0.05) decreased luciferase activity after treatment with TA is indicated.
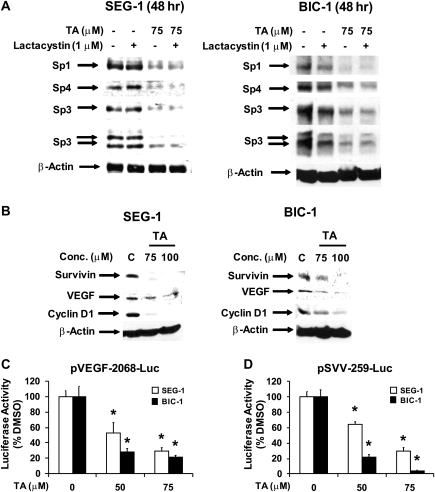
Previous studies in pancreatic cancer cells indicate that TA induced proteasome-dependent degradation of Sp1, Sp3 and Sp4 proteins (30,32) and, therefore, we investigated the effects of the proteasome inhibitor lactacystin on Sp levels in esophageal cancer cells treated with 75 μM TA. Treatment of SEG-1 and BIC-1 cells with TA decreased Sp1, Sp3 and Sp4 protein expression. Lactacystin alone had minimal effect on these proteins (some decrease in Sp1 in BIC-1 cells) and, in cells cotreated with TA plus lactacystin, the proteasome inhibitor did not affect the TA-dependent decrease of Sps (Figure 3A and B). Thus, TA represses Sp expression and promoter activity, suggesting a transcriptional mechanism for this response in esophageal cancer cells.
Fig. 3.
Role of proteasomes in TA-dependent repression of Sps and effects of TA on Sp-dependent genes. Effect of lactacystin in SEG-1 (A) and BIC-1 (B) cells treated with TA. Cells were treated with dimethyl sulfoxide (DMSO), lactacystin, TA or TA plus lactacystin for 24 h and whole-cell lysates were analyzed by western blots as described in the Materials and Methods. (C) TA decreases expression of Sp-dependent proteins. SEG-1 and BIC-1 cells were treated with dimethyl sulfoxide or TA for 24 h and whole-cell lysates were analyzed by western blots as described in the Materials and Methods. (D) TA decreases transactivation. SEG-1 and BIC-1 were transfected with pVEGF131-2068-luc or pSVV-259-luc and treated with dimethyl sulfoxide or TA, and luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SEs for three replicate determinations for each treatment group and a significant (*P < 0.05) decrease in activity is indicated.
Compounds such as TA, curcumin and betulinic acid that decrease Sps also downregulate expression of several Sp-dependent genes and proteins and these include survivin, VEGF and cyclin D1 (29–32). Results in Figure 3C show that treatment of SEG-1 and BIC-1 cells with TA for 24 h decreased expression of VEGF, cyclin D1 and survivin proteins and the latter response was consistent with the observed induction of apoptosis (Figure 1C). TA also decreased luciferase activity in SEG-1 and BIC-1 cells transfected with pVEGF131-luc and pSVV-269-luc which contain the −131 to +50 and −269 to +49 regions of the VEGF and survivin promoters, respectively (Figure 3D). Thus, TA-dependent downregulation of Sp expression is accompanied by a parallel decrease in Sp-dependent genes as reported previously for other compounds that exhibit similar activities (29–32).
