Abstract
“Smart-pooling,” in which test reagents are multiplexed in a highly redundant manner, is a promising strategy for achieving high efficiency, sensitivity, and specificity in systems-level projects. However, previous applications relied on low redundancy designs that do not leverage the full potential of smart-pooling, and more powerful theoretical constructions, such as the Shifted Transversal Design (STD), lack experimental validation. Here we evaluate STD smart-pooling in yeast two-hybrid (Y2H) interactome mapping. We employed two STD designs and two established methods to perform ORFeome-wide Y2H screens with 12 baits. We found that STD pooling achieves similar levels of sensitivity and specificity as one-on-one array-based Y2H, while the costs and workloads are divided by three. The screening-sequencing approach is the most cost- and labor-efficient, yet STD identifies about twofold more interactions. Screening-sequencing remains an appropriate method for quickly producing low-coverage interactomes, while STD pooling appears as the method of choice for obtaining maps with higher coverage.
Genome projects have enabled the development of a variety of large-scale functional genomics and proteomics projects. Some aim at identifying relatively rare events, such as mapping of binary protein–protein interactions (PPIs), protein–DNA interactions, or genetic interactions (for example, Yu et al. 2008, Deplancke et al. 2006, and Tong et al. 2004, respectively). These projects typically face three issues: reducing the cost and the number of assays (efficiency), recognizing false-positives that reflect technical artifacts (specificity), and avoiding false-negatives (sensitivity). Performing individual tests multiple times remains the gold standard for data quality but is often prohibitively costly and time-consuming. A frequently used alternative consists in assaying pools and then identifying the positives in a second step. For example, in the yeast two-hybrid (Y2H) screening-sequencing approach (Screen-Seq), first a set of preys are pooled and screened for interaction with a specific bait, then the positive clones are sequenced to identify the interactions (Rual et al. 2005). Alternatively, if a positive is detected in a pool, then all the constituents of the positive pool can be retested individually (Zhong et al. 2003; Stelzl et al. 2005). These methods improve the efficiency, and false-positives can be limited by subsequent experiments such as stringent retests, but false-negatives in the initial screen cannot be recovered.
Another method called “smart-pooling” (or “group testing”) aims to further increase efficiency, accuracy, and coverage in high-throughput screening projects. Smart-pooling has been used for screening clone libraries (Bruno et al. 1995) and was recently employed for Y2H (Jin et al. 2006, 2007) and yeast one-hybrid screens (Vermeirssen et al. 2007). It consists of assaying well-chosen pools of “items” (for example preys in Y2H) such that each item is present in several pools, hence tested several times (Thierry-Mieg 2006b). The goal is to construct the pools so that the positive items can be identified from the pattern of positive pools, even despite the occurrence of false-positive and false-negative pools.
Central to the smart-pooling procedure is the choice of the pooling design, and key parameters are both the “redundancy” and the “extra redundancy” of the design. We call “redundancy” the number of pools that contain any given item. Part of the redundancy is necessary to identify a single positive item, and the remaining “extra redundancy” allows the system to deal with noise (false-positive and false-negative pools) and multiple positive items (within a particular batch of pools).
Previous smart-pooling systems biology studies (Jin et al. 2006, 2007; Vermeirssen et al. 2007) have established the experimental feasibility of the smart-pooling concept in this field, but they have relied on designs that do not leverage the method's full potential. The PI-deconvolution approach (“pooling with imaginary tags”) (Jin et al. 2006, 2007) uses a variant of the classic grid design, where items are arrayed on an imaginary grid and a pool is constructed for each row and each column. Specifically, in PI deconvolution the grid is extended to N dimensions but restricted to a side length of 2 (for example with N = 3 it becomes a 2 × 2 × 2 cube, and each pool is defined by one of the six possible 2 × 2 × 1 slices). From a theoretical point of view, this design can be improved at two levels. First, it has a single degree of freedom: Choosing the redundancy imposes the pool size and the number of preys per batch, whereas one would like to set these three characteristics independently. Second, all of its redundancy is required for identifying a single positive in a noiseless experiment, leaving no extra redundancy to deal with multiple positives within a batch or correct for noise. Consequently, decoding is highly ambiguous when multiple positives, false-positives, or false-negatives occur. The second study (Vermeirssen et al. 2007) relied on a more sophisticated design derived from a Steiner system. The design has a redundancy of 3, and two pools uniquely identify a prey, leaving an extra redundancy of 1. As shown (Vermeirssen et al. 2007), this provides improved performance compared to PI deconvolution, but the noise-correction capabilities remain modest when multiple positives occur. The authors overcame this limitation by resorting to sequencing for confirmation or identification of positives, and by “uniplexing” known highly connected transcription factors: these were excluded from the smart-pool design and were tested individually instead, thus reducing the occurrence of multiple positives in a smart-pool batch. This is a useful strategy, but as a consequence it is difficult to interpret the obtained results in terms of success of the smart-pooling per se.
