Abstract
Next-generation sequencers have sufficient power to analyze simultaneously DNAs from many different specimens, a practice known as multiplexing. Such schemes rely on the ability to associate each sequence read with the specimen from which it was derived. The current practice of appending molecular barcodes prior to pooling is practical for parallel analysis of up to many dozen samples. Here, we report a strategy that permits simultaneous analysis of tens of thousands of specimens. Our approach relies on the use of combinatorial pooling strategies in which pools rather than individual specimens are assigned barcodes. Thus, the identity of each specimen is encoded within the pooling pattern rather than by its association with a particular sequence tag. Decoding the pattern allows the sequence of an original specimen to be inferred with high confidence. We verified the ability of our encoding and decoding strategies to accurately report the sequence of individual samples within a large number of mixed specimens in two ways. First, we simulated data both from a clone library and from a human population in which a sequence variant associated with cystic fibrosis was present. Second, we actually pooled, sequenced, and decoded identities within two sets of 40,000 bacterial clones comprising approximately 20,000 different artificial microRNAs targeting Arabidopsis or human genes. We achieved greater than 97% accuracy in these trials. The strategies reported here can be applied to a wide variety of biological problems, including the determination of genotypic variation within large populations of individuals.
There are many biological applications wherein the sequence of a small region must be determined from many independent specimens and wherein the link between the specimen and the sequence must be preserved. Examples might include studies of variation within specific regions from a large population, looking for correlations between variants and phenotype in areas that might have been narrowed by prior linkage analysis, or even medical diagnostics. We were faced with the practical matter of determining clone identity and quality in a very large collection of arrayed clones derived from a pooled oligonucleotide mixture. Use of next-generation sequencing technologies (NGST) for interrogating sequence for any of these tasks is hampered if libraries must be constructed independently from each sample or if different barcodes must be applied to each specimen prior to sequencing, particularly if specimen numbers run into many thousands.
To circumvent such limitations, we have developed a novel multiplexing framework in which the identity of individual specimens is not encoded directly by an appended barcode sequence. Instead, specimen identities are encoded by positional information created by an optimized series of pooling schema. In this manner, more than 100,000 different samples can be analyzed using only a few hundred barcodes (e.g., 384).
Results
Conceptual framework for the strategy
Over the past several years, we have worked to produce collections of shRNAs and artificial microRNAs that correspond to all genes in the human, mouse, rat, and Arabidopsis genomes. Our approach relies on highly parallel oligonucleotide synthesis (Cleary et al. 2004) to generate mixtures of approximately 22,000–55,000 different species that are adapted and ligated into expression vectors. Previously, individual clones from this mixed library were picked, arrayed, and conventionally sequenced both to determine their identity and to verify their accuracy. We sought to reduce the cost of this step by moving from capillary sequencing platforms to NGST. The essential barrier was that while NGSTs generate very large numbers of sequence reads, there was no straightforward way to link reads back to the sample, in this case, the clone, from which it was derived. Simple barcoding strategies could address this issue (Craig et al. 2008; Cronn et al. 2008); however, each clone would have to be individually coded, meaning that the number of barcoding oligos and the number of PCRs to generate sequencing templates would scale linearly with the number of distinct clones that we sought to analyze.
As an alternative, we envisioned a multiplexing framework that relied on encoding the identity of the source of each sequence read within a pooling scheme rather than by directly barcoding individual samples. In such a scheme, the number of pools is much smaller than the number of specimens. Thus, the number of barcodes required and the number of sequencing templates are greatly reduced.
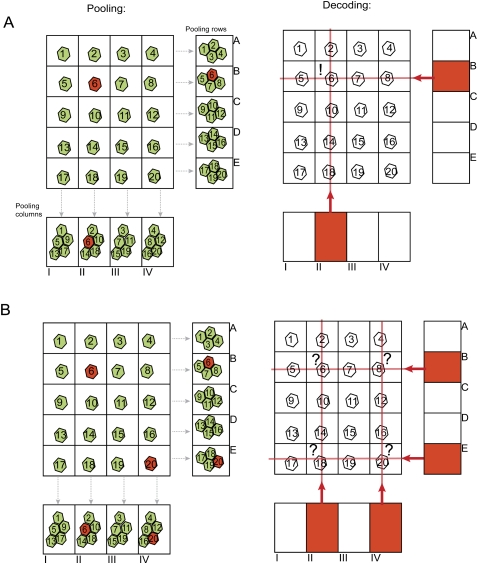
While we envisioned many simple pooling patterns, these proved insufficiently robust, and uncertainties in linking clones to their source wells mounted quickly as the numbers of specimens increased. For example, one simple approach viewed a clone collection as a simple matrix of rows and columns (Fig. 1). Pooling all the clones in each row and pooling all clones in each column yielded pools equivalent to  , where N is the number of clones (if the array was symmetrical). A specific sequence would appear coupled to a particular row identifier and also with a particular column identifier. This should point to a unique position in the clone array. The approach breaks down immediately if not every sequence in the library is unique. Of course, in our practical application and in most uses of such a strategy, sequences appear multiple times. Therefore, if such strategies were applied to multiplexed genotyping, individual alleles appearing many times within the overall population could not be correctly linked to their parent specimen. We therefore needed a pooling pattern that would reveal unique specimen identities even in the face of redundancy. We envisioned the possibility of integrating information from different pooling patterns to determine specimen identity. While each individual pooling pattern might yield multiple solutions to the link between sequence and specimen source, the combination of all pooling patterns would provide sufficient constraints that only a single, high-confidence solution would emerge for the vast majority of samples. Many elements of this approach were reminiscent of seeking the solution to a Sudoku puzzle, which led us to dub this strategy “DNA Sudoku.”
, where N is the number of clones (if the array was symmetrical). A specific sequence would appear coupled to a particular row identifier and also with a particular column identifier. This should point to a unique position in the clone array. The approach breaks down immediately if not every sequence in the library is unique. Of course, in our practical application and in most uses of such a strategy, sequences appear multiple times. Therefore, if such strategies were applied to multiplexed genotyping, individual alleles appearing many times within the overall population could not be correctly linked to their parent specimen. We therefore needed a pooling pattern that would reveal unique specimen identities even in the face of redundancy. We envisioned the possibility of integrating information from different pooling patterns to determine specimen identity. While each individual pooling pattern might yield multiple solutions to the link between sequence and specimen source, the combination of all pooling patterns would provide sufficient constraints that only a single, high-confidence solution would emerge for the vast majority of samples. Many elements of this approach were reminiscent of seeking the solution to a Sudoku puzzle, which led us to dub this strategy “DNA Sudoku.”
Figure 1.
