Abstract
MicroRNAs (miRNAs) are known to post-transcriptionally regulate target mRNAs through the 3′-UTR, which interacts mainly with the 5′-end of miRNA in animals. Here we identify many endogenous motifs within human 5′-UTRs specific to the 3′-ends of miRNAs. The 3′-end of conserved miRNAs in particular has significant interaction sites in the human-enriched, less conserved 5′-UTR miRNA motifs, while human-specific miRNAs have significant interaction sites only in the conserved 5′-UTR motifs, implying both miRNA and 5′-UTR are actively evolving in response to each other. Additionally, many miRNAs with their 3′-end interaction sites in the 5′-UTRs turn out to simultaneously contain 5′-end interaction sites in the 3′-UTRs. Based on these findings we demonstrate combinatory interactions between a single miRNA and both end regions of an mRNA using model systems. We further show that genes exhibiting large-scale protein changes due to miRNA overexpression or deletion contain both UTR interaction sites predicted. We provide the predicted targets of this new miRNA target class, miBridge, as an efficient way to screen potential targets, especially for nonconserved miRNAs, since the target search space is reduced by an order of magnitude compared with the 3′-UTR alone. Efficacy is confirmed by showing SEC24D regulation with hsa-miR-605, a miRNA identified only in primate, opening the door to the study of nonconserved miRNAs. Finally, miRNAs (and associated proteins) involved in this new targeting class may prevent 40S ribosome scanning through the 5′-UTR and keep it from reaching the start-codon, preventing 60S association.
MicroRNA is commonly thought to have two functions in post-transcriptional regulation, mRNA degradation and inhibition of mRNA translation (Pillai 2005; Engels and Hutvagner 2006; Valencia-Sanchez et al. 2006; Filipowicz et al. 2008). Although partial complementary sequences between miRNA and its targets (mostly in animal) are associated with translational repression, there is substantial evidence of decreased mRNA levels even with imperfect base-pairing (Bagga et al. 2005; Jing et al. 2005; Lim et al. 2005; Giraldez et al. 2006; Sood et al. 2006), suggesting a miRNA role in mRNA destabilization and a nonexclusive relationship between degradation and repression. It has been suggested a miRNA may target ∼200 mRNAs (Krek et al. 2005), with varying degrees of protein repression (Baek et al. 2008; Selbach et al. 2008), though the number of predicted targets can range in the thousands. A major reason for these false positives lies in the partially complementary matches between miRNA and its targets.
miRNA target sites are known to lie in 3′-UTR in animals. The first discovered miRNA, lin-4 in Caenorhabditis elegans, was found to regulate developmental timing by targeting multiple sites in 3′-UTR of lin-14 (Wightman et al. 1993). Following the discovery of miRNAs in other animals, an early computational study identified some of the 5′-ends of miRNAs as complementary to 3′-UTR sequences (Pillai 2005). In the meanwhile, many miRNAs predicted due to highly conserved sequences across species were mostly confirmed (Suh et al. 2004). This high conservation in conjunction with 8-mer perfect complementary sequences of miRNA led to a genome-wide highly conserved 8-mer UTR motif study which found significant miRNA target sites only in the 3′-UTR through the 5′-end of miRNA (Xie et al. 2005). The significance of this 5′-end of miRNA targeting 3′-UTR sites was recently confirmed by a proteomics study which showed superior protein inhibition capacity for the 3′-UTR sites over those in the coding region (Baek et al. 2008).
There are several gaps in previous studies. Genome-wide motif studies have focused on highly conserved motifs, not including less conserved or human-specific motifs (Xie et al. 2005). This approach has the intrinsic limitation of not identifying species-specific sequences, including nonconserved miRNAs. A later study looking for mRNA regions of interaction restricted its search to the 5′-end of miRNAs (Baek et al. 2008), while previous studies suggested a role for the 3′-end of a miRNA in either complementing a seed match or compensating for an imperfect one (Doench and Sharp 2004; Kiriakidou et al. 2004; Kloosterman et al. 2004; Grimson et al. 2007). Most miRNA target verification experiments have only used 3′-UTR interaction sites since many studies had shown miRNA effects with only portions of 3′-UTR. Even the first lin-4 and lin-14 experiments used not the whole mRNA, but rather the 3′-UTR together with coding region (Wightman et al. 1993). On the other hand, a few experiments have indicated possible target sites in the 5′-UTR (Jopling et al. 2005; Lytle et al. 2007; Orom et al. 2008).