Other theoretical pooling designs that offer higher extra redundancy have been described (for review, see Thierry-Mieg 2006a) but lack experimental validation. In particular, we previously proposed a powerful and flexible algorithm for designing smart-pools: the Shifted Transversal Design (STD) (Thierry-Mieg 2006a). STD was shown to significantly outperform other published combinatorial designs in terms of flexibility and/or efficiency under a standard combinatorial model (the so-called “guarantee requirement,” where given bounds on the numbers of positives and erroneous observations, i.e., false-positives and false-negatives, a design must guarantee the identification of all positives). In STD, a large redundancy can be chosen and the extra redundancy is maximized, therefore providing high noise-correction capabilities. However, this power comes at a price: Despite its clean mathematical construction, the design is complex and difficult to visualize. In addition, interpreting experimental results is straightforward with simpler designs whose noise-correction abilities are intrinsically limited (Jin et al. 2006, 2007; Vermeirssen et al. 2007), but it becomes a difficult computational problem with highly redundant designs such as STD. Recently, this has been addressed by developing a new exact and efficient algorithm for interpreting smart-pooling results: interpool (Thierry-Mieg and Bailly 2008).
Here we experimentally evaluate the STD-based smart-pooling method in the context of interactome mapping by Y2H. We screened 12 Caenorhabditis elegans SH3 domains as baits against a C. elegans ORFeome library comprising 12,675 preys. We employed two STD designs adapted to different array densities, as well as the labor-intensive one-on-one array-based Y2H method (Uetz et al. 2000) (referred to as 1-on-1 hereafter). Additionally, six of these 12 baits were screened with the well-established Screen-Seq approach (Rual et al. 2005). All screens were performed with two repeats, and every positive from each method underwent pairwise retest in quadruplicate. Since all experiments in this study used the same reagents, false-negatives mainly contribute to “sampling sensitivity” in the recently proposed framework (Venkatesan et al. 2009), i.e., they should be identifiable by every method, given enough repeats. Our results show that STD Y2H is highly specific. Compared with 1-on-1, STD is much more cost- and labor-efficient, yet it remains very competitive in terms of sensitivity. While the Screen-Seq method is the most efficient in terms of costs and workload, STD Y2H appears approximately twice as sensitive. STD smart-pooling emerges as a method of choice for obtaining high-coverage interactomes, and could prove effective in a wide range of high-throughput experiments.
Results
Building STD pools for the worm activation domain ORFeome library
To study the application of STD pooling in proteome-scale Y2H, we assembled a set of reagents for array-based Y2H analysis (Uetz et al. 2000). Our prey array consists of 12,675 activation domain (AD) proteins represented in C. elegans ORFeomes v1.1 and v3.1 (Reboul et al. 2003; Lamesch et al. 2004). The baits consisted of 12 worm SH3 domains (Supplemental Table 1), a class of peptide recognition modules that often mediate PPIs by binding proline-rich peptide sequences (Ren et al. 1993; Tong et al. 2002).
We built and tested two STD designs with different pool sizes, adapted to different array densities. For any pooling design, the pool size is a major parameter. For example, larger pools improve the efficiency because fewer pools are required, but this may compromise the sensitivity due to dilution of the AD ORFs within the pools. Before designing the STD arrays, we performed dilution tests at two different densities, 384 and 1536 spots per plate, in order to identify the largest pool sizes that enable detection of positive controls (Supplemental Fig. 1; Supplemental Table 2). In the less dense 384 format, more yeast diploids can be obtained and the yeast colonies can grow larger, enabling more sensitive Y2H analysis, and thus allowing a larger pool size. In conjunction with a preliminary pilot experiment (Supplemental Note; Supplemental Fig. 2) and further simulations performed with interpool (data not shown), these initial tests led us to choose pool sizes of 78 for the 384 format and 26 for the 1536-format arrays.
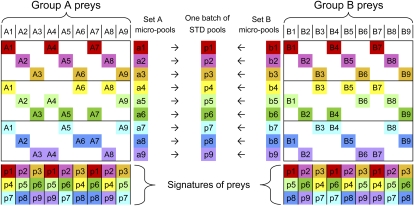
To limit the cost of building the STD pools and to increase their flexibility, we took advantage of inherent STD symmetries by designing and building small intermediary micropools. In Figure 1, a simple example illustrates this process: 18 preys are pooled according to a small STD design. Initially, the 18 preys are split into two groups of nine preys (groups A and B), and each group is pooled independently according to its corresponding STD subdesign to obtain two sets of micropools (sets A and B). Each micropool contains three different preys (pool size of 3), and each prey is contained in three different micropools (redundancy of 3), which form this prey's unique signature. In fact, any two micropools are sufficient to uniquely identify a prey, so these micropools have an extra redundancy of 1. Finally, each pair of same-numbered micropools from sets A and B are superposed to obtain one batch of STD pools (p1–p9). These STD pools still possess a redundancy of 3, but their pool size is now 6, and the nine STD pools accommodate all 18 preys. Each prey still has a unique signature, although the extra redundancy is now 0 because all three pools are required to identify each prey uniquely.