The problem of nonunique genotypes in simple row/column strategies. (A,B) Pooling and arraying of 20 specimens. Letters indicate pools of rows, and Roman numerals indicate pools of columns. (A) The (red) mutant state is unique and appears only in specimen #6 (left). After sequencing the pools (right), this genotype is found in pool II and B, a pattern that can only be associated with specimen #6, demonstrating successful decoding. (B) The mutant state appears in #6 and #20. After decoding there is an ambiguity of whether the mutant state is associated with specimens #6 and #20, or #8 and #18, or #6, #8, #18, and #20.
The key to the success of our strategy lay in optimizing the scheme of pooling patterns. To maximize the information that we could encode within a series of pooling patterns, we based our strategy on the Chinese remainder theorem (CRT). This approach is scalable, robust to both redundancy and sequence error (missing information), and is easy to debug. The framework that we designed is firmly rooted in information-theoretic nonadaptive group testing methodology (for review, see Du and Hwang 2000, 2006a). This body of knowledge deals with efficient pooling designs that reduce the number of test procedures, while maintaining the identity of the source specimen. This framework has applications in various fields, including data storage (Kautz and Singleton 1964) and sensor networks (Hong and Scaglione 2004). It has also been used in the past for biological applications such as finding sequencing tagged sites (STS) in YAC libraries (Bruno et al. 1995). While taking advantage of these theoretical tools, we were mindful of the physical overhead of creating pools and the costs that factor into pool analysis, namely, next-gen sequencing. We therefore established compromises that minimized cost and labor within the range of states and specimens that we sought to analyze, specifically multiple tens of thousands.
In general, our scheme starts with N specimens that are arrayed in source wells, for instance, a series of 96-well plates. During the pooling step, aliquots of specimens are moved from the source wells to T destination wells, each of which accommodates a specific pool. Each pool, rather than each specimen, is tagged with a unique barcode identifier. The pools themselves are further mixed into one or a small number of samples that are then analyzed by NGST. Data from the sequencing run are fed to a decoder, which solves the DNA Sudoku and generates a list of individual specimens and their corresponding inferred sequences.
The ability to achieve a solution in DNA Sudoku requires tracking specimens within the context of each pooling pattern and relating results generated by different pooling patterns. Since specimens may give rise to many sequences (e.g., fragments of a genome) and since specimens may be nonuniform at any sequence position (e.g., mixed samples or heterozygosity), we refer to each specimen as having a state, which is inherent in its sequence. The information content of each pooling pattern is strongly related to the diversity of states among the specimens. Considering extreme examples, if states are identical among all specimens, then assignment of the common state is trivial. Alternatively, if only one specimen has a state that differs from the others, it is straightforward to track its context in the different pooling patterns and work back to identify the source that gave rise to that state; in this case, even the simple row–column pooling pattern outlined above would be sufficient. If states are very common, for example, if the specimens contain an equal mixture of two states, then it is extremely difficult to link contextual information from each pooling pattern to a specimen source. Thus, the strategy upon which we embarked works best for linking relatively rare, albeit redundant, states to their source specimens. This is well suited to our goal of identifying clones representing individual oligonucleotides from high-complexity mixtures. This logic also dictates that our strategy would be suitable for identifying relatively rare polymorphisms within large populations.
Mathematical analysis of potential approaches
In essence, the number of specimens that share the same state determines the ease in which identity can be encoded within pooling schema. As a reflection of this, we denote redundant states with d0. In a case with many states in the population, d0 enumerates the largest group of specimens with the same state. In cases in which an allele exists in binary states, d0 denotes the number of carriers for the rarer of the variants.
Overall, the lowest adverse impact of d0 is achieved by minimizing the number of times any two specimens reside in the same pool within a series of pooling patterns. The pooling design can be represented by a pooling matrix M, which denotes the specimens residing in each pool. The entries of the matrix are binary, either 0 or 1; the rows represent pools, and the columns represent the specimens. Thus, M is a T × N matrix. For example, if the first row is (1, 0, 1, 0, 1,…), it specifies that the 1st, 3rd, 5th, … specimens together occupy pool #1. The trivial case of one specimen, one barcode would yield the identity matrix I(N). C(i) and C(j) are two column vectors in M, corresponding to the pools associated with the ith and the jth specimens; W(i) denotes the weight of C(i), namely, the number of pools in which a specimen is participating, essentially the number of 1s in the vector.  is the dot-product of the two vectors. The dot product reveals the number of intersecting pools that hold two particular specimens whose behavior is represented by the vectors.
is the dot-product of the two vectors. The dot product reveals the number of intersecting pools that hold two particular specimens whose behavior is represented by the vectors.
The minimal weight, Wmin, and the maximal intersection, λmax, of the pooling matrix are critical properties of the pooling design, defining our ability to identify a specimen whose state appears multiple times within the collection of samples. In information theory, this property is called d-disjunction and refers to the ability of a pooling design to convey sufficient information to guarantee fully correct decoding despite the presence of up to d specimens with the same state. In their seminal paper, Kautz and Singleton (1964) proved that a matrix with:
 |
is d-disjunct. Our goal was to minimize the difference between d0 and d and when possible find a design where d > d0. This can be achieved by increasing the weight or decreasing λmax. In practical terms, increasing the weight has operational costs, and we therefore focused on minimizing λ, namely, increasing the orthogonality between the column vectors. We wished to accomplish this without adding more rows (pools) to the matrix, which would increase costs associated with pool preparation and sequencing. This task has been studied extensively in information theory (see, for example, Johnson 1962; Kautz and Singleton 1964) and is related to the design of efficient error-correcting codes. The difficulty of the task can be appreciated by considering λmax = 0, which denotes maximum orthogonality, and reducing the weight to the minimum for all columns, W = 1. Basic linear algebra dictates that M must be equivalent to the identity matrix T = N, which implies the one-specimen one-barcode framework. The theory of group testing suggests highly efficient methods that compromise W and λ to yield T ∼ ln (N), essentially giving logarithmic growth of the number of barcodes in comparison to the number of specimens (Du and Hwang 2000, 2006a; Eppstein et al. 2007; Mezard et al. 2007).
Practical considerations for efficient pooling designs
In practice, reducing the number of barcodes to the minimum is not the only requirement for effective multiplexing, since costs associated with labor and sequencing must also be considered. These are proportional to the weight, W. The weight determines the number of times each specimen is pooled and is related to the number of robotic steps, and thus cost in time and reagents, for pooling. It is also preferable to have a weight-balanced pooling design, in which all the specimens are sampled the same number of times. The more times each specimen is added to a pooled library, the more the library size increases. Overall, the required sequencing coverage does not depend on the number of pools, but only on the weight of the design, which is a major deviation from the theory of group testing, where the only cost is associated with the number of pools.
Several additional biological constraints must also be taken into account, and these are related to the compression level R(t), the number of specimens that are participating in the tth pool. High compression levels can reduce the efficiency and the reliability of the test, for instance, PCR on a pool with thousands of specimens is prone to representation biases, elevating the likelihood that some specimens will not be sufficiently sampled in the sequencing step. Experience has highlighted sufficient sequencing coverage as a key to the success of our decoding strategy (see below).
