Abstract
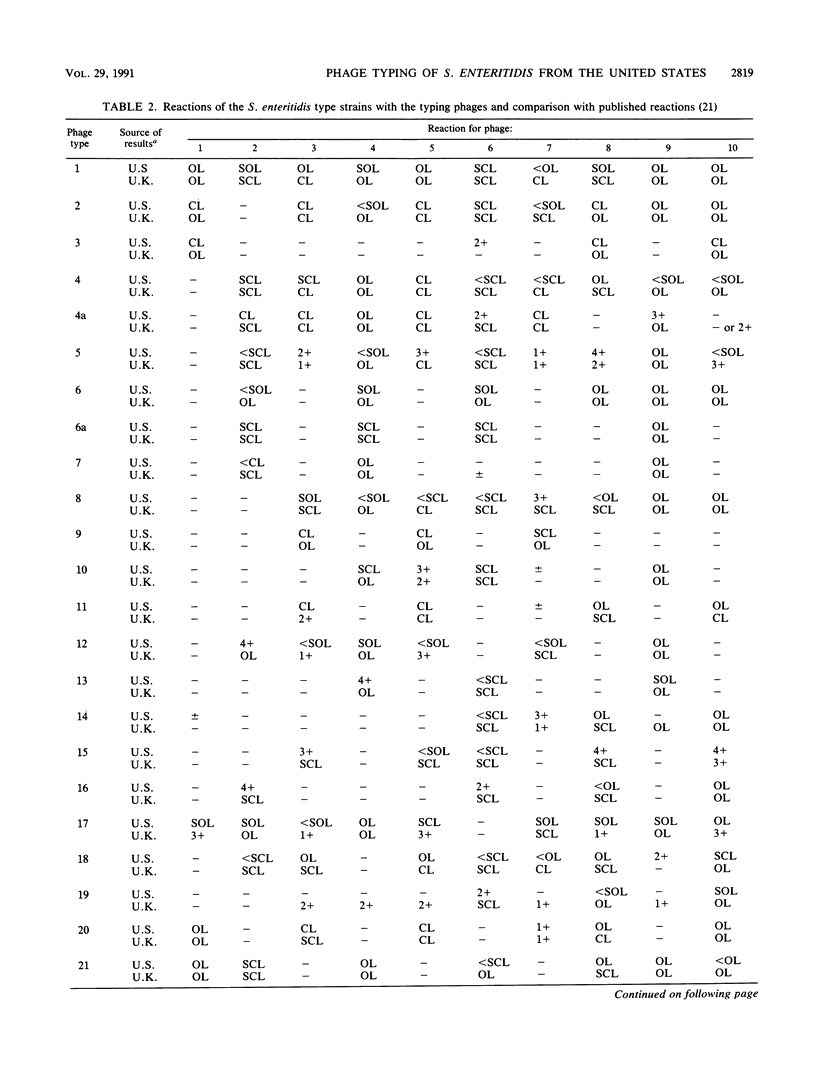
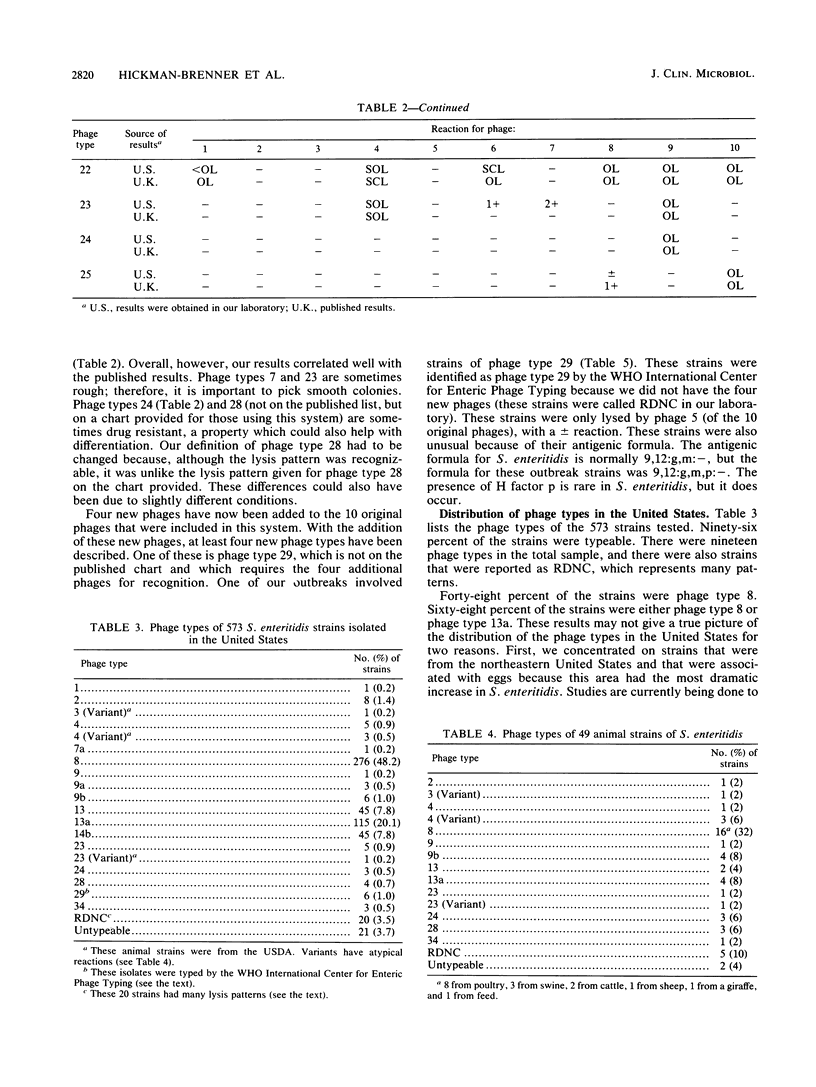
The number of reported isolates of Salmonella enteritidis has increased dramatically in the last 10 years. For many years phage typing has been a useful epidemiologic tool for studying outbreaks of S. typhi and S. typhimurium. In 1987, Ward et al. (L. R. Ward, J. De Sa, and B. Rowe, Epidemiol. Infect. 99:291-294, 1987) described a phage typing scheme for S. enteritidis. This system differentiated 27 phage types by use of 10 typing phages. With these phages, we typed 573 strains of S. enteritidis from humans (42 outbreaks), animals, food, and the environment. Ninety-six percent of the strains were typeable. The most common phage types were 8 (48.2%), 13a (20.1%), 13 (7.8%), and 14b (7.8%). Most of the strains were specifically collected from egg-related outbreaks in the northeastern United States in 1988 and 1989, probably accounting for the distribution of the four most common types in this sample. This system was particularly useful for differentiating a group of animal strains that had a number of diverse phage types. For 49 animal strains typed, 16 different patterns were obtained. Phage type 8 represented 32% of these strains, but no other phage type represented more than 8% of these strains. One-half of the 16 animal strains that were phage type 8 were from poultry. This phage typing system will be useful for comparing phage types found in the United States with those types encountered worldwide and for determining whether virulent strains of phage type 4 are entering the United States. Additional phage typing systems as well as molecular techniques are being studied to determine whether they can differentiate strains of phage types 8 and 13a.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANDERSON E. S., WILLIAMS R. E. Bacteriophage typing of enteric pathogens and staphylococci and its use in epidemiology. J Clin Pathol. 1956 May;9(2):94–127. doi: 10.1136/jcp.9.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altwegg M., Hickman-Brenner F. W., Farmer J. J., 3rd Ribosomal RNA gene restriction patterns provide increased sensitivity for typing Salmonella typhi strains. J Infect Dis. 1989 Jul;160(1):145–149. doi: 10.1093/infdis/160.1.145. [DOI] [PubMed] [Google Scholar]