We report here, based on both hybridization energy and sequence matches, many endogenous motifs within human 5′-UTRs specific to the 3′-ends of miRNAs. Rather than suggesting possible miRNA interactions with other regions of mRNA, we report combinatory interactions between a single miRNA and both end regions of an mRNA, based on our finding that many miRNAs contain significant interaction sites with mRNA 5′-UTR and 3′-UTR motifs through their 3′- and 5′-end sequences, respectively. As a model system, we experimentally verified that hsa-miR-34a function depends on both UTR sites of AXIN2. We propose here a new miRNA target class containing simultaneous 5′- and 3′-UTR interaction sites. Since we identify such sites in genes showing large-scale protein changes upon deletion or overexpression of all four miRNAs used in Baek and colleagues' study (Baek et al. 2008), this target class can serve as an efficient screening tool for identifying real targets, especially in the case of nonconserved miRNAs or target sites.
Results
Presence of miRNA interaction sites in human 5′-UTR
We checked for genome-wide miRNA interaction motifs in human 5′-UTR and 3′-UTR. Xie et al. (2005) have reported conserved miRNA motifs in the 3′-UTR but not in the 5′-UTR or in coding sequences. We used the same UTR motif data set sent to us by the authors but defined new conservation classes C = 0 (nonconserved but human-enriched), 1 (minimally conserved and human-enriched), and ≥10 (highly conserved). To determine seed and nonseed region effects, all mature miRNAs were downloaded from miRBase (release 11.0) (Griffiths-Jones et al. 2008) and split into their respective 5′- and 3′-ends, making miRNA halves. Following thermodynamic searches for half miRNA–UTR motif interactions using RNAhybrid (Kruger and Rehmsmeier 2006), we treated only consecutively matched sequences as signals. To calculate significance, total numbers of pairwise interactions between half-miRNAs and UTR motifs were compared with the numbers of interactions with shuffled UTR motifs generated 1000×.
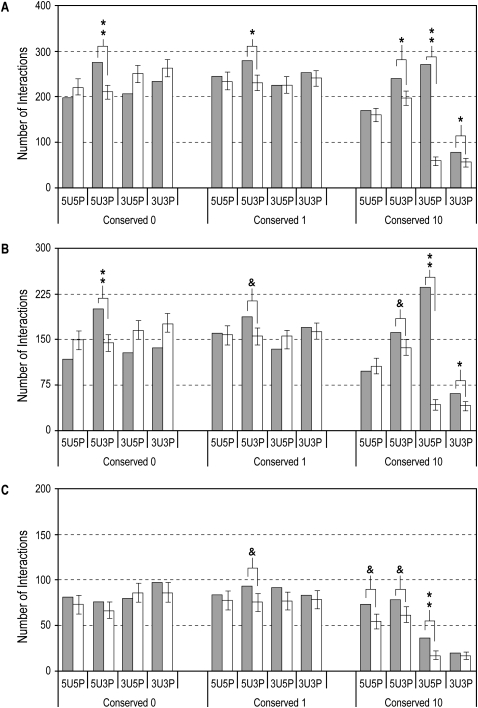
In these analyses, we identified 5′-UTR motifs (5U) which interact significantly with the miRNA 3′-end (3P) in all conservation categories (5U3Ps in Fig. 1A), most significantly in the case of C = 0. 3′-UTR motifs (3U), on the other hand, show significant interactions with miRNA only in the case of highly conserved 8-mers (C ≥ 10) consistent with previous reports (Conserved 10: 3U5P and 3U3P in Fig. 1A). Besides the most significant and well-known interaction of 3U5P, our identification of 3U3P interaction is in accordance with previous findings that the 3′-end of a miRNA may either complement a seed match or compensate for an imperfect one (Doench and Sharp 2004; Kiriakidou et al. 2004; Kloosterman et al. 2004; Grimson et al. 2007). Our new finding of 5U3P interaction was also observed with human-enriched 5′-UTR motifs when we followed the conservation score of Xie et al. (2005) (Methods and Supplemental Fig. 1).
Figure 1.
Analysis of predicted interactions between 8-mers from different conservation classes and miRNAs. Closed bars indicate number of predicted interactions between 5′-UTR or 3′-UTR 8-mer sequences (indicated by 5U or 3U, respectively) and 5′- or 3′-ends (indicated by 5P or 3P, respectively) of a full set of mature miRNAs (A), of conserved miRNAs (B), and of nonconserved miRNAs (C). Open bars correspond to mean number of interactions after 1000 shuffling iterations, and error bars indicate standard deviations. (**) P < 5 × 10−5; (*) P < 5 × 10−3; (&) P < 0.05.