Using combinatorial arguments, we demonstrated that only pooling schemes with  can minimize sequencing and robotic costs while still maintaining decoding robustness (d-disjunction) for clones with nonunique states (see Supplemental material). For comparison, T ∼ ln (N) methods (Du and Hwang 2006b; Eppstein et al. 2007) that confer similar decoding robustness typically have weights that are at least twofold higher than the achievable weight of
can minimize sequencing and robotic costs while still maintaining decoding robustness (d-disjunction) for clones with nonunique states (see Supplemental material). For comparison, T ∼ ln (N) methods (Du and Hwang 2006b; Eppstein et al. 2007) that confer similar decoding robustness typically have weights that are at least twofold higher than the achievable weight of  methods. In addition, the compression level of the T ∼ ln (N) methods is greater, creating more complex pools and potentially compromising performance in a biological setting. Given our aim of genotyping thousands of specimens that can be nonunique in one Illumina GAII run, we focused on constructing a
methods. In addition, the compression level of the T ∼ ln (N) methods is greater, creating more complex pools and potentially compromising performance in a biological setting. Given our aim of genotyping thousands of specimens that can be nonunique in one Illumina GAII run, we focused on constructing a  method.
method.
The multiplexing scheme
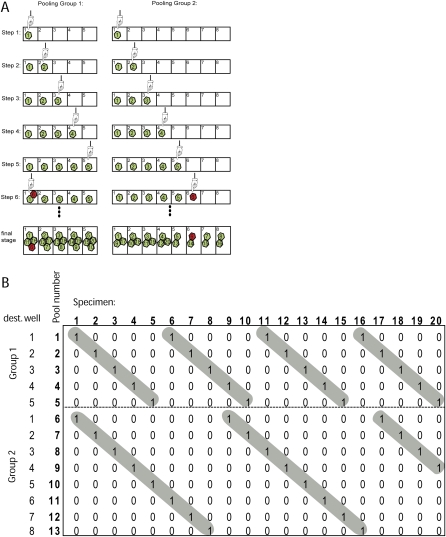
In essence, we chose to treat a pooling rule as a modular equation. For instance, in Figure 2A, we illustrate pooling of 20 specimens according to the following two pooling rules:
 |
For each pooling rule we create a destination plate, which accommodates the pools that are created based on the rule. More generally, each pooling rule has the following format:
which brings aliquots of the r, r + x, r + 2x, … specimens to the rth well in the corresponding destination plate, where 0 < r ≤ x, and x is called the “pooling window.” Let the “pooling group” be a set of all pools that are created according to a given x pooling window. Our general scheme uses a system of W pooling rules:
 |
The total number of wells in the pooling groups gives the overall number of pools:
For instance, in our example above, we see that T = 5 + 8 = 13.
Figure 2.
An example of pooling design. (A) The steps involved in pooling 20 specimens with two pooling patterns: (left) n ≡ r2 (mod 5); (right) n ≡ r2 (mod 8). Note that each pattern corresponds to a pooling group and to a set of destination wells. For simplicity, the destination wells are labeled 1–5 and 1–8 instead of 0–4 and 0–7. In this example, we choose specimen #6 to be mutant (red, green = wild type) among the 20 specimens. (B) The corresponding pooling matrix. The matrix is 13 × 20 and partitioned into two regions (broken line) that correspond to the two pooling patterns. The (highlighted in gray) staircase pattern in each region is typically created in our pooling scheme. The weight of the matrix is 2, the maximal intersection between each set of two column vectors is 1, and the maximal compression rate is 4.
This representation of the pooling scheme by the pooling matrix, M, has the following characteristics. First, the rows of the matrix are partitioned to W regions, so that the first region has x1 rows, the second region x2 rows, with the pattern repeating until the number of pools is reached. In every region, the first row has a “1” entry in the first column. “1” appears again after every increment that corresponds to the size of the corresponding pooling window; in the second row, the “1” entry starts at the second column. In this way, we get a staircase pattern in every region (Fig. 2B). Our pooling design has a weight of W, since every specimen is sampled only once in every pooling window, and the matrix is weight-balanced. In addition, the total number of “1” entries in every region of the matrix is the same. Thus, the sequencing capacity that is needed for each pooling group is the same.
The crucial element in maximizing the efficiency of such a design is the choice of the pooling windows: x1, x2, …, xw. For this purpose, we turned to the Chinese remainder theorem, one of the most ancient and fundamental in number theory (Andrews 1994; Ding et al. 1996; Cormen et al. 2001). The CRT has been studied over the past few years as an encoding system for computer science applications (Goldreich et al. 2000; Guruswami et al. 2000) and airborne radars (Stimson 1998).
Our pooling rules were designed to satisfy two basic requirements. First, the size of every pooling window was chosen to be greater than the square root of the number of specimens. Second, every set of pooling intervals was designed, according to the CRT, to be co-prime with respect to each other. This means that no pooling windows can have a common factor other than 1. For instance, (7, 9, 10) are co-prime, but (6, 9) are not co-prime because 3 is a common factor. These two requirements are denoted by the following equations:
When these two conditions are applied, The CRT asserts that any two column vectors in the pooling matrix intersect at most one time. Thus,
gives the minimal possible intersection for any pooling design of T < N and confers the highest likelihood of detecting the state of each specimen. From Equations 1 and 8 the decoding robustness of our pooling design is maximal and equals
Example 1 follows these two requirements since (5, 8) are co-prime, and  , and d = 1, guaranteeing correct decoding when all states are unique.
, and d = 1, guaranteeing correct decoding when all states are unique.
Equation 5 implies that the maximal compression level is:
providing a very sparse compression that will never exceed 230 specimens for most situations (N < 50,000). In addition, we can derive the total number of pools in our design from Equation 5:
This construction is very close to the lower theoretical bound shown in the Supplemental material (a formal proof will be given elsewhere).
In our case, we chose to determine states by sequencing the output of one pooling rule in one lane of an Illumina GAII. Thus, we were able to reuse the same set of barcodes/oligonucleotides in each pooling group, reducing the number of barcodes needed to around  for small W. Tools to construct this pooling design can be found at http://hannonlab.cshl.edu/dna_sudoku/main.html.
for small W. Tools to construct this pooling design can be found at http://hannonlab.cshl.edu/dna_sudoku/main.html.