- Beltran P., Musser J. M., Helmuth R., Farmer J. J., 3rd, Frerichs W. M., Wachsmuth I. K., Ferris K., McWhorter A. C., Wells J. G., Cravioto A. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chart H., Row B., Threlfall E. J., Ward L. R. Conversion of Salmonella enteritidis phage type 4 to phage type 7 involves loss of lipopolysaccharide with concomitant loss of virulence. FEMS Microbiol Lett. 1989 Jul 1;51(1):37–40. doi: 10.1016/0378-1097(89)90073-6. [DOI] [PubMed] [Google Scholar]
- Farmer J. J., 3rd, Hickman F. W., Sikes J. V. Automation of Salmonella typhi phage typing. Lancet. 1975 Oct 25;2(7939):787–790. doi: 10.1016/s0140-6736(75)80004-3. [DOI] [PubMed] [Google Scholar]
- Frost J. A., Ward L. R., Rowe B. Acquisition of a drug resistance plasmid converts Salmonella enteritidis phage type 4 to phage type 24. Epidemiol Infect. 1989 Oct;103(2):243–248. doi: 10.1017/s0950268800030594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman M. Phage typing system for Salmonella enteritidis. Appl Environ Microbiol. 1976 Jul;32(1):190–191. doi: 10.1128/aem.32.1.190-191.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman M. Single phage-typing set for differentiating salmonellae. J Clin Microbiol. 1977 Mar;5(3):302–314. doi: 10.1128/jcm.5.3.302-314.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman F. W., Farmer J. J., 3rd Salmonella typhi: identification, antibiograms, serology, and bacteriophage typing. Am J Med Technol. 1978 Dec;44(12):1149–1159. [PubMed] [Google Scholar]
- Holmberg S. D., Wachsmuth I. K., Hickman-Brenner F. W., Cohen M. L. Comparison of plasmid profile analysis, phage typing, and antimicrobial susceptibility testing in characterizing Salmonella typhimurium isolates from outbreaks. J Clin Microbiol. 1984 Feb;19(2):100–104. doi: 10.1128/jcm.19.2.100-104.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- László V. G., Csórián E. S., Pászti J. Phage types and epidemiological significance of Salmonella enteritidis strains in Hungary between 1976 and 1983. Acta Microbiol Hung. 1985;32(4):321–340. [PubMed] [Google Scholar]
- Reeves M. W., Evins G. M., Heiba A. A., Plikaytis B. D., Farmer J. J., 3rd Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989 Feb;27(2):313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue D. C., Tauxe R. V., Rowe B. International increase in Salmonella enteritidis: a new pandemic? Epidemiol Infect. 1990 Aug;105(1):21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L. R., de Sa J. D., Rowe B. A phage-typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987 Oct;99(2):291–294. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]