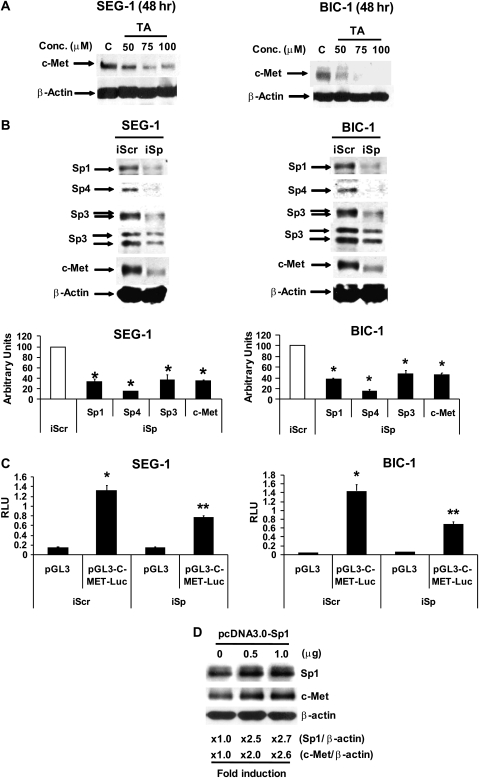
TA represses expression of Sp-dependent genes including c-Met
The hepatocyte growth factor receptor c-Met is overexpressed in esophageal tumors compared with non-tumor tissue and there is evidence that this receptor plays an important role in esophageal cancer growth and angiogenesis (33–35). BIC-1 and SEG-1 cells overexpress c-Met (33), and Figure 4A shows that TA decreased expression of c-Met protein in both cell lines. The c-Met promoter contains GC-rich Sp-binding sites (36,37), and therefore we further investigated the role of TA-dependent repression of Sps as a mechanism for downregulation of c-Met. Esophageal cancer cells were transfected with iSp, a cocktail containing small inhibitory RNAs for Sp1, Sp3 and Sp4 [described previously (28,29)]. In SEG-1 cells transfected with iSp, there was a decrease in expression of Sp1, Sp3 and Sp4 proteins and similar results were observed in BIC-1 cells (Figure 4B). We also showed that in EC cells transfected with iSp, there was a significant decrease in c-Met protein expression (Figure 4B), thus confirming that the TA-dependent decrease in c-Met expression (Figure 4A) is due to repression of Sps by this compound (Figure 2). In addition, Figure 4C illustrates that luciferase activity in SEG-1 and BIC-1 cells transfected with a c-Met promoter construct [p-c-Met-luc(−703/+60)] was significantly decreased after transfection with iSp, confirming that expression of the c-Met promoter is also Sp dependent. The role of Sp1 in regulating c-Met expression was confirmed in SEG-1 cells transfected with Sp1 expression plasmid for 36 h which not only increased levels of Sp1 protein but also c-Met protein (Figure 4D).
Fig. 4.
Characterization of c-Met as an Sp-responsive gene in esophageal cancer cells. (A) TA decreased c-Met expression. SEG-1 and BIC-1 cells were treated with dimethyl sulfoxide or TA for 48 h and whole-cell lysates were analyzed by western blots as described in the Materials and Methods. (B) Sp knockdown decreases c-Met. SEG-1 and BIC-1 cells were transfected with iSp containing siRNA for Sp1, Sp3 and Sp4, and whole-cell lysates were analyzed by western blots as described in the Materials and Methods. (C) Sp knockdown decreases c-Met promoter activity. Cells were transfected with iSp or iCtl (non-specific) and the p-c-Met promoter construct, and luciferase activity was determined as described in the Materials and Methods. Results are expressed as means ± SEs for three replicate determinations for each treatment group and significantly (*P < 0.05) decreased activity in cells transfected with iSp is indicated. (D) Sp overexpression induced c-Met. SEG-1 cells were transfected with Sp1 expression plasmid (in pcDNA3.0) and, after 36 h, whole-cell lysates were analyzed by western blots. Expression of Sp1 and c-Met relative to β-actin (loading control) in cells transfected with the empty vector (1.0) or Sp1 expression plasmid is indicated. The results were similar in duplicate experiments.