For the number of specimens that we envisioned (N < 50,000), we analyzed the differences in the number of barcodes required for our  scheme to the theoretically oriented T ∼ ln (N) method of Eppstein et al. (2007) (Table 1). We chose Eppstein's construction as a reference for several reasons. Their method has the maximal reduction in pool number and shows the best general performance among information theoretic methods. Moreover, their construction is also based on the CRT, but without asserting that the number of pooling windows be greater than the square root of the number of specimens. We assume that each pooling group is sequenced in one Illumina lane and that the number of barcodes needed is equivalent to the maximal number of pooling windows in each method.
scheme to the theoretically oriented T ∼ ln (N) method of Eppstein et al. (2007) (Table 1). We chose Eppstein's construction as a reference for several reasons. Their method has the maximal reduction in pool number and shows the best general performance among information theoretic methods. Moreover, their construction is also based on the CRT, but without asserting that the number of pooling windows be greater than the square root of the number of specimens. We assume that each pooling group is sequenced in one Illumina lane and that the number of barcodes needed is equivalent to the maximal number of pooling windows in each method.
Table 1.
Comparison between the best-known theoretic method and DNA Sudoku
The difference in pools, weight, and maximal number of barcodes in a comparison between the best-known information theoretic method (Eppstein et al. 2007) and DNA Sudoku.
As expected, Eppstein's method performed better in reducing the number of pools. However, this came at the cost of heavily increasing the weight of the design, and thus sequencing and robotic costs. For example, our analysis of 40,000 specimens (see below) required less than one Illumina GAII flow cell (five lanes), whereas Eppstein's method would consume more than two full flow cells (17 lanes). The increase in the number of barcodes required was not dramatic, about 150 in the worst case.
Solving the DNA Sudoku: General principles
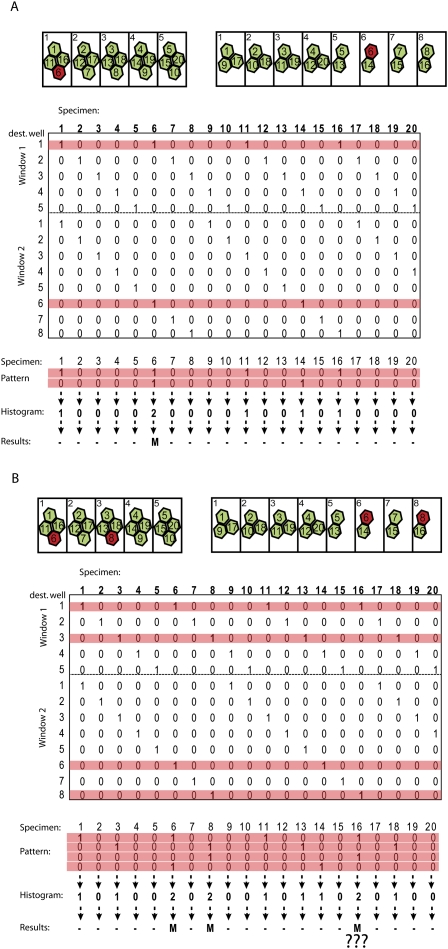
Solving the DNA Sudoku is a matter of determining which pooling patterns for a particular state coincide with the experimentally determined content of the pools, which we created. This can be understood most easily by considering a case of binary states, wherein each specimen is either a wild type (WT) or a rare mutant. In this case, the task is to find the rare mutants. Considering the example in Figure 3A, the only data available to the decoder are the following pattern: well #1 in window #1 and well #6 in window #2 contain one or more specimens with the mutant state. In order to uncover the configuration that led to the observed pattern, the decoder eliminates specimens whose association with the mutant state would violate the observed pattern. This is achieved by scanning the pooling matrix along the pools (rows) that contain a mutant state and summing the columns of those rows, creating a histogram of specimen versus the number of pooling windows in which it appeared. If a specimen is mutant, the mutant state should appear in all windows harboring the specimen. Thus, any specimen with a level below w in the histogram is excluded. This histogram essentially displays the number of constraints fulfilled by associating the mutant state with each specimen. We refer to this overall procedure as “pattern consistency decoding.”
Figure 3.
Pattern consistency decoder. (A) Successful decoding. (Red) Specimen #6 is the only mutant in the library (top panel). After pooling and sequencing, the only data available for the decoder are the following pattern: mutant is in the first pool of the first pooling window and in the sixth pool of the second pooling window (middle panel; see corresponding red rows in the matrix). Summing along the columns of the pattern (bottom panel) creates a histogram that represents the number of windows in which a specimen was found. Notice that the scores of the histograms range from 0 to the weight of the matrix. The pattern consistency decoder asserts that only specimens that appeared in all windows will be assigned to the mutant state. Since only specimen #6 has a score of 2 in the histogram meaning it appeared in all possible windows, it is associated with the mutant state, and the decoder reports the correct result. (B) Failure in the decoding. This pooling matrix has decoding robustness of d = 1, and the correct decoding of d0 = 2. Thus, in the example, correct decoding is not guaranteed. Specimens #6 and #8 are mutants (top), and the pattern that the decoder encounters is indicated in the matrix (middle). Consequently, associating specimen #16 with the mutant state is consistent, and it gets a score of 2 in the histogram, and is reported as a mutant.
Pattern consistency decoding definitively provides correct configuration when the number of mutant specimens, d0, is less than or equal to d (Du and Hwang 2006a). When d0 is higher than d (Fig. 3B), several configurations of specimen identity become consistent with the pattern. Hence, many specimens may falsely be reported as mutant. In practice, when the difference between d0 and d is not high, pattern consistency decoding still reveals the correct specimen identities with high probability.
Pattern consistency decoding for multiple states takes a similar approach. In each round, the decoder takes one state and analyzes it as if it were the binary case, excluding its association with specimens that would contradict the observed pattern of that state. At the end of the round, the decoder adds the interrogated state to their list of possible states for specimens that were found to be consistent. At the end of a completely successful decoding, each specimen is associated with exactly one possible state. When d0 of the interrogated state is too high, this state may appear as a false-positive associated inappropriately with a number of specimens. Since their true state will also be associated with the same specimens, they are ultimately reported by the decoder as “ambiguous.”
Simulated data sets: Decoding clone collections and identifying variants within a population
We tested the performance of DNA Sudoku with pattern consistency decoding for two simulated scenarios that matched our general interests. In the first, we sought to identify and verify shRNA inserts harbored by a large number of bacterial clones. In the second, we sought to identify carriers of a rare genetic disease among a large number of individuals. In both cases, we limited the number of pooling windows to eight, so that sequencing could be carried out on one Illumina GAII flow cell.