Figure 1.
A simple STD pooling design. Two groups of nine preys (group A: A1–A9 and group B: B1–B9) are separately pooled into nine color-coded micropools (set A: a1–a9 and set B: b1–b9), according to subdesigns of a larger STD design (which can accommodate all 18 preys). Each micropool contains three preys (e.g., a1 contains A1, A4, and A7), and each prey is present in a unique combination of three micropools (a unique signature, e.g., a1|a5|a9 for prey A4). The pairs of same-numbered micropools from sets A and B can then be superposed to generate STD pools (one batch: p1–p9), each containing six preys, and each prey still has a unique signature in the STD pools.
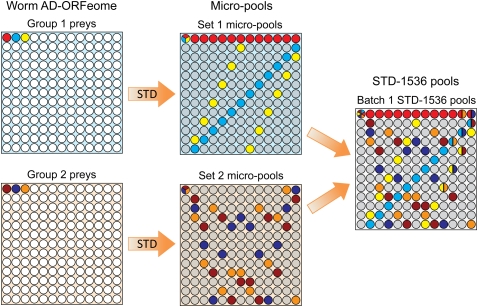
We built the worm STD pools in a similar manner but on a higher scale (Figs. 2, 3). The prey library, which contains 12,675 unique AD ORFs, was conceptually split into 75 groups of 169 preys (75 × 169 = 12,675). Each group was STD-pooled independently to obtain a set of 169 worm micropools containing 13 preys each (micropool size: 13). These micropools possess a redundancy of 13, including an extra redundancy of 11. All 12 sets of micropools were built according to subdesigns of a larger STD design, so that the micropools are superposable to generate larger STD pools (see Fig. 2 and Methods). Based on the chosen 1536- and 384-format pool sizes, the micropools were either superposed in pairs (as shown in Fig. 2), to produce the STD-1536 pools containing 26 preys per pool, or superposed in sextuplets, to generate the STD-384 pools with 78 preys per pool. In the resulting STD pooling designs, the 12,675 preys are either split into 40 batches of STD-1536 pools with up to 338 preys per batch, or 13 batches of STD-384 pools with up to 1014 preys per batch. The STD-1536 and STD-384 batches each contain 169 pools. All batches within an STD design are arrayed as colonies on a series of plates, but the batches are disjoint and decoded independently. Both designs possess an extra redundancy of 10, which provides high noise-correction capabilities.
Figure 2.
STD pooling design used in this paper. Worm AD-ORFeome preys (12,675) were split into 75 groups, each containing 169 preys. Each group was STD-pooled into a set of 169 micropools. Each micropool contains 13 preys (micropool size: 13), and each prey is contained in a unique combination of 13 micropools (a unique signature; redundancy: 13), as illustrated with three color-coded preys in sets 1 or 2. Two preys co-occur in at most one micropool, so that each prey is uniquely identified by any two of the 13 pools that contain it; therefore, these micropool designs have an extra redundancy of 11. In addition, the micropool signatures of preys with identical AD-ORFeome coordinates from groups 1 and 2 are very different (e.g., light red in set 1 and dark red in set 2): Every two sets of micropools can be superposed to obtain one batch of STD-1536 pools, such that two preys from different groups co-occur in at most two common pools. Consequently, in STD-1536 each prey is uniquely identified by any three of the 13 pools that contain it: STD-1536 pools possess an extra redundancy of 10.
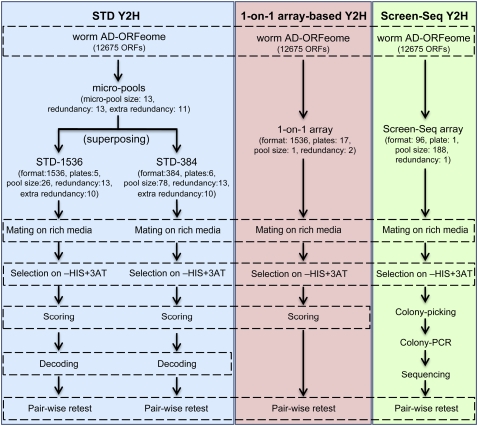
Figure 3.
Flowchart of Y2H approaches used in this paper, comparing the main steps of STD Y2H, 1-on-1 array-based Y2H, and Screen-Seq Y2H.