In vivo carcinogenicity studies and immunohistochemistry of human esophageal tumors
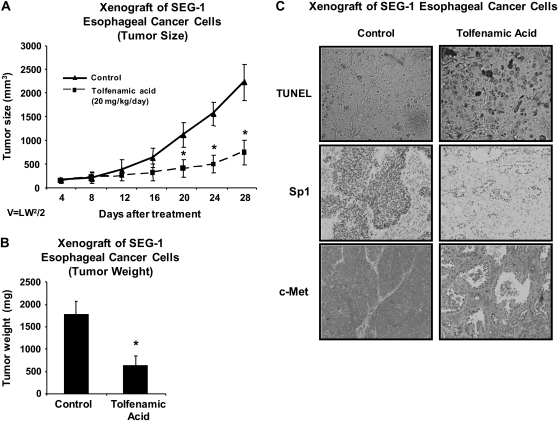
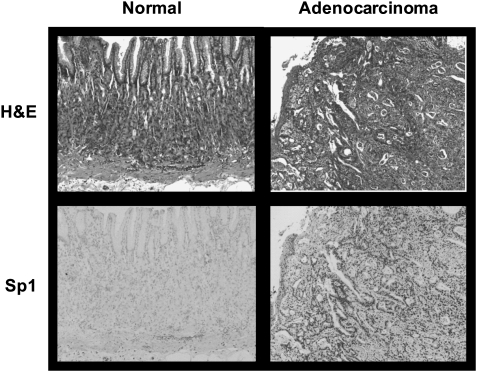
We also examined the effects of daily administration of TA (20 mg/kg/day) on tumor growth in athymic nude mice bearing SEC-1 cells as xenografts. The results show that this dose of TA significantly inhibited tumor growth and tumor weight (Figure 5A and B). In addition, we also used TUNEL staining of tumors from control and TA-treated animals to show a marked increase in apoptosis in tumors from TA-treated mice (Figure 5C) and this correlated with downregulation of Sp and c-Met proteins in tumors from treated versus control animals. These results demonstrate that TA is a highly effective antitumor drug that inhibits growth of esophageal tumors in a mouse xenograft model and the effective dose (20 mg/kg/day) is significantly lower than reported previously for inhibition of pancreatic tumor growth in an orthotopic model (25 mg/kg/day) (32). Serial sections from biopsies obtained at the gastroesophageal junction of two patients, one with normal histology (proximal and distal margins free of neoplasia, left column) and the other with esophageal tumor (poorly differentiated adenocarcinoma of the gastroesophageal junction, right column), were examined (Figure 6). Upper panels were stained with hematoxylin and eosin and lower panels were immunoperoxidase stained with rabbit polyclonal antibody to Sp1. Note the intense nuclear Sp1 staining in the adenocarcinoma in contrast to the section from the biopsy identified as free of neoplasia. Width of each field is 940 μM. These results indicate higher expression of Sp1 in tumor versus non-tumor tissue and ongoing studies are using similar procedures for investigating Sp1, Sp3 and Sp4 expression in esophageal tumors (and non-tumor tissue) from a large number of patients.
Fig. 5.
Antitumorigenic activity of TA. Inhibition of tumor volume (A) and weight (B) after treatment with TA. Athymic nude mice bearing SEG-1 cells as xenografts were treated with corn oil (control) or TA (20 mg/kg/day) in corn oil by oral gavage and tumor volumes were determined every 4 days as described in the Materials and Methods. Tumor weights were obtained after killing and a significant (*P < 0.05) decrease in tumor volumes or weights after treatment with TA is indicated. (C) TUNEL assay and immunostaining for Sp1 and c-Met proteins. Tumor sections from control and TA-treated mice were stained for apoptosis using the TUNEL as described in the Materials and Methods. Slides of tumor sections from control and TA-treated mice were stained for Sp1 and c-Met proteins as described in the Materials and Methods.
Fig. 6.
Hematoxylin and eosin (H&E) and immunohistochemistry of esophageal tumor and non-tumor tissue. Hematoxylin and eosin and immunohistochemical staining of esophageal tissue from a cancer patient and cancer-free patient were carried out as described in the Materials and Methods.
Discussion
Sp1 was the first transcription factor identified (38) and is one of 25 Sp/KLF genes that have been characterized (39). Sp1 and other Sp family members are critical for early embryonic development as evidenced in gene ablation studies in mice and Sp/KLF transcription factors play a key role in regulation of genes required for cell growth, differentiation and angiogenesis (39,40). However, there is evidence that tissue and organ levels of Sp1 and other Sps decrease with age (41,42), and ongoing studies in this laboratory show that only trace to non-detectable levels of Sp1, Sp3 and Sp4 proteins are expressed in normal mouse tissue (29,32) (data not shown). In contrast, Sps are overexpressed in tumors derived from multiple sites (16–22), and recent studies have shown that Sp1, Sp3 and Sp4 are highly expressed in several breast cancer cell lines but only minimal levels are expressed in non-transformed mammary cells (23). The overexpression of Sps in tumors, coupled with their role in regulation of genes associated with cancer and tumor cell proliferation, survival and angiogenesis, suggests that these transcription factors are potential mechanism-based drug targets for cancer chemotherapy (43).