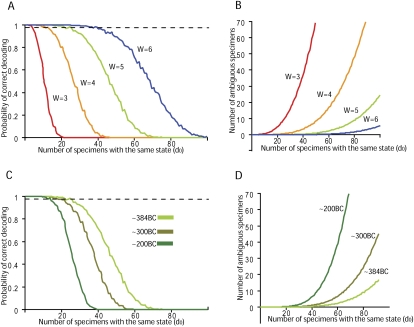
In the shRNA scenario, 40,320 simulated bacterial clones carried one of 12,000–17,000 possible inserts, a number consistent with array-based oligonucleotide library synthesis (Cleary et al. 2004). Based on prior experience, we estimated d0 to be no more than 40–50, even in the presence of biased cloning. Given these parameters, we simulated the effect of number of pooling windows (Fig. 4A,B) and the number of barcodes on the decoding performance (Fig. 4C,D). Evaluating the probability of achieving unique assignments as a function of d0 (Fig. 4A,C), we noted strong transitions when d0 passed a certain threshold, depending on the pooling design. We also determined the number of ambiguous assignments for different values of d0 (Fig. 4B,D). We used these measurements in a relatively qualitative manner to understand the effect of adding pooling windows or increasing the number of barcodes used within our scheme. We found that each additional pooling window reduced the remaining uncertainty substantially, while adding barcodes had modest effects. These observations could be used to construct a rough heuristic to determine the number of barcodes and windows required to analyze a set of samples of a particular size and complexity. In comparison to the random k-set design method suggested by Bruno et al. (1995), our number of unrelated specimens for different inputs is typically smaller by 25% (data not shown). This supports the value of using their analytical equations as an upper bound for the optimization process instead of exhaustively simulating large numbers of instances. As a stringent test, we also validated the performance of the decoder when faced with an input set of 40,320 specimens distributed among only 1000 different states, which corresponds to d0 ≈ 60 (Supplemental Fig. 1).
Figure 4.
Simulations of decoder performance in different CRT pooling designs with 40,320 specimens. (A,B) The effect of increasing the weight with 384 barcodes. (C,D) The effect of increasing the number of barcodes with five pooling windows. (A,C) The probability of correct decoding as a function of d0 with different pooling designs. (Gray line) 99%. (B,D) The expected number of ambiguous specimens due to decoding errors as a function of d0 and different pooling designs.
To estimate the value of DNA Sudoku for the analysis of rare variants in individuals, we presented the process with a population in which two prevalent cystic fibrosis (CF) risk alleles were segregating, CFTR mutations ΔF508 (MIM: 602421.0001) and W1282X (MIM: 602421.0009). We set the carrier frequency of each allele according to levels found in a large cohort analyzed by the Israeli CF Screening Unit (Orgad et al. 2001), ∼1.8%. This relatively high frequency of risk alleles challenges DNA Sudoku, testing the method against a difficult genetic screening problem. The simulation included 18,000 individuals that were either homozygous wild-type or heterozygous carriers of risk alleles, translating to a d0 of ∼320. We simulated seven pooling windows with approximately 384 barcodes. We randomly sampled the pooled data and created artificial sequence reads that recapitulate Illumina GAII sequencing error profiles, which are dominated by substitutions of C ↔ A and G ↔ T (Erlich et al. 2008). We used two methods to simulate sequencing errors. The first was based on an analysis of more than 30 million sequences of Phi-X from three different runs, and the other was based on a recent study of Druley et al. (2009) (human data set; see Methods).
In the case of ΔF508, we simulated 4000 reads for each pool, which corresponds to 1.5 million reads per lane, a relatively low number for a GAII instrument. Before applying pattern consistency decoding, we filtered reads that did not perfectly match to the WT or risk allele. In both error profiles, Phi-X-based and human-based, the sensitivity was 100%, and the specificity was 98.2%. The case of W1282X was more complex since pools that essentially contain no carriers may report a presence of a carrier because of sequencing error. In order to distinguish between sequencing error and the presence of a true carrier in the pool, we introduced a probabilistic threshold procedure to the pattern consistency decoder. This determined the expected ratio of carrier reads and the standard deviation for each pool if exactly one carrier were present in the pool. The threshold was set as three standard deviations below the expected value, and pools that passed the threshold were marked as containing at least one carrier. With the error profile based on Phi-X and 10,000 reads per pool (about 4 million per lane), we reached a sensitivity of 100% and a specificity of 98.3%. Setting the threshold to two standard deviations below the expected value reduced the sensitivity to 98.1% and did not affect the specificity, meaning that false-positives are not due to the sequencing errors but are due instead to a limitation of the pooling procedure in dealing with large d0 values. The simulation based on error profiles derived from human data revealed a similar picture. The sensitivity was 100%, and the specificity was 97.9% with approximately 10,000 valid reads per pool.
We tested the effect of dramatically reducing sequence coverage using the human error profile. We sampled 500 reads per pool (approximately 200,000 reads per lane), corresponding to only five reads per chromosome in each pool. Under those conditions, the sensitivity dropped to 98.7% and the specificity to 94.4%; changing the threshold to one standard deviation restored the specificity to 98.9% but reduced the sensitivity to 73.8%. These results not only highlight the importance of sufficient sequencing coverage in DNA Sudoku, especially for medical genetics applications, but also demonstrate the robustness of the procedure under nonideal conditions.
Using DNA Sudoku to decode physical clone collections
We next tested the ability of the DNA Sudoku procedure to identify and validate shRNA clones derived from two different multiplexed oligonucleotide synthesis runs. In the first case, we synthesized 17,500 oligonucleotides representing artificial microRNAs (amiRNAs) targeting Arabidopsis genes. These were cloned via a two-step procedure into a pGreen derivative (data not shown). In this, 40,320 bacterial colonies were picked into 96-well culture blocks using a Genetix QPIX (see Methods). The goal was to determine and verify both the passenger and the guide strand of each of the greater than 40,000 clones.
Creating the patterns of pooled samples is perhaps the most time-consuming part of DNA Sudoku. We therefore tuned our procedure for use on a Tecan liquid handling robot with a fixed-tip head that has eight individually addressable pins. Use of washable pins extends robotic run times but eliminates the otherwise substantial cost for disposable tips. This type of system is in relatively common usage, but the logic underlying the optimization is sufficiently flexible to allow adaptation to other robotic platforms.
Considering practical constraints placed on the procedure, it required the liquid handling robot ∼45 min to move specimens from one source plate to one destination plate, with the majority of time being spent on tip washing. Thus, each additional pooling window added substantially to the overall pooling operation. At the present level of optimization, the creation of five pooling windows from approximately 40,000 clones requires ∼7–9 d, which we consider reasonable. Most liquid handling stations can dispense into plates with up to 384 wells, placing this as an upper limit on the number of barcodes that could be conveniently used. Considering these practical constraints, the simulation results, and the expected d0 level, we decided to use a five-window combination of pooling intervals: 384, 383, 381, 379, and 373, which provided a compromise between higher inference rates and tolerable robot time (see Methods).