C. elegans ORFeome-wide experiments
We screened both STD-1536 and STD-384 with the 12 selected worm SH3 domains (Supplemental Table 1). In order to thoroughly evaluate the STD method, we also screened the same baits using the 1-on-1 Y2H approach (Uetz et al. 2000), where each prey is present individually in duplicate on the array in 1536 format. In addition, six of the baits were screened following the established Screen-Seq protocol (Rual et al. 2005). Finally, to address whether high plate density affected the STD method, we screened the three most connected baits against STD-SL, an STD array with small batch and pool size and low density (namely the STD-1536 pools assembled in 384 format instead of 1536). All screens were performed twice to evaluate each method's repeatability. Figure 3 compares the steps of the Y2H approaches used in this study.
Each method produced a list of candidate PPIs. All candidate PPIs underwent pairwise retest in quadruplicate, leading to the definition of three classes of hits: core, the strong and reproducible positives; FP, false-positive, when the retest fails; and noncore, when the retest results are unclear (e.g., quadruplicate retesting results in two positives and two negatives, see Methods for details). The noncore data set is small (Supplemental Table 3) and was not included in our subsequent analyses. Our classification relies only on the retest results and is independent of the hit's origin (number of detection methods, confidence scores, etc.); therefore, it allows an objective assessment of each method. Pairwise retests are conceptually similar to 1-on-1, but they are performed in a low density format using larger volumes of fresh cultures; this explains why interactions missed in the ORFeome-wide 1-on-1 screens still often retest successfully.
Comparison of Y2H methods
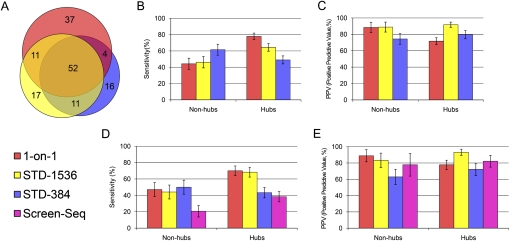
We retrieved a total of 156 core PPIs (Supplemental Table 4); 148 of these core PPIs were identified from 1-on-1, STD-1536, or STD-384, the three methods used to screen all 12 baits. Many were recovered by all three methods, but a significant number was also found exclusively by each method (Figure 4A). In particular, although 1-on-1 finds the most PPIs, it misses many PPIs that were found by STD-1536 or STD-384, which indicates that the 1-on-1 screens are not saturating even when repeated twice.
Figure 4.
Comparison of Y2H results. (A) Area-proportional Venn diagram of PPIs found by 1-on-1, STD-1536, and STD-384 (generated by http://venndiagram.tk/). (B) Sensitivity (the percentage of core hits from each method in the whole core set) and (C) positive predictive value (PPV; the percentage of each method's hits that successfully pass pairwise retest and end up in core) of 1-on-1, STD-1536, and STD-384, restricted either to hub baits or to nonhub baits. (D) Sensitivity and (E) PPV of 1-on-1, STD-1536, STD-384, and Screen-Seq, restricting all data sets to the four nonhub and two hub baits screened in Screen-Seq. Error bars indicate standard error.
Highly connected baits are known to be challenging for smart-pooling, as they can jeopardize the decoding of results. More precisely, a given smart-pool design can only identify a limited number of positives within a batch, and to increase this number would require a different design with more pools. Consequently, the problem is expected to be less pronounced with STD-1536 than STD-384, which has more preys per batch. Among the 12 baits, eight interact with at most 12 preys (which we grouped as nonhub baits) while the remaining four have between 19 and 35 interactors (grouped as hub baits). This is a high cutoff for defining nonhubs and hubs. It was chosen because the baits used in this study are highly connected overall, and a lower cutoff would have resulted in a nonhubs group with too few data points for a reliable analysis. Using the current cutoff, we estimate our nonhubs category would include the vast majority of proteins in the proteome.
We first compared STD-384 and STD-1536 with 1-on-1 in terms of sensitivity (the percentage of core hits from each method in the whole core data set). When considering nonhub baits, the STD pooling approach performed very well: STD-1536 is as sensitive as 1-on-1, and STD-384 is even significantly more sensitive (61.5% versus 44.2%, P-value 0.004 calculated using a binomial distribution; Fig. 4B). This shows that, even when preys are arrayed individually, a significant number of false-negatives occur and cannot be recovered in 1-on-1 analysis. Naturally, false-negative spots are also frequent in STD arrays, but due to the high extra redundancy the STD designs often succeed in coping with them. On the other hand, when considering hub baits, 1-on-1 is the most sensitive followed by STD-1536 and STD-384 (77.9%, 64.4%, and 49.0%, respectively). With hubs, the advantage conferred by the high STD redundancy is expected to be offset by the large number of positives, which can saturate the STD designs such that some interactions cannot be deciphered. Such saturation was evident in the pilot experiment (Supplemental Note) but did not clearly occur with STD-1536 or STD-384, where other factors must have come into play.