TA, betulinic acid and curcumin are anticancer drugs that inhibit proliferation of pancreatic, prostate and bladder cancer cells in culture and tumor growth in murine xenograft or orthotopic models, respectively (29–32). These compounds also decrease expression of Sp1, Sp3 and Sp4 proteins and several Sp-dependent genes and proteins such as VEGF, VEGFR1, survivin, cyclin D1 and bcl-2, and these responses contribute to their anticancer activity. Moreover, in ongoing studies, several other genes such as epidermal growth factor receptor and cyclooxygenase are also Sp dependent, and their role in mediating the activity of TA, curcumin and betulinic acid in cancer cells is currently being investigated. In this study, we used TA as a model to investigate the potential applications of targeting Sp transcription factors for treatment of esophageal cancer. TA or Clotam™ is a relatively non-toxic non-steroidal anti-inflammatory drug that has been used for treatment of migraine headaches and rheumatoid arthritis in humans and as a veterinary drug product for treating pain and stress responses in animals (44–46). TA decreased proliferation of SEG-1 and BIC-1 cells (Figure 1A and B) and also induced Annexin V staining (Figure 1C and D) which is an early marker of apoptosis. The half-maximal growth inhibitory concentration values for growth inhibition were 36 and 48 μM in SEG-1 and BIC-1 cells, respectively, and these were lower than observed in previous studies in pancreatic cancer cells (32).
Figure 2 summarizes the time- and concentration-dependent effects of TA on Sp1, Sp3 and Sp4 proteins and the results demonstrate that the three transcription factors were highly expressed in both cell lines and that concentrations as low as 25–50 μM TA decreased Sps after treatment for up to 72 h. This data, coupled with decreased luciferase activity in cells treated with TA and transfected with Sp1 or Sp3 promoter constructs (Figure 2C and D), demonstrate that TA repressed expression of Sp transcription factors in esophageal cancer cells. In contrast to studies with TA in pancreatic cancer cells (32), TA-dependent downregulation of Sp1, Sp3 and Sp4 in SEG-1 and BIC-1 cells was not affected by the proteasome inhibitor lactacystin. Studies in this laboratory also show that methyl 2-cyano-3,11-dioxo-18β-olean-1,12-dien-30-oate, a derivative of glycyrrhetinic acid (a phytochemical in licorice), decreased Sps through downregulation of microRNA-27a and upregulation of ZBTB10, a Sp repressor (23,47). However, TA did not affect microRNA-27a or ZBTB10 expression in esophageal cancer cells (data not shown) and current studies are investigating other mechanisms for mediating TA-dependent downregulation of Sps.
The effects of TA on Sp-dependent genes such as survivin, VEGF and cyclin D1 (29,31,32) were also determined in SEG-1 and BIC-1 cells (Figure 3C) and, not surprisingly, expression of all three proteins was decreased. Sp transcription factors regulate expression of several genes required for cancer cell growth, survival and angiogenesis and these include VEGF, survivin and cyclin D1, which are also decreased by TA, betulinic acid and curcumin in pancreatic, prostate and bladder cancer cells, respectively (29–32). Previous studies in prostate cancer cells and in various murine kidney cell lines showed that c-Met expression was also regulated by Sp1 or Sp1/Sp3 (36,37), suggesting that this gene may also be regulated by Sps in esophageal cancer cells. Both BIC-1 and SEG-1 cells overexpress c-Met (33,47) and a small molecular weight inhibitor specific for c-Met kinase differentially affected various responses in esophageal cancer cell lines (BIC-1, SEG-1 and Flo-1) but decreased viability of all esophageal cancer cell lines (48). Results illustrated in Figure 4A indicate that TA decreased c-Met protein expression in BIC-1 and SEG-1 cells. Since Sp1, Sp3 and Sp4 are highly expressed in BIC-1 and SEG-1 cells, we hypothesized that the effects of TA on c-Met expression were due to repression of Sps by this compound (Figure 2). This was further investigated in esophageal cancer cells by RNA interference which showed that simultaneous knockdown of Sp1, Sp3 and Sp4 was accompanied by decreased expression of c-Met protein (Figure 4B), confirming that c-Met expression in BIC-1 and SEG-1 cells is Sp dependent. Moreover, overexpression of Sp1 in SEG-1 cells also induced c-Met protein expression (Figure 4D). These results suggest that drugs such as TA that repress Sp transcription factors in esophageal cancer represent an alternate approach for targeting c-Met. We have also confirmed that other compounds such as betulinic acid that repress Sps in cancer cells (31) are active in esophageal cancer cells and also decrease c-Met expression (data not shown).