We sequenced each pooling group in a separate lane of an Illumina GAII flow cell, obtaining a total of 25 million sequence reads. We parsed the sequence reads and annotated the barcodes. Several practical challenges emerged from the sequencing data. First, the number of reads were nonuniformly distributed among the different lanes (Supplemental Fig. 2), with some lanes exceeding 10 million reads, and others reaching as low as 1 million reads, meaning 25 reads per specimen on average. A second challenge was the low numbers of reads from specific pools, in particular, the thirteenth pool and repeatedly every 48 pools thereafter in most of the pooling groups (Supplemental Figs. 3–7). This reflected inefficiencies in preparation of the sequencing templates in those wells. A third practical challenge, that was not simulated previously, was low-level contamination by sequences that appeared in up to several hundred pools and thus introduced significant “noise” into the decoding process. Finally, we were faced with a large number of sequencing errors. We found that the library had in total more than 800,500 possible states (i.e., distinct sequence reads), which exceeds by 20-fold the number of sequences used to program oligonucleotide synthesis (Supplemental Fig. 8). Since the whole procedure was deployed to distinguish clones with synthesis errors versus correct clones, we could not revert these sequence reads simply by aligning them to the design file.
In the face of these challenges, the pattern consistency decoder performed remarkably well, providing 28,000 specimens with potential state calls, of which 20,500 specimens were unambiguous. To test our performance, we compared the results of the decoding to a set of about 3000 specimens whose state we had determined by conventional sequencing. This revealed that 95% of the 20,500 assignments were correct.
We were encouraged by the high decoding rate, but we sought approaches to minimize the number of specimens with empty or ambiguous lists, without sacrificing veracity. Based on experience, we developed various heuristics. First, we increased tolerance for missing states when the sequencing depth for a particular pool was low and introduced penalties for states with low read numbers when sequencing depth was otherwise high. We incorporated a priori knowledge of d0 and penalized possible assignments that match to large numbers of specimens.
Thus, instead of a hard consistency requirement, we created a decoder that measures the discrepancy of the pattern from an ideal one and registers that value. After evaluating all possible states, the decoder scans through specimens and assigns the state whose pattern exhibits the minimal discrepancy. In addition, it reports an assignment quality score that is defined as the difference between the discrepancies of the best state and the second best state (see Supplemental material for a detailed description of the decoder).
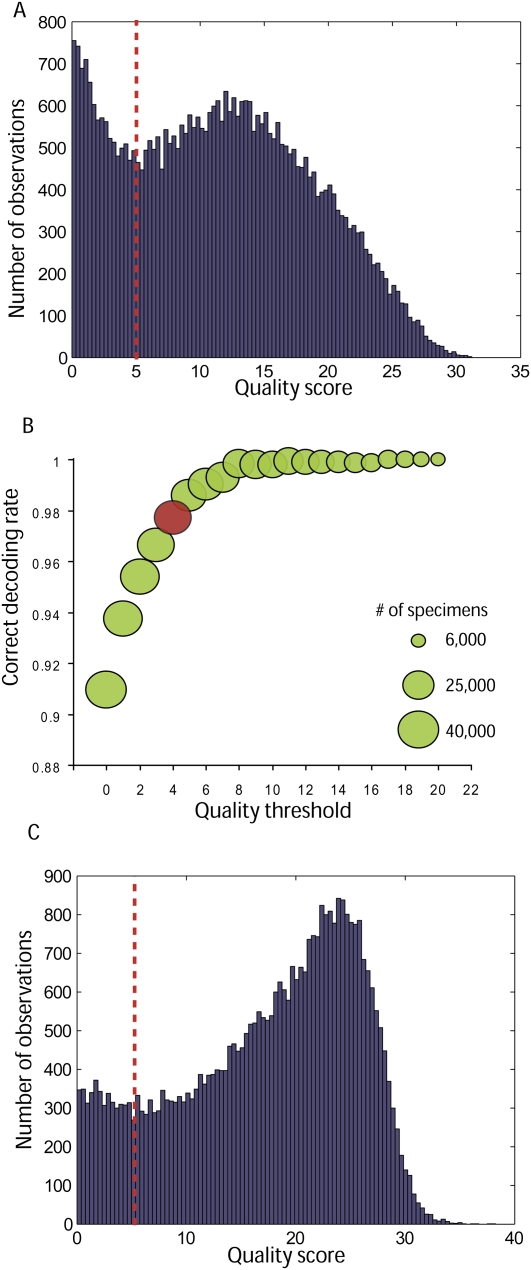
Analyzing the distribution of assignment quality scores revealed a bimodal distribution (Fig. 5A), and we tested whether this corresponded to mediocre versus high-quality decoding. We found an informative correlation between the quality scores and the correct decoding rates (Fig. 5B), which resembled the bimodal distribution with a strong transition around the value of 5. We therefore filtered specimens with quality scores below 5 and compared the remaining 31,000 specimens to the capillary reference. Correct decoding rates jumped to 98.5%. When we removed quality filtration, the correct decoding rate dropped to 91% for all 40,320 specimens. Thus, the improvement we achieved using the minimal discrepancy decoder over the pattern consistency decoder was substantial.
Figure 5.
Performance of the minimal discrepancy decoder. (A, red line) The distribution of quality scores for the Arabidopsis library is bimodal with transition around a quality score of 5. We speculate that this corresponds to two decoding regimes—noisy on the left and error-free on the right. (B) Analyzing the correlation between filtering above different quality scores and the correct decoding rate. The size of the bubbles corresponds to the number of specimens passing the threshold. The increase in the correct decoding rate with the threshold of the quality validates the predictive value of the quality scores. (Red bubble) The strong drop in the correct decoding rates below a quality score of 5 confirms our speculations about the two decoding conditions. (C) The quality score distribution of the human library. The noisy area, left of a quality score of 5, contains fewer specimens than observed in the Arabidopsis library. The shift of the distribution to the right suggests better decoding performance presumably owing to higher sequencing depth. After filtering specimens with a quality score above 5, the correct decoding rate was estimated to be 98.2% for 36,000 specimens and indicates the robustness of the quality score. Decoding all specimens regardless of the quality score revealed a correct decoding rate of 97.3%.
We tested DNA Sudoku and the minimal discrepancy decoder on a human shRNA library comprising 12,500 different fragments in 40,320 bacterial clones. We optimized the sequencing procedure to yield higher coverage (67 million reads total) and used longer barcodes of 10 nt. Consistency checks on barcode representation revealed substantial background contamination in the Sudoku pools. Decoding the library with the minimal discrepancy decoder revealed an enhancement in the quality score distribution with a peak around the value of 25 (Fig. 5C). We filtered specimens with quality scores below 5 and retained 36,000 out of the 40,320 clones. To evaluate decoding accuracy, we compared our assignments to conventional sequencing of 384 clones. In this case, the correct decoding rate was 98.2%. When we eliminated quality filtering, we had correct decoding rates of 97.3% for the 40,320 specimens. These results confirm that the quality scores are robust and have predictive value in identifying correct clones.