Viewed in terms of conserved and nonconserved miRNAs, interactions with conserved miRNAs show a trend similar to the one above, differing only in the levels of significance (Fig. 1B). Interestingly, 5U3P interactions with nonconserved miRNAs lack significance for C = 0 motifs (Fig. 1C), the 5U3P signal in C = 0 in Figure 1A coming from that of conserved miRNAs. We also observed significant interactions between highly conserved 5′-UTR motifs and the 5′-end (5U5P) of nonconserved miRNAs (Fig. 1C).
In conjunction with the significant interaction between the seed region of a miRNA and the 3′-UTR, the preferential 5′-UTR interaction with the 3′-end of miRNA raises the question whether a common miRNA may target both UTRs of an mRNA by interacting with different ends of the miRNA. Based on the significant data in Figure 1A, 37 common miRNAs identified between 5U3P (C = 0 and 1: total 250 miRNAs) and 3U5P (C = 10: total 116 miRNAs) cases are listed in Supplemental Table 1. When these kinds of motifs exist in a single gene, will they be regulated by a single miRNA?
hsa-miR-34a targets AXIN2 through both UTRs
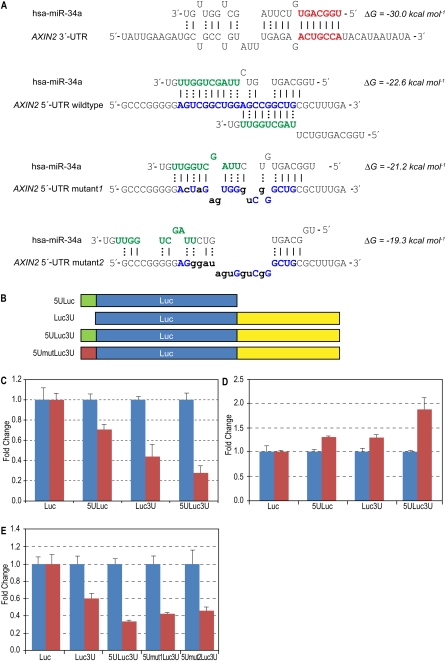
A highly conserved human miRNA, hsa-miR-34a, has such interaction sites in the human gene AXIN2 (Fig. 2A). Though miR-34a is not in Supplemental Table 1, the 5′-end was predicted to interact with three highly conserved (and one nonconserved) AXIN 3′-UTR sites, and the 3′-end with two overlapping 5′-UTR sites (Fig. 2A) present only in human and mouse but enriched in human 5′-UTRs (Supplemental material). We used hsa-miR-34a and AXIN2 as a model system to verify simultaneous UTR interactions. Since interactions between miRNA and 3′-UTR are well-established, we focused on the 5′-UTR interaction sites, using only minimal interaction sequences of 36-mer in the experimental constructs. As shown in Figure 2A, the hsa-miR-34a effects on this 36-mer should mostly come from the 3′-end. Note that the full 3′-UTR inserted in the construct is 1408 nucleotides (nt) long.
Figure 2.
Human miRNA hsa-miR-34a and target AXIN2. (A) Predicted interactions between hsa-miR-34a and AXIN2 UTR sequences. Extended seed match between the 5′-end of miR-34a and one of the 3′-UTR binding sites is shown in bold red. All predicted 3′-UTR sites are marked in the Supplemental material. Overlapping interactions between the 3′-end of miR-34a and the 5′-UTR inserted sequences are shown in bold blue. Energy was calculated using RNAhybrid. (B) Schematic showing vector constructs containing firefly luciferase reporter gene used in transfection experiments. The 5′-UTR and 3′-UTR inserts are indicated as 5U and 3U, respectively. (C) Luciferase expression fold change with miR-34a (red bars) normalized with negative control RNA oligo (blue bars). Firefly luciferase protein expression was normalized with Renilla luciferase protein. (D) Reporter constructs were co-transfected with anti-miR-34a oligo (red bars, Ambion, product ID, AM11030) and normalized with negative control RNA oligo (blue bars). (E) Effect of mutations in the 5′-UTR site—luciferase protein levels when reporter constructs were co-transfected with miR-34a (red bars) or negative control (blue bars). Error bars in panels C–E represent standard deviation from triplicate experiments.