The in vivo anticarcinogenic activity of TA was investigated in athymic nude mice bearing SEG-1 cells as xenografts. At a dose of 20 mg/kg/day, TA significantly inhibited tumor growth and weight and increased TUNEL staining (apoptosis) in tumor sections from treated versus control animals and this was also accompanied by decreased Sp1 and c-Met staining in tumors from treated mice (Figure 5). Clinical studies with TA for chronic treatment of rheumatoid arthritis use doses of TA as high as 10 mg/kg/day which is only 2-fold lower than required for inhibition of esophageal tumor growth in the xenograft mouse model (Figure 5). These results demonstrate that TA is a highly effective anticancer agent for both in vitro and in vivo models of esophageal cancer and this complements results of previous studies in pancreatic cancer cells and tumors (32). The mechanism of action of TA as a tumor growth inhibitor is due, in part, to repression of Sp transcription factors and Sp-dependent genes, many of which are overexpressed in tumors and cancer cell lines (43). Moreover, in esophageal cancer cells, we demonstrate for the first time that TA also decreases c-Met expression, and RNA interference studies confirm that c-Met is also a Sp-dependent gene. This is particularly relevant for the potential efficacy of TA for treatment of esophageal cancer since c-Met is overexpressed in esophageal cancer cells and tumors and is a target for esophageal cancer chemotherapy (33–35). We also examined Sp1 expression in esophageal tumor and non-tumor tissue and the results (Figure 6) show that Sp1 is highly overexpressed in tumor tissue. Current studies are focused on the relative expression of Sp1, Sp3 and Sp4 in esophageal tumors and their prognostic significance and also on the development of TA and related compounds as a novel class of drugs for clinical treatment of esophageal cancer.
Funding
National Institutes of Health (CA108178); Texas A&M AgriLife.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- c-Met
hepatocyte growth factor receptor
- DMEM
Dulbecco's modified Eagle's medium
- EC
esophageal cancer
- FBS
fetal bovine serum
- β-gal
β-galactosidase
- PBS
phosphate-buffered saline
- siRNA
small inhibitory RNA
- Sp
specificity protein
- TA
tolfenamic acid
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- VEGF
vascular endothelial growth factor
References
- 1.Crew KD, et al. Epidemiology of upper gastrointestinal malignancies. Semin. Oncol. 2004;31:450–464. doi: 10.1053/j.seminoncol.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 2.Devesa SS, et al. The rising incidence of gastric cardia cancer. J. Natl. Cancer Inst. 1999;91:747–749. doi: 10.1093/jnci/91.9.747. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, et al. Cancer statistics, 2008. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 4.Talback M, et al. Cancer survival in Sweden 1960-1998—developments across four decades. Acta Oncol. 2003;42:637–659. doi: 10.1080/02841860310013391. [DOI] [PubMed] [Google Scholar]
- 5.Blot WJ, et al. The changing epidemiology of esophageal cancer. Semin. Oncol. 1999;26:2–8. [PubMed] [Google Scholar]
- 6.Enzinger PC, et al. Esophageal cancer. N. Engl. J. Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 7.Pera M, et al. Epidemiology of esophageal adenocarcinoma. J. Surg. Oncol. 2005;92:151–159. doi: 10.1002/jso.20357. [DOI] [PubMed] [Google Scholar]