In terms of specificity (Fig. 4C), all three methods display very satisfactory Positive Predictive Values (PPVs, i.e., the percentage of candidate hits found by a given method that passed pairwise retest and ended up in core), averaging at 75%, 78%, and 91% for 1-on-1, STD-384, and STD-1536, respectively. 1-on-1 is the only one to significantly differ between nonhubs and hubs, decreasing from 88.5% to 71.7%, but this change is not surprising because it correlates with the increased 1-on-1 sensitivity.
The higher sensitivity of STD-384 over STD-1536 for nonhubs (Fig. 4B) presumably results from the lower density. For example, weakly positive spots are easier to score in 384 format: this allows identification of genuine Y2H-weak interactions, although it also results in slightly lower PPV for STD-384 (Fig. 4C). Additionally, miniaturization increases the influence of random fluctuations, making it harder to have consistent optimal conditions in 1536 format: Small variations in factors that result in lower signal or higher background have a stronger influence. In particular, in preliminary experiments we noticed that the amount of cells transferred to the target plates is an important parameter. Since the 1536-format pins are 0.7 mm in diameter compared to 1 mm for 384-format pins, they transfer fewer cells and the effect of experimental variability is greater. This observation is not limited to STD pooling but potentially applies to all high density experiments. Indeed, this explains why the two 1536-format assays used here, 1-on-1 and STD-1536, obtain sensitivities that are similar and significantly lower than STD-384 when screening nonhub baits. This is not contradictory with the high repeatability rates that we observed (see below), because our duplicated screens were designed to study the variability intrinsic to each method rather than that due to external parameters: The duplicates were performed in parallel, using the same batches of source and target plates and very similar experimental conditions.
The six baits screened with the Screen-Seq approach include two hub baits and four nonhub baits (Supplemental Table 1). Sensitivities (Fig. 4D) and PPVs (Fig. 4E) were calculated by restricting each data set to the Screen-Seq baits. Screen-Seq displays a high PPV similar to that of the other methods. In terms of sensitivity, Screen-Seq was surpassed by STD-1536 and STD-384 by factors of 1.8 and 1.1 for hubs, and it was surpassed by factors of 2.1 and 2.4 for nonhubs. Furthermore, the selected Screen-Seq baits were among the most connected in both the hubs and nonhubs groups. This explains the reduced sensitivity of STD-384 when restricting it to these six baits (Fig. 4D versus 4B), and biases the comparison in favor of Screen-Seq. Since nearly all baits in a proteome are nonhubs, we estimate that, in a large-scale interactome mapping project where the selected method is applied once or twice, STD would be at least twice as sensitive as Screen-Seq.
We then examined STD-SL, where three hub baits were screened against the STD-1536 pools arrayed in 384-format. STD-SL was not more sensitive than STD-1536 (Supplemental Fig. 3A), indicating that high plate density did not impact STD-1536. Concerning specificity, a single STD-SL candidate failed pairwise retest, entailing an almost perfect PPV (98.2%; Supplemental Fig. 3B). This shows that, with the STD-SL design, the problem of false-positives is virtually eliminated.
Based on our two replicates of each screening, we studied the repeatability of each method for core and FP hits (Supplemental Fig. 4). Core hits are largely repeatable for all methods: The fraction of PPIs identified in both replicates ranges from 65% for STD-1536 up to 86% for 1-on-1 and STD-SL. Concerning FP hits, they are almost never found in both repeats of STD data sets, as expected. However, they are surprisingly repeatable in 1-on-1 and also in Screen-Seq, although this is less significant since there were only seven Screen-Seq false-positives. This may be partly due to localized problems such as cross-contamination in the 1-on-1 master array, which could lead to repeatable false-positives in 1-on-1. Localized problems in the STD arrays would not have such a pronounced effect, because each prey is present in 13 pools that are distributed across the array: STD appears particularly robust with regards to contamination.
Due to its high redundancy, STD can provide valuable information in terms of error rates. False-positive spots were rare in our hands, but false-negatives were frequent, and the false-negative rate appeared largely variable between interactions. Furthermore, interactions yielding a strong signal in one STD series were often strong in other series: The “Y2H strength” of an interaction appears mostly reproducible. However, our data set is too small to draw conclusions on specific baits or preys: Application of highly redundant smart-pooling on a larger scale would be necessary to identify poorly performing baits and preys in a proteome.
Discussion
We have demonstrated that STD-based smart-pooling is a feasible and flexible strategy for mapping PPIs by Y2H at the scale of a complete ORFeome, and we have shown that the method can take advantage of high density 384 and 1536 formats. We have compared it with the established Screen-Seq high-throughput method (Rual et al. 2005), and with the labor-intensive “gold standard” one-on-one array-based Y2H (1-on-1) (Uetz et al. 2000).