Reporter gene assay of MCF-7 cells revealed that miR-34a down-regulated luciferase expression in constructs containing either the 5′-UTR (5ULuc) or 3′-UTR (Luc3U) alone. When both AXIN2 UTR sites were present (5ULuc3U), luciferase expression was further repressed by miR-34a (Fig. 2C). In order to identify endogenous miRNA effects in addition to those exogenously induced, we blocked endogenous miR-34a using inhibitor antisense RNA oligo. 5ULuc3U expression was greater than that of 5ULuc or Luc3U, suggesting that the 5′-UTR of AXIN2 together with the 3′-UTR are functional target sites for miR-34a in the cells (Fig. 2D). In addition, the fold change of 5ULuc3U 1.88 is greater than with the addition of 5ULuc and Luc3U 1.61. Considering the many interaction sites in the 3′-UTR, the synergetic 5′-UTR effect on endogenous miRNA function is remarkable. These data suggest that, in conjunction with the 3′-UTR, the 5′-UTR of AXIN2 plays a role in miRNA-mediated repression in human cells beyond fine-tuning. In order to confirm the sequence specificity of 5′-UTR effects, we created two constructs with sites mutated (5Umut1Luc3U and 5Umut2Luc3U). Separate luciferase experiments inducing hsa-miR-34a showed reduction of repression when the 5U interaction sites are mutated (Fig. 2E).
hsa-miR-34a interacts with WNT1 in a 5′-UTR sequence-specific way
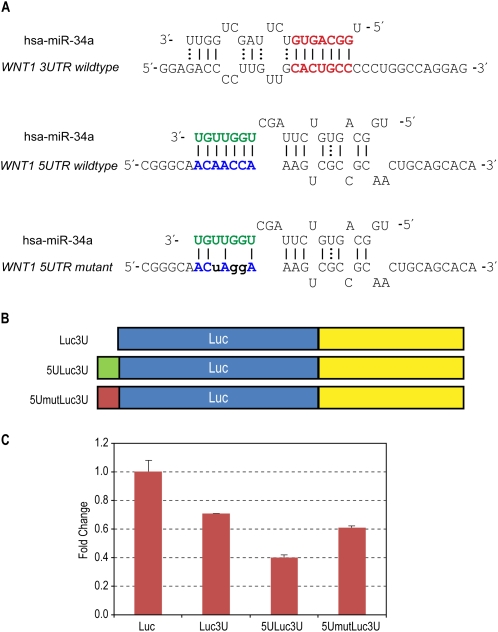
We were interested in other genes presenting similar sequence specificity of 5′-UTR interaction sites. An interesting interaction occurs between the 5′-UTR of WNT1 and the 3′-end of hsa-miR-34a, there being 7-mer consecutive matches without any GU wobbles and a target A-nucleotide corresponding to the end miRNA (Fig. 3A), mirroring TargetScan's t1A interaction in the 3′-UTR (Lewis et al. 2005). In order to establish 5′-UTR interaction site specificity, we again used only 39-mer sequences containing this interaction site but at a different interaction position from the AXIN2 case, while using entire 3′-UTR sequences. Here we changed only three sequences to disrupt the 7-mer canonical Watson–Crick base-pairing (Fig. 3A).
Figure 3.
Human miRNA hsa-miR-34a and target WNT1. (A) Predicted interactions between hsa-miR-34a and WNT1 UTR sequences. Extended seed match between the 5′-end of miR-34a and the 3′-UTR binding site is shown in bold red. Interactions between the 3′-end of miR-34a and the 5′-UTR inserted sequences are shown in bold blue. (B) Schematic of vector constructs containing firefly luciferase reporter gene used in transfection experiments. 5′-UTR and 3′-UTR inserts are indicated as 5U and 3U, respectively. (C) Effect of 5′-UTR site in wild-type and mutant form on luciferase expression when treated with miR-34a. Renilla-normalized luciferase expression was normalized with negative control RNA oligo. Error bars represent standard deviation from triplicate experiments.
Reporter gene assay of MCF-7 cells showed that miR-34a down-regulated constructs containing both WNT1 UTR sites (5ULuc3U) to a greater extent than those constructs containing 3′-UTR (Luc3U) alone (Fig. 3C). This additional repression was relieved when the 5U interaction sites were mutated (Fig. 3C), demonstrating that miRNA interaction occurs in a sequence-specific manner. We observed similar miR-34a functional dependency on the 5′-UTR sequences in the other gene (data not shown).