- 8.Holmes RS, et al. Epidemiology and pathogenesis of esophageal cancer. Semin. Radiat. Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Casson AG, et al. Epidemiology and molecular biology of Barrett esophagus. Semin. Thorac. Cardiovasc. Surg. 2005;17:284–291. doi: 10.1053/j.semtcvs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Flejou JF. Barrett's oesophagus: from metaplasia to dysplasia and cancer. Gut. 2005;54(suppl. 1):i6–i12. doi: 10.1136/gut.2004.041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinberg L, et al. Chemoradiation in the management of esophageal cancer. J. Clin. Oncol. 2007;25:4110–4117. doi: 10.1200/JCO.2007.12.0881. [DOI] [PubMed] [Google Scholar]
- 12.Ku GY, et al. Esophageal cancer: adjuvant therapy. Cancer J. 2007;13:162–167. doi: 10.1097/PPO.0b013e318074dbe7. [DOI] [PubMed] [Google Scholar]
- 13.Syrigos KN, et al. Targeted therapy for oesophageal cancer: an overview. Cancer Metastasis Rev. 2008;27:273–288. doi: 10.1007/s10555-008-9117-z. [DOI] [PubMed] [Google Scholar]
- 14.Ekman S, et al. Activation of growth factor receptors in esophageal cancer—implications for therapy. Oncologist. 2007;12:1165–1177. doi: 10.1634/theoncologist.12-10-1165. [DOI] [PubMed] [Google Scholar]
- 15.Shah MA, et al. Multicenter phase II study of irinotecan, cisplatin, and bevacizumab in patients with metastatic gastric or gastroesophageal junction adenocarcinoma. J. Clin. Oncol. 2006;24:5201–5206. doi: 10.1200/JCO.2006.08.0887. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin. Cancer Res. 2003;9:6371–6380. [PubMed] [Google Scholar]
- 17.Yao JC, et al. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin. Cancer Res. 2004;10:4109–4117. doi: 10.1158/1078-0432.CCR-03-0628. [DOI] [PubMed] [Google Scholar]
- 18.Shi Q, et al. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143–4154. [PubMed] [Google Scholar]
- 19.Zannetti A, et al. Coordinate up-regulation of Sp1 DNA-binding activity and urokinase receptor expression in breast carcinoma. Cancer Res. 2000;60:1546–1551. [PubMed] [Google Scholar]
- 20.Chiefari E, et al. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer. 2002;2:35. doi: 10.1186/1471-2407-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosoi Y, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int. J. Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- 22.Jiang NY, et al. Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 2008;17:1648–1652. doi: 10.1158/1055-9965.EPI-07-2791. [DOI] [PubMed] [Google Scholar]
- 23.Mertens-Talcott SU, et al. The oncogenic microRNA-27a targets genes that regulate specificity protein (Sp) transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 24.Lou Z, et al. Down-regulation of overexpressed Sp1 protein in human fibrosarcoma cell lines inhibits tumor formation. Cancer Res. 2005;65:1007–1017. [PubMed] [Google Scholar]
- 25.Abdelrahim M, et al. Cyclooxygenase-2 inhibitors decrease vascular endothelial growth factor expession in colon cancer cells by enhanced degradation of Sp1 and Sp4 proteins. Mol. Pharmacol. 2005;68:317–329. doi: 10.1124/mol.105.011825. [DOI] [PubMed] [Google Scholar]
- 26.Abdelrahim M, et al. Small inhibitory RNA duplexes for Sp1 mRNA block basal and estrogen-induced gene expression and cell cycle progression in MCF-7 breast cancer cells. J. Biol. Chem. 2002;277:28815–28822. doi: 10.1074/jbc.M203828200. [DOI] [PubMed] [Google Scholar]
- 27.Abdelrahim M, et al. Role of Sp proteins in regulation of vascular endothelial growth factor expression and proliferation of pancreatic cancer cells. Cancer Res. 2004;64:6740–6749. doi: 10.1158/0008-5472.CAN-04-0713. [DOI] [PubMed] [Google Scholar]
- 28.Higgins KJ, et al. Regulation of vascular endothelial growth factor receptor-2 expression in pancreatic cancer cells by Sp proteins. Biochem. Biophys. Res. Commun. 2006;345:292–301. doi: 10.1016/j.bbrc.2006.04.111. [DOI] [PubMed] [Google Scholar]