We separately analyzed “nonhub” baits that were involved in at most 12 interactions, and the more highly connected “hub” baits. This cutoff was chosen because overall the baits used here have many interactions, but from a broader perspective it is quite a high cutoff. For example, only 42 Saccharomyces cerevisiae proteins, representing <1% of the proteome, are involved in more than 12 interactions in the “Y2H-Union” data set (Yu et al. 2008), which merges the three proteome-wide S. cerevisiae Y2H data sets published to date (Uetz et al. 2000; Ito et al. 2001; Yu et al. 2008). The nonhubs in this study are therefore representative of the vast majority of proteins in an organism and can serve as a useful guide for choosing an approach, while the hubs are informative in that they represent the worst-case scenario for smart-pooling methods.
Every candidate interaction underwent pairwise retest in quadruplicate. This showed that all methods were highly specific in our hands: At least 75% of each method's candidates retested successfully (91% for STD-1536). Screen-Seq is the least sensitive by a factor of up to 2.4 (for nonhubs versus STD-384). When considering nonhub baits, STD-384 and to a lesser extent STD-1536 were very successful: Their sensitivity attains or even exceeds that of 1-on-1, with a 39% increased sensitivity for STD-384 compared to 1-on-1, despite being much more cost- and labor-efficient. This demonstrates the advantage conferred by highly redundant STD pools. As anticipated, STD pooling performed less well with the highly connected baits, yet it remained satisfactory: Sensitivity was intermediate between Screen-Seq and 1-on-1, and as expected due to its smaller batch size, STD-1536 was more resilient to hubs than STD-384. Because the large majority of proteins in C. elegans and other organisms are not PPI hubs (Barabasi and Oltvai 2004; Gandhi et al. 2006), and because of the previously discussed large cutoff value used for defining nonhubs in this study, STD could be very useful as a highly sensitive and efficient first pass for large-scale interactome mapping. Any exceptionally strong hubs could be subsequently screened more deeply using another method such as 1-on-1, or by sequencing positive colonies that cannot be decoded unambiguously in the STD screen.
STD-1536 and STD-384 require five and six plates, respectively, while Screen-Seq fits on a single plate and 1-on-1 needs 17 plates. We have shown that, due to its high redundancy, the STD method is not affected by de novo autoactivators, which arise by acquiring mutations during the screening process, and the Screen-Seq step of cycloheximide counter-selection (Rual et al. 2005) can be safely skipped. Additionally, positives are directly identified in STD pooling, whereas Screen-Seq resorts to colony picking and sequencing (Fig. 3). Altogether, we estimate that the STD workload and costs are approximately three times higher than those of Screen-Seq, while coverage is increased at least twofold. In contrast, performing three repeats of Screen-Seq only improves coverage by 30% relative to single-pass Screen-Seq (Yu et al. 2008).
Comparing now with 1-on-1, since the Y2H screening steps are identical, the STD approach is approximately three times more cost- and labor-efficient, while being in fact more sensitive except for the few strong hub baits. STD also requires an initial investment to build the STD pools, but this is a one-time expenditure, as the pools can be copied and used many times. In addition, we designed and built intermediate micropools, which can be simply superposed to generate larger STD pools of various sizes, such as STD-1536 and STD-384 used here. This strategy minimizes the costs of building STD pools and provides greatly increased flexibility: The complex cherry-picking step for building micropools is performed a single time, and building STD pools of diverse pool and batch sizes is then a quick and cheap procedure, allowing adaptation of the pooling design to specific assay conditions.
Two other smart-pooling methods have been recently used to map PPIs (Jin et al. 2006, 2007) and protein–DNA interactions (Vermeirssen et al. 2007). However, they relied on designs that lack flexibility and possess an extra redundancy of at most one. This limits their ability to deal with the high false-positive and false-negative rates that are common in many assays, so that identifying the positives in these studies required sequencing positive colonies or retesting many ambiguous candidates. In contrast, STD is very flexible and one can choose a high extra redundancy if desired, for example 10 as used in this study. This allows us to successfully deal with high levels of noise, without any need for sequencing and without generating large numbers of low-confidence candidates, as shown by the high PPV values obtained with our STD designs.
In summary, we showed the application of the STD pooling strategy in ORFeome-wide Y2H screening and compared it with established high-throughput approaches, one-on-one array-based Y2H (Uetz et al. 2000) and Screen-Seq (Rual et al. 2005). Screen-Seq remains an appropriate method for quickly producing low-coverage interactomes, while STD appears as the method of choice for obtaining maps with higher coverage. STD pooling is also more powerful and flexible than other recently employed pooling designs (Jin et al. 2006, 2007; Vermeirssen et al. 2007). We expect that STD-based smart-pooling can be applied in other large-scale functional genomics experiments that rely on a basic yes-or-no test to identify rare positive events, provided that pools can be tested and yield a positive signal if they contain at least one positive, such as yeast one-hybrid, drug screening (e.g., Kainkaryam and Woolf 2008), or PCR- or hybridization-based analyses (e.g., Wu et al. 2008).