lin-4-like artificial miRNA interacts with lin-28-like 5′-UTR sites in a sequence-specific way
To establish that our finding of sequence-specific 5′-UTR interaction in addition to 3′-UTR interaction was not limited to a single miRNA or cell line, we chose the C. elegans lin-4 and lin-28 pair for human cell line validation experiments. The 3′-UTR of lin-28 contains a single canonical target site conserved in the lin-28 homologs of human, mouse, and chimpanzee whereas the single 5′-UTR site predicted to bind with the 3′-end of the miRNA is lacking in all of the homologs. Considering the lower physiological temperature of C. elegans and lack of homologs in human, we prepared artificial 5U3P (5′-UTR and the 3′-end miRNA) sequence pairs containing 8-mer consecutive matches without GU wobbles, while keeping 3U5P interaction the same as in the lin-4 and lin-28 case (5ULuc3U in Fig. 4A). The construct having the 5′-UTR sequences of lin-28 was used as the 5′-UTR mutant site (5UmutLuc3U) corresponding to lin-4-like artificial miRNA (lin-4-like) to verify 5′-UTR interaction site specificity. Note that the length of the inserted sequences and interaction site position are different from the miR-34a experiments to establish the sequence specificity.
Figure 4.
Effect of 5′-UTR interaction site with lin-4-like on reporter expression levels. (A) Predicted interactions between lin-4-like and lin-28-like UTR sequences. The functional strand of the lin-4-like contains an intact cel-lin-4 seed region (red) while the 3′-end is modified (green). There is an extended seed match between the 5′-end of lin-4-like and the wild-type lin-28 3′-UTR binding site (bold red). The 3′-end of lin-4-like is complementary to the artificial lin-28-like 5′-UTR binding site created by introducing a few GC base pairs (bold italics) to form a perfect match. The wild-type lin-28 5′-UTR presents an imperfect match (bold blue). Structure and energy calculations were carried out using RNAhybrid. (B) Schematic showing vector constructs containing firefly luciferase reporter gene used in transfection experiments. lin-28-like 5′-UTR segments containing 8-mer perfectly matched and mutated sites are indicated as 5ULuc3U and 5UmutLuc3U, respectively. (C) Fold changes of Renilla-normalized firefly luciferase expression levels upon co-transfection with lin-4-like (red bars) with respect to nonspecific hsa-miR-16 (blue bars). Error bars represent standard deviation recorded from eight pooled replicates.
Reporter gene assay of HEK293 cells showed that luciferase expression reduction due to lin-4-like was diminished when the 5′-UTR interaction site was mutated (Wilcoxon rank-sum test P < 0.005 for 5ULuc3U and 5UmutLuc3U in Fig. 4C). It is clear that mismatches in the 5′-UTR corresponding to the 3′-end of lin-4-like disrupt interaction.
Translationally repressed genes contain both UTR sites
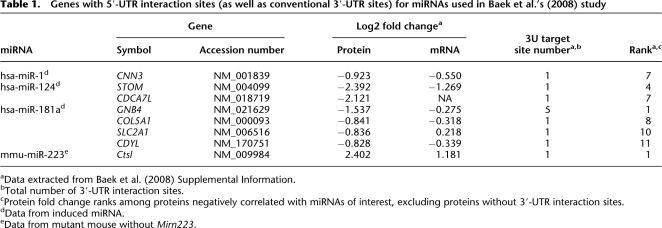
To what extent is this finding generalizable in endogenous miRNAs? A recent study measured thousands of protein levels in response to miRNA changes (Baek et al. 2008). One of the experiments involved removing endogenous Mirn223, while the rest induced additional miRNAs, finding hundreds of proteins negatively correlated to each of the four miRNAs used, many of the proteins having conventional 3′-UTR sites. We wondered if 5′-UTR interaction sites exist for the 3′-end of miRNAs in genes showing large protein- fold changes and having the 3′-UTR sites identified in Baek and colleagues' study (Baek et al. 2008). For each miRNA, we checked the 10 genes whose protein levels were most changed and found both UTR sites present in at least one case for all miRNAs. These miRNAs and targets are in Table 1, including CDYL, which ranks 11th among hsa-miR-181a targets in terms of fold change. Not only do all targets in Table 1 have highly significant fold changes; all protein fold changes greatly exceed mRNA fold changes, showing significant translation repression. Note that mmu-miR-223 targeting both UTRs of Ctsl and hsa-miR-181a targeting GNB4 are cases of maximal protein fold change, while GNB4 mRNA change is only modest. We thus propose a new miRNA target class containing simultaneous interactions of 5U3P and 3U5P.
Table 1.