- 29.Chadalapaka G, et al. Curcumin decreases specificity protein expression in bladder cancer cells. Cancer Res. 2008;68:5345–5354. doi: 10.1158/0008-5472.CAN-07-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelrahim M, et al. Regulation of vascular endothelial growth factor receptor-1 (VEGFR1) expression by specificity proteins 1, 3 and 4 in pancreatic cancer cells. Cancer Res. 2007;67:3286–3294. doi: 10.1158/0008-5472.CAN-06-3831. [DOI] [PubMed] [Google Scholar]
- 31.Chintharlapalli S, et al. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Res. 2007;67:2816–2823. doi: 10.1158/0008-5472.CAN-06-3735. [DOI] [PubMed] [Google Scholar]
- 32.Abdelrahim M, et al. Tolfenamic acid and pancreatic cancer growth, angiogenesis, and Sp protein degradation. J. Natl. Cancer Inst. 2006;98:855–868. doi: 10.1093/jnci/djj232. [DOI] [PubMed] [Google Scholar]
- 33.Herrera LJ, et al. The HGF receptor c-Met is overexpressed in esophageal adenocarcinoma. Neoplasia. 2005;7:75–84. doi: 10.1593/neo.04367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuynman JB, et al. Met expression is an independent prognostic risk factor in patients with oesophageal adenocarcinoma. Br. J. Cancer. 2008;98:1102–1108. doi: 10.1038/sj.bjc.6604251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren Y, et al. Hepatocyte growth factor promotes cancer cell migration and angiogenic factors expression: a prognostic marker of human esophageal squamous cell carcinomas. Clin. Cancer Res. 2005;11:6190–6197. doi: 10.1158/1078-0432.CCR-04-2553. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Sp1 and Sp3 transcription factors synergistically regulate HGF receptor gene expression in kidney. Am. J. Physiol. Renal Physiol. 2003;284:F82–F94. doi: 10.1152/ajprenal.00200.2002. [DOI] [PubMed] [Google Scholar]
- 37.Verras M, et al. The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res. 2007;67:967–975. doi: 10.1158/0008-5472.CAN-06-3552. [DOI] [PubMed] [Google Scholar]
- 38.Dynan WS, et al. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 39.Bouwman P, et al. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 40.Black AR, et al. Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 41.Ammendola R, et al. Sp1 DNA binding efficiency is highly reduced in nuclear extracts from aged rat tissues. J. Biol. Chem. 1992;267:17944–17948. [PubMed] [Google Scholar]
- 42.Oh JE, et al. Downregulation of transcription factor, Sp1, during cellular senescence. Biochem. Biophys. Res. Commun. 2007;353:86–91. doi: 10.1016/j.bbrc.2006.11.118. [DOI] [PubMed] [Google Scholar]
- 43.Safe S, et al. Sp transcription factor family and its role in cancer. Eur. J. Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Hansen PE. Tolfenamic acid in acute and prophylactic treatment of migraine: a review. Pharmacol. Toxicol. 1994;75(suppl. 2):81–82. doi: 10.1111/j.1600-0773.1994.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 45.Rejholec V, et al. A comparative, double-blind study on tolfenamic acid in the treatment of rheumatoid arthritis. Scand. J. Rheumatol. Suppl. 1979:13–16. [PubMed] [Google Scholar]
- 46.Lascelles BD, et al. Nonsteroidal anti-inflammatory drugs in cats: a review. Vet. Anaesth. Analg. 2007;34:228–250. doi: 10.1111/j.1467-2995.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 47.Chintharlapalli S, et al. Oncogenic microRNA-27a is a target for anticancer agent methyl 2-cyano-3,11-dioxo-18β-olean-1,12-dien-30-oate in colon cancer cells. Int. J. Cancer. 2009 doi: 10.1002/ijc.24530. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson GA, et al. Inhibition of c-Met as a therapeutic strategy for esophageal adenocarcinoma. Neoplasia. 2006;8:949–955. doi: 10.1593/neo.06499. [DOI] [PMC free article] [PubMed] [Google Scholar]