Methods
Details on the STD designs
In theory, with a redundancy of 13 and a design comprising 169 pools per batch (as we used in our STD pooling), STD can make pools for up to 1313 preys per batch, although the pool size increases proportionately with the number of preys per batch. Going down from 1313, the extra redundancy starts at zero and increases by one each time the exponent decreases. For example, between 14 (131 + 1) and 169 (132) preys per batch, any two preys co-occur in at most one common pool (leaving an extra redundancy of 11, as in the worm micropools); while between 170 (132 + 1) and 2197 (133) preys per batch, a pair of preys co-occurs in at most two pools (leaving an extra redundancy of 10, as in STD-1536 or STD-384).
Every 12 sets of worm micropools (169 preys per set) is a collection of subdesigns of an STD design with 2028 preys per batch (whose extra redundancy is 10). They can therefore be superposed to obtain the original STD design. Each individual set of micropools is also isomorphic to a smaller STD design, and can be used as an STD pooling batch in its own right, with an extra redundancy of 11. When two or six consecutive micropool sets are superposed to obtain STD-1536 or STD-384, the resulting designs are again isomorphic to an STD design with 228 or 1014 preys per batch, respectively, and therefore they both have an extra redundancy of 10. More specifically, worm micropools are subdesigns of STD(2028;13;13) isomorphic to STD(169;13;13), and were superposed to obtain designs isomorphic to STD(338;13;13) for STD-1536 and STD(1014;13;13) for STD-384 (see Thierry-Mieg 2006a for details).
Building STD pools
The sources were all Worm ORFeome v1.1 and v3.1 AD plates (11001 to 11114 and 31001 to 31022). The source plates were thawed at room temperature, inoculated in 96-format deep well plates containing SD-Trp liquid media, and incubated at 30°C for 2 d. After resuspension by shaking, micropools were assembled in 96-format deep well plates by cherry-picking using a Tecan Freedom EVO liquid handling robot (Tecan Group Ltd.). The robot was programmed directly in GWL (Supplemental Data), which optimized the process. STD-1536 and STD-384 pools were generated in 384-format and 96-format (STD-384 only) by superposing the appropriate micropool plates with a Tecan Aquarius MultiChannel Pipetting robot (Tecan Group Ltd.). All pools were frozen and stored at −80°C with 20% glycerol.
Handling 1-on-1 and STD arrays
Before Y2H screening, 1-on-1 or STD arrays glycerol stock plates were thawed at room temperature, mixed thoroughly with a plate shaker and transferred to SD-Trp agar plates with a “BM3-SC+Carousel” robot (S&P Robotics). After incubation, this set of “master” agar plates was replicated into multiple copies (up to eight), which could each be used either for screening or as a source for further replications. However, fresh arrays should still be occasionally remade from glycerol stock, because the STD arrays begin losing representation after more than five sequential replications (data not shown). The arrays appear fully functional after being stored at 4°C for at least 2 mo, although in this study we used them within 1 wk after replication to avoid confounding factors and guarantee the highest data quality.
Y2H screening with 1-on-1 and STD arrays
The Y2H screening with 1-on-1 and STD arrays was performed using a “BM3-SC+Carousel” robot (S&P Robotics) following the previously reported protocol (Uetz et al. 2000), except that the diploid selection step on SD-Leu-Trp was skipped: Preliminary experiments showed that, in our hands, including this step did not result in any improvements, perhaps because the robotic replication step may not be fully effective in transferring all components of a colony spot, so that the additional replication step compensates any gains from the diploid selection. We used pins of 1 mm diameter for 384-format and 0.7 mm for 1536-format. 1-on-1 spots were scored manually using the in-house ColonyImager image-processing program (H Ding and C Boone, unpubl.) as positive or negative. Each prey is present in duplicate on the 1-on-1 arrays; a 1-on-1 hit obtained a confidence score of Weak if it was positive in a single spot and Strong if it was positive in both duplicate spots. STD spots were scored similarly, except we used four discrete levels for each spot: strong (clear positive) or weak (smaller than strong but well above background) for positives, and none (no detectable signal) or faint (barely above background, most likely negative) for negatives. These results were transformed into a suitable XML format with Perl scripts, and decoded with interpool (Thierry-Mieg and Bailly 2008). The “distance” parameter δ was chosen to fit our experimental conditions. It turned out that false-positives were relatively rare while false-negatives were common, leading us to use a very sensitive distance (δNONE = 2, δFAINT = 1, δWEAK = 4, δSTRONG = 6). Clearly this choice did not strongly compromise specificity, as shown by the STD PPV values obtained after pairwise retesting (Fig. 4C). All relevant scripts, programs, and data files are available (Supplemental Data). A confidence score was attributed to each STD hit, depending solely on the number of putative false-negative spots for the hit. Specifically, “none” spots carry a cost of 2 and “faint” spots 1, and summing over all false-negatives for a hit yields a total cost; if this total cost is at most 4 the confidence score is 5, if it is up to 8 the score is 4, and so on until reaching the lowest confidence score of 1 if the total cost is between 17 and 20. All results were imported into a custom database for further analysis.