Genes with 5′-UTR interaction sites (as well as conventional 3′-UTR sites) for miRNAs used in Baek et al.'s (2008) study
aData extracted from Baek et al. (2008) Supplemental Information.
bTotal number of 3′-UTR interaction sites.
cProtein fold change ranks among proteins negatively correlated with miRNAs of interest, excluding proteins without 3′-UTR interaction sites.
dData from induced miRNA.
eData from mutant mouse without Mirn223.
miBridge targets
This new miRNA target class can greatly diminish the number of predicted targets. While the number of conserved miRNA targets predicted for conserved 3′-UTR sites can reasonably be checked, there has been no practical way to study nonconserved miRNA targets due to the thousands of target predictions. Encouraged by the large protein fold change genes containing potential interaction sites in both their UTRs, we provide here potential targets containing both UTR sites, such as simultaneous interactions between 5′-UTR and the 3′-end together with 3′-UTR and the 5′-end of a miRNA (miBridge). While 3′-UTR interaction sites have been extensively studied, rules for 5′-UTR interaction need to be newly established. As a first attempt, we basically follow current 3′-UTR interaction rules except for the position restriction. Following initial interaction searches between halves of miRNAs and corresponding UTRs using RNAhybrid, we considered sequence matches. We set a −13 kcal/mol energy cutoff in the RNAhybrid parameter when there are consecutive sequence matches without GU wobbles, the same condition for 3′-UTR interaction (based on the interaction energy between the 5′-end of miR-1 and 3′-UTRs). We additionally constrained 3′-UTR interactions to match positions 2–7 from the 5′-end. Note that the calculation includes no conservation information. Predictions of miRNAs targeting genes with both 5′-UTR and 3′-UTR sites are available at http://sitemaker.umich.edu/miBridge (Supplemental Data Set).
The highly reduced target search space of miBridge now allows us to identify a human-specific miRNA target. SEC24D is one of our genes of interest due to its transport function. TargetScan (Lewis et al. 2005) predicts 102 miRNA families as regulating SEC24D (a combination of conserved and nonconserved family info files downloaded from http://www.targetscan.org/cgi-bin/targetscan/data_download.cgi?db=vert_42), while miBridge predicts two miRNA, miR-524-5p and miR-605, common to TargetScan. Since TargetScan treats miR-520d-5p and miR-524-5p as one family having the same targets, we investigate miR-605 for ease of comparison. The number of predicted targets of miR-605, a miRNA currently identified only in primate, is 4952 by TargetScan (from their nonconserved family info file). Based on TargetScan's prediction, miR-605 is highly unlikely to turn up as regulating SEC24D. However, we could easily select miR-605 and SEC24D as a miRNA functional pair based on miBridge target prediction. RNAhybrid calculated the interaction energy between the 3′-end of miR-605 and the 5′-UTR of SEC24D as −16.1 kcal/mol. The 5′-end of miR-605 interacts with the 3′-UTR at −16.7 kcal/mol with 9-mer matches without GU (Fig. 5A). Endogenous SEC24D levels in HeLa cells were measured using miR-605 induction or by blocking endogenous miR-605 in comparison with control miRNAs. Induction of miR-605 reduced SEC24D mRNA and protein levels, while anti-miR-605 increased SEC2D mRNA and protein levels, confirming miR-605's effect on endogenous SEC24D (Fig. 5). Therefore, initial candidate miR-605 turns out to be a true regulating miRNA.
Figure 5.
Human miRNA hsa-miR-605 and SEC24D. (A) Predicted sites on the 5′-UTR and 3′-UTR targeted by the 3′-end and 5′-end, respectively, of hsa-miR-605. (B) Western blot analysis of SEC24D. Protein extract (40 μg) from three days post-transfected HeLa cells was separated by SDS-PAGE and probed with anti-SEC24D monoclonal antibody, with actin (ACTB) as control. (C) Densitometric analysis of the Western blots. X-ray films were scanned with HP Scanjet 3570c (Hewlett-Packard) and quantified using NIH image software. (D) SEC24D mRNA expression fold change in HeLa cells two days post-transfection with Negative Control-1, Pre-mir-605, or anti-miR-605. Error bars represent standard deviation from a triplicate experiment.
Discussion
Translation repression has been reported to occur when a 3′-UTR target site for endogenous let-7a in HeLa cells is moved to the 5′-UTR (Lytle et al. 2007). We now show there exist many endogenous target sites in 5′-UTR for endogenous miRNAs, so that these 5′-UTR sites can contribute to miRNA function. The data in Figure 1A are intriguing in that (1) significant miRNA interactions in the 5′-UTR occur only with the 3′-end of miRNA (5U3P), and (2) such 5U3P significance seems to arise in highly conserved 8-mers and spread into less conserved but highly human present motifs (C = 0 and 1). Nonconserved sites have been explored under the assumption that each species or genome might employ them to attribute specificity in some manner (Farh et al. 2005). Considering that the 3′-end of miRNA family members (intraspecies) and those of some miRNAs across species differ, the 3′-end of miRNAs may contribute to gene- or species-specific target site recognition of the 5′-UTR. Dividing miRNAs into conserved and nonconserved ones, it seems that human-specific 5U motifs interact with preexisting miRNAs (Fig. 1B) and that human-specific miRNAs interact with preexisting 5U motifs (Fig. 1C). The significant 5U5P presence in the highly conserved UTR motifs and nonconserved miRNAs (Fig. 1C) may reflect an emergent feature of human-specific miRNAs, wherein miRNA and 5′-UTR are actively evolving in response to each other.