Discussion
The goal of this study was to transform a random NGST sequencing method into one that allowed an association of sequence reads with the specimens from which they were derived. For this purpose, we present a multiplexing framework that encodes the identity of specimens within a pattern that is established by a pooling procedure. We show that we can effectively reduce the number of barcodes required and the experimental overhead of pool production to levels acceptable for the analysis of many tens of thousands of specimens. Both the pooling and decoding methods can be tuned to address different specimen scenarios, different robotic pooling platforms, and different NGST technologies. We have investigated the trade-offs in pooling strategies and validated our procedure both via in silico simulation and through two real case studies. We find that our procedure is largely insensitive to the types of sequencing errors that plague NGST instruments but is vulnerable to low sequencing depth and biochemical failures in template preparation. We developed an improved algorithm to address those issues and achieved high decoding rates of >97% on average for all the clones, and robust quality score measurements that enable evaluation of the veracity of state assignments.
Our framework raises the possibility of a new approach to the use of next-generation sequencers for personal genomics and population genetics. By analogy to the goal of the $1000 genome (Mardis 2006), it would be highly beneficial to reduce the cost of sequencing defined subsets of genes that are responsible for severe genetic diseases or for determining histocompatiblity. Our strategy is presumably applicable to both of those examples since in severe genetic diseases, one of the alleles (state) is generally very rare and the HLA locus is highly polymorphic. However, these tasks present additional challenges, including inferring the state of a diploid genome and the difficulty of reconstructing complex haplotypes from short reads. No doubt, future improvements in effective read lengths and in the bulk output of NGSTs will help us to address these challenges and further boost the power of the approaches we describe. Specifically, such advances may pave the way to use our strategy for finding common genetic variations among large numbers of individuals. While common genetic variations are not easily decodable, extended sequencing lengths will provide additional “hitchhiker” variations, which in total will provide enough polymorphism to detect the state of common variants in individual specimens. Recent studies have shown that autosomal heterozygosity is about 1/1000 nt (Bentley et al. 2008), implying that 5000–10,000-nt fragments may give enough polymorphism to detect common genetic variations in large samples using our strategy. Given the advent of what has been dubbed next-next-generation sequencing technologies (Eid et al. 2009), this may become possible in the near future.
Methods
Comparing DNA Sudoku to Eppstein's pooling method
Eppstein's pooling method construction was based on the Python backtracking algorithm. Our method is based on the C program, which performs an exhaustive search of co-prime numbers such that their sum is minimal and they are above a certain threshold. Running both programs took up to several seconds on a regular desktop computer.
Simulation of decoding performance for the shRNA library
W = 3, 4, 5, and 6 correspond to the following pooling windows: (384, 383, 381), (384, 383, 381, 379), (384, 383, 381, 379, 373), (384, 383, 381, 379, 371). BC = 200, 300, and 384 correspond to the following barcode combinations: (209, 205, 203, 202, 201), (307, 305, 303, 302, 299), (384, 383, 381, 379, 373). We simulated each pooling design for 1000 random specimen arrays on Cold Spring Harbor Laboratory's High Performance Computing Cluster (HPCC). The system is an IBM E1350 blade-based cluster running Linux. It comprised 500 compute nodes, but our simulation used 100 nodes. Each compute node contained two dual-core Opteron 64 processors running at 2.0 GHz, a local 73 GB SFF SCSI disk, and memory from 2 to 8 GB.
Simulation of decoding performance for cystic fibrosis–associated mutations
The error profile of Illumina GAII using Phi-X data was constructed using three runs with Illumina standard base-calling versions >1.0. The sequence reads were aligned to the Phi-X reference genome (NC_001422.1) using BLAT (Kent 2002) with a tolerance of up to five mismatches. The error profiles were calculated by enumerating the differences between the reference and the observed sequence read for each cycle. Ns were discarded. This procedure provided 36 4 × 4 confusion matrices. Each row was normalized to 1 to get probabilities. The error profile based on human data was set to 0.05% for all possible substitutions in the first 12 cycles according to the data in Druley et al. (2009).
The aforementioned noise profiles were added to the following sequences: Δ508, ATCATCTTTGGTGTTTCCTATGATGAA (WT); ATCATTGGTGTTTCCTATGATGAATAT (M); G1282X, TGGAGGAAAGCCTTTG (WT); TGAAGGAAAGCCTTTG (M).
The pooling design used the following windows: 383, 382, 381, 379, 377, 373, 371.
Arabidopsis chip construction
A plasmid library representing 17,553 Arabidopsis synthetic miRNAs based on the miR319a precursor was synthesized on an Agilent microarray. The DNA was eluted and cloned into a pGreen derivative (data not shown).
GIPZ chip construction
A plasmid library representing 12,784 human synthetic shRNAs based on the miR30 precursor was synthesized on an Agilent microarray. The DNA was eluted and cloned into a GIPZ derivative (Open Biosystems).
Pooling bacterial clones
Plasmid DNA was transformed into a DH10B Escherichia coli suspension, and transformants were selected on a series of Q-tray 20 × 20-cm Petri dishes with LB + spectinomycin at 25 μg/mL medium; 40,320 bacterial colonies were arrayed into 105 384-well SBS microplates (Genetix) using a Genetix QPix colony picker. A Tecan Genesis liquid handling robot carried the pooling using eight fixed tips. Three microliters of bacterial suspension was taken from each source well and delivered to each destination well. This was followed by washing the tips by aspirating 30 μL of 70% ethanol from two different ethanol baths and rinsing with milli-Q water. For destination plates, we used five 384-deep-well plates (Nunc). The pooling procedures were carried out at room temperature.
The pooling design for Arabidopsis was 384, 383, 381, 379, 373. A calculation error caused us to use a window size of 381, which is not co-prime with 384. However, we found that this would have an impact only if the number of specimens was above 48,768 (least common multiple of 384 and 381).
The pooling design for the human library was 384, 383, 379, 377, 373. In addition, we treated the clones so as the first one is #9887, the second is #9888, and so on. The reason behind that was to avoid a situation in which the same specimen encountered the same barcode in all pooling windows. For instance, in the ordinary design specimens, #1 pooled to the first well of each pooling plate, and was marked with the barcode #1. In case of a failure in the synthesis process of this barcode, specimen #1 will be lost. When labeling the specimens with larger numbers (e.g., starting with 9887), they encounter different barcodes in the pools and the risk is averaged.
Barcoding and sequencing: Arabidopsis
Five microliters of diluted bacterial culture was taken from each destination well for the PCR amplification. The PCR conditions were 2 μL of 10× KOD buffer, 0.8 μL of 25 mM MgSO4, 6 μL of bacterial DNA, 750 nM of each primer, 2 μL of 2 mM dNTPs, 0.4 μL of KOD hot start polymerase; in total 20 μL. The reactions were cycled as follows: 5 min at 95°C (hot start), (35 sec at 95°, 35 sec at 57°C, and 20 sec at 68°C) for 22 cycles. We used 384 primer set for each destination plate. The primers were synthesized by Operon and purchased in four 96-well deep plates wet (50 μM × 200 μL). The reverse primers had the following structure: 5′-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT********GCGACTCGGTATTTGGATGAATGAGTCG-3′, where the stars indicate 8-nt barcodes that were constructed using a subset of the Hamady et al. (2008) barcode set. This subset was constructed by lexicographically sorting the set and choosing every fourth entry.