Y2H screening with the Screen-Seq approach
The 188 preys from every two worm ORFeome plates were pooled together. All resulting pools were assembled into one 96-well plate to generate the so-called “superpool” plate. Each bait was screened against the superpool plate using the method reported before (Rual et al. 2005). At most three positive colonies were picked from each spot, and prey inserts were amplified by colony PCR and sequenced for identification.
Pairwise retest
Retests were performed in quadruplicate by scoring a single phenotype of the HIS reporter in 96-format on agar plates, using 5 μL of bait and 5 μL of prey fresh cultures from archival stocks. Each retest was scored as negative, weak, or strong (0, 1, or 2, respectively). Summing over the four replicates, we obtained a retest score between 0 and 8 for each hit. Core hits are those whose retest score was at least 6, while hits with scores at most 2 were classified as FP and the remaining hits with intermediate retest scores were classified as noncore.
Author contributions
X.X., D.E.H., M.V., C.B., and N.T.M. conceived the project. X.X. and N.T.M. designed the experiments. J.F.R., T.H.K., and N.T.M. performed the pilot project experiments. N.T.M. designed the STD arrays, and X.X., D.E.H., and N.T.M. built the STD arrays. X.X. performed the ORFeome-wide Y2H experiments and scored the plates. X.X. and N.T.M. performed the computational analyses, produced the figures, and wrote the manuscript.
Acknowledgments
We thank Haiyuan Yu, Jingjing Li, and Olivier Francois for help with the statistical analysis. This work was supported by a Canadian Cancer Society grant awarded to C.B., and grant R01-HG001715 from the National Human Genome Research Institute of the National Institutes of Health awarded to M.V. M.V. is a “Chercheur Qualifié Honoraire” from the “Fonds de la Recherche Scientifique” (FRS-FNRS, French Community of Belgium).
Footnotes
[Supplemental material is available online at www.genome.org. The protein interactions from this publication have been submitted to the IMEx (http://imex.sf.net) Consortium through IntAct (PMID 17145710) and assigned the identifier IM-11695.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.090019.108.
References
- Barabasi AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Bruno WJ, Knill E, Balding DJ, Bruce DC, Doggett NA, Sawhill WW, Stallings RL, Whittaker CC, Torney DC. Efficient pooling designs for library screening. Genomics. 1995;26:21–30. doi: 10.1016/0888-7543(95)80078-z. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction data sets. Nat Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Hazbun T, Michaud GA, Salcius M, Predki PF, Fields S, Huang J. A pooling-deconvolution strategy for biological network elucidation. Nat Methods. 2006;3:183–189. doi: 10.1038/nmeth859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Avramova L, Huang J, Hazbun T. A yeast two-hybrid smart-pool-array system for protein-interaction mapping. Nat Methods. 2007;4:405–407. doi: 10.1038/nmeth1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainkaryam RM, Woolf PJ. poolHiTS: A shifted transversal design based pooling strategy for high-throughput drug screening. BMC Bioinformatics. 2008;9:256. doi: 10.1186/1471-2105-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Milstein S, Hao T, Rosenberg J, Li N, Sequerra R, Bosak S, Doucette-Stamm L, Vandenhaute J, Hill DE, et al. C. elegans ORFeome version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 2004;14:2064–2069. doi: 10.1101/gr.2496804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, et al. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein–protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg N. A new pooling strategy for high-throughput screening: The shifted transversal design. BMC Bioinformatics. 2006a;7:28. doi: 10.1186/1471-2105-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg N. Pooling in systems biology becomes smart. Nat Methods. 2006b;3:161–162. doi: 10.1038/nmeth0306-161. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg N, Bailly G. Interpool: Interpreting smart-pooling results. Bioinformatics. 2008;24:696–703. doi: 10.1093/bioinformatics/btn001. [DOI] [PubMed] [Google Scholar]
- Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Deplancke B, Barrasa MI, Reece-Hoyes JS, Arda HE, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Brent MR, et al. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat Methods. 2007;4:659–664. doi: 10.1038/nmeth1063. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu L, Close TJ, Lonardi S. Deconvoluting BAC-gene relationships using a physical map. J Bioinform Comput Biol. 2008;6:603–622. doi: 10.1142/s0219720008003564. [DOI] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Zhang H, Stanyon CA, Tromp G, Finley RL., Jr A strategy for constructing large protein interaction maps using the yeast two-hybrid system: Regulated expression arrays and two-phase mating. Genome Res. 2003;13:2691–2699. doi: 10.1101/gr.1134603. [DOI] [PMC free article] [PubMed] [Google Scholar]