We used 36-mer sequences for the AXIN2 5′-UTR construct, which interacts mostly with the 3′-end of miR-34a. In contrast to 3′-UTR sites, which are well-dispersed across 1408 nt, making additive miRNA effects possible, the two 5′-UTR sites overlap, leaving no opportunity for additive effects. We expect to see four times higher 3′-UTR effects than with 5′-UTR, assuming the 5′-end represses translation in the 3′-UTR just as the 3′-end does in the 5′-UTR. Therefore, the contribution of AXIN2 5′-UTR sites in protein repression by hsa-miR-34a induction is no less than that of each site in the 3′-UTR (Fig. 2C). Of some interest are the endogenous miRNA effects on both UTRs in this pair (Fig. 2D). Not only is the inserted 5′-UTR site effect similar to that of the whole 3′-UTR (∼40× longer than the inserted 5′-UTR sequences), but the presence of both UTRs has a synergetic effect on miRNA function. Exogenous hsa-miR-34a effects on top of endogenous hsa-miR-34a function may lead to saturation of repression capacity with 5ULuc3U in Figure 2C, while repression of Luc3U is more easily achieved with exogenous miR-34a. Previous studies with exogenous miRNAs may have found significant effects where exogenous miRNAs compete minimally with endogenous miRNAs.
The endogenous miRNA effect on targets with both 5′-UTR and 3′-UTR is highlighted in Table 1. The largest fold change protein CTSL, due to mmu-miR-223 deletion, is identified as having both UTR sites for mmu-miR-223. Most salient in Table 1 is the low translational efficiency when both UTR interaction sites are present. In order to fully understand miRNA function, therefore, we advise the insertion of both 5′- and 3′-UTR sequences in miRNA functional experiments, which has rarely been done before. Previous miRNA functional experiments using 3′-UTR alone usually achieved ∼40%–60% protein reduction (Lim et al. 2005). We may see more protein reduction with 5′-UTR inclusion where interaction sites exist, as seen in Figures 2 and 3.
Considering the large number of potential targets with conventional 3′-UTR sites, prioritizing searches using miBridge targets can expedite real target identification, as shown in the case of miR-605 and SEC24D, where miBridge yielded two miRNAs rather than TargetScan's 102. We believe miBridge targets thus provide an avenue for exploring nonconserved miRNAs. Moreover, this new class of miRNAs and targets may fall into the class of translation blockers prior to the 40S ribosome reaching the translation start region, preventing 60S association (Wang et al. 2008), one possible miRNA mechanism of translation repression. These 5′-UTR-interacting miRNAs associated with proteins will provide large steric hindrance against ribosome scanning. A recent study reported that miRNA function was abolished when the interaction site context was changed from within the 3′-UTR to within the extended coding region through stop codon change, while siRNA function was retained (Gu et al. 2009). Middle-bulged miRNA interaction may require 80S ribosome-free 3′-UTR for interaction, while the 3′-end of miRNA interaction may be strong and/or bulky enough to compete with a smaller subunit 40S ribosome. It is also possible that the 3′-ends of miRNAs interact with 5′-UTR-associated proteins other than Argonaute proteins.
In this study, all interaction was calculated with miRNA halves. On the other hand, Smalheiser and Torvik previously provided computational evidence for the existence of long interactions (>10 nt) that do not arise preferentially from the 5′-end of miRNAs and are not biased toward the 3′-UTRs of putative targets (Smalheiser and Torvik 2004). Longer seed interaction deserves to be revisited. Finally, in order to establish interaction site specificity, we did not use full-length 5′-UTR. Studies with full 5′-UTR sequences may reveal further miRNA functions within this new target class.
Methods
Pre-processing of microRNA data
Mature human miRNA sequences were downloaded from miRBase, version 11.0. These were separated into two categories, conserved and nonconserved. We define a conserved miRNA as one that has a similarly named counterpart in at least one other species regardless of the percentage identity. For example, miR-34a exists in humans as well as mouse and many others whereas miR-1178, a nonconserved miRNA by our definition, exists only in humans. Following this, miRNAs were split into their respective 5′ and 3′-end halves.