The forward primer was 5′- AATGATACGGCGACCACCGAGATCTACACCTCGGACGCATATTACACATGTTCATACAC-3′.
Five microliters of each amplified product was taken, and products of the same pooling group were unified. We gel-purified each pooling group of ∼250 bp. Each one of the pooling groups was sequenced on a distinct lane of Illumina GAII using the standard paired-end protocol of 36 cycles. Pooling window 384 was sequenced for additional time as the first time the number of reads was 300,000. We merged the two runs.
Barcoding and sequencing: Human GIPZ library
We used a similar approach to the one indicated in the Arabidopsis set. The PCR primers were Forward, 5′-ATCGTTGCCTGCACATCTTGGAAAC-3′; Reverse, 5′-CGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT**********ACATCTGTGGCTTCACTA-3′, where the stars indicates 10-nt barcodes with 5-nt Hamming distance (data not shown; barcode sequences available upon request).
Decoding
Prototype programs of the decoding procedures were written in Perl and used a MySQL database. The pattern consistency decoder typically took 5–10 min on a desktop computer (CPU: QX6850, 3 Gb RAM, Linux Ubuntu). However, the minimal discrepancy decoder was very demanding and took ∼20 h on average. We expect that future implementation improvements will reduce decoding time. All programs are available for academic and nonprofit usage by request from the authors.
In order to benchmark the decoding results of the Arabidopsis library, we capillary-sequenced the specimens of the 1st, 11th,… plates, up to the 91st plate. Then, we filtered sequences that did not contain the sequences flanking HindIII and EcoRI restriction sites. In total, we were left with sequences from 2760 specimens.
More than 4-nt mismatches between the results of the decoder to the capillary sequences were considered as decoding errors, and 4-nt and fewer mismatches were considered as sequencing errors on one (or both) of the sequencers. Only 4% of the correct decoding result had sequencing errors. We probed the cause of the sequencing errors by comparing the decoder sequences and the capillary sequences to the reference design. Remarkably, we found that in 70% of cases, the sequence reported by the decoder appeared in the reference design, and in only 25% of the cases was the capillary sequence in the design reference. This indicates that most of the sequencing errors stem from the capillary sequencer and not from the decoder.
For benchmarking the decoding results of the human library, we sequenced an entire 384 plate using a capillary platform (Agencourt) and filtered reads that did not match the constant loop region of the shRNA design.
Acknowledgments
We thank Partha Mitra for useful discussions. Troy Moore and Blake Simmons were instrumental in creating library pools. We thank Danea Rebolini, Laura Cardone, Melissa Kramer, and Richard McCombie for Illumina sequencing. Y.E. is a Goldberg-Lindsay Fellow and ACM/IEEE Computer Society High Performance Computing Ph.D. Fellow of the Watson School of Biological Sciences. G.J.H. is an investigator of the Howard Hughes Medical Institute. This work was supported by grants from the NIH (GJH) and by a kind gift from Kathryn W. Davis.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.092957.109.
References
- Andrews GE. Number theory. Dover; New York: 1994. Solving congruences; pp. 58–75. [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno WJ, Knill E, Balding DJ, Bruce DC, Doggett NA, Sawhill WW, Stallings RL, Whittaker CC, Torney DC. Efficient pooling designs for library screening. Genomics. 1995;26:21–30. doi: 10.1016/0888-7543(95)80078-z. [DOI] [PubMed] [Google Scholar]
- Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- Cormen TC, Leiserson CE, Rivest RL, Stein C. Introduction to algorithms. 2nd ed. MIT Press; Cambridge, MA: 2001. Number theoretic algorithms; pp. 849–905. [Google Scholar]
- Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Pei D, Salomaa A. Chinese Remainder Theorem: Applications in computing, coding, cryptography. World Scientific Publishing; Singapore: 1996. [Google Scholar]
- Druley TE, Vallania FL, Wegner DJ, Varley KE, Knowles OL, Bonds JA, Robison SW, Doniger SW, Hamvas A, Cole F, et al. Quantification of rare allelic variants from pooled genomic DNA. Nat Methods. 2009;6:263–265. doi: 10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Hwang FK. Combinatorial group testing and its applications. 2nd ed. World Scientific Publishing; Singapore: 2000. [Google Scholar]
- Du D, Hwang FK. Pooling designs and nonadaptive group testing. World Scientific Publishing; Singapore: 2006a. [Google Scholar]
- Du D, Hwang FK. Pooling designs and nonadaptive group testing. World Scientific Publishing; Singapore: 2006b. pp. 58–59. [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Eppstein D, Goodrich MT, Hirschberg DS. Improved combinatorial group testing for real-world problem sizes. SIAM J Comput. 2007;36:1360–1375. [Google Scholar]
- Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: A self-optimizing base caller for next-generation sequencing. Nat Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldreich O, Ron D, Sudan M. Chinese remaindering with errors. IEEE Trans Inf Theory. 2000;46:1330–1338. [Google Scholar]
- Guruswami V, Sahai A, Sudan M. “Soft-decision” decoding of Chinese remainder codes; Proceedings of the 41st Annual Symposium on Foundations of Computer Science; Washington, DC: IEEE Computer Society; 2000. p. 159. [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. 1. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YW, Scaglione A. IEEE Inf Theory Workshop. 2004. On multiple access for distributed dependent sensors: A content-based group testing approach. [DOI] [Google Scholar]
- Johnson S. A new upper bound for error-correcting codes. IRE Trans. Inf. Theory. 1962;8:203–207. [Google Scholar]
- Kautz W, Singleton R. Nonrandom binary superimposed codes. IEEE Trans Inf Theory. 1964;10:363–377. [Google Scholar]
- Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Anticipating the $1,000 genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezard M, Tarzia M, Toninelli C. Statistical physics of group testing. J Phys: Conf Ser. 2007;95:012019. doi: 10.1088/1742-6596/95/1/012019. [DOI] [Google Scholar]
- Orgad S, Neumann S, Loewenthal R, Netanelov-Shapira I, Gazit E. Prevalence of cystic fibrosis mutations in Israeli Jews. Genet Test. 2001;5:47–52. doi: 10.1089/109065701750168725. [DOI] [PubMed] [Google Scholar]
- Stimson GW. Introduction to airborne radar. 2nd ed. SciTech Publishing; Mendham, NJ: 1998. [Google Scholar]