Bioinformatic analysis
Xie et al. (2005) kindly provided us with data on conservation of all possible 8-mer sequences from aligned 5′-UTRs and 3′-UTRs among human, mouse, rat, and dog. Each 8-mer was listed along with the number of occurrences conserved in all four species (C), the number of occurrences in the human sequence (N), and the conservation rate (R) given by the ratio C/N, where 0 ≤ R ≤ 1. We created five motif conservation categories: (1) C = 0, nonconserved 8-mers ordered on decreasing N, (2) C = 1, 8-mers with exactly one conserved occurrence, ordered on decreasing N, (3) C ≥ 10, 8-mers with at least 10 conserved occurrences ordered on decreasing C and decreasing R, (4) positive MCS, and (5) negative MCS described below. Briefly, the motif conservation score (MCS, from Xie et al. [2005]) is reported as a Z-score calculated using binomial probability, MCS = (C – Np0)/[Np0(1 – p0)]1/2, where C is the number of conserved instances, N the number of occurrences in human, and p0 the estimated rate of conservation. We calculated p0 as the average conservation rate of all 65,536 8-mers. The top 540 highest scoring 5′-UTR and 3′-UTR 8-mers from each category above were then used for further analysis. RNAhybrid (Kruger and Rehmsmeier 2006) was used to search for potential interactions between the UTR motifs and each miRNA. As Doench and Sharp demonstrated the correlation between binding energy and fold repression (Doench and Sharp 2004), we set an energy threshold of −14 kcal/mol based on the RNAhybrid binding energy prediction for the CXCR4 siRNA seed region and the corresponding target site used in Doench and Sharp's paper. The results were then filtered for consecutive 8-mer matches with GU wobbles between the 8-mers and miRNA ends.
Statistical analysis
Shuffled 8-mers derived from the corresponding conservation category were used as controls to assess the significance of the number of interactions between motifs and miRNAs. The control data sets were generated 1000× and the number of interactions calculated as an average over these iterations. We assumed the distribution of number of interactions to be normal and calculated P-value using the Z-test.
Reporter gene constructs and assay for miR-34a experiments
The luciferase coding sequences were PCR-amplified and inserted between HindIII and BamHI sites of pcDNA3.1-Hyg(+), a mammalian expression vector (Invitrogen), to generate luciferase expression constructs. To make 3′-UTR constructs, the 3′-UTR of AXIN2 (NM_004655; +1 ∼ +1059) and WNT1 (NM_005430; +1 ∼ +1056) were amplified from genomic DNA of MCF-7 cells and cloned into the BamHI and NotI sites. The synthetic oligonucleotide containing 5′-UTR sequences targeted by miR-34a of AXIN2 (5′-GCCCGGGGGAGTCGGCTGGAGCCGGCTGCGCTTTGA, corresponding to +44 ∼ +79 among 314 nt of the 5′-UTR) and that of WNT1 (5′-CGGGCAACAACCAAAGTCGCCGCAACTGCAGCACAGAGC-3′, positions +141 ∼ +179 among 209 nt of the 5′-UTR) were inserted into NheI and HindIII sites upstream of luciferase vectors. The two AXIN2 5′-UTR mutants are 5′-GCCCGGGGGACTAGAGTGGGUCGGGCTGCGCTTTGA-3′ and 5′-GCCCGGGGGAGGGATAGTGGUCGGGCTGCGCTTTGA-3′ and WNT1 5′-UTR mutant is 5′-CGGGCAACTAGGAAAGTCGCCGCAACTGCAGCACAGAGC-3′. For the miR-34a induction experiments, each reporter construct (1–5 ng) was co-transfected with 20 pmol of negative control RNA oligo (Ambion, AM17110) or miR-34a precursor RNA oligo (Ambion, product ID PM11030) using Lipofectamine 2000 (Invitrogen) for 48 h. In experiments inhibiting endogenous hsa-miR-34a, 1–5 ng of each construct was co-transfected with 40 pmol of anti-miR-34a inhibitor (Ambion, product ID AM11030) or anti-miR negative control (Ambion, product ID AM17010). Fold change by miR-34a or miR-34a inhibitor was measured by a dual-luciferase assay kit (Promega), and the firefly luciferase activity normalized relative to a simultaneously transfected 1 ng of SV40-driven Renilla luciferase expression plasmid. Experiments were performed in triplicates.
Reporter gene constructs for lin-28-like
The synthetic oligonucleotides for lin-28-like UTR sequences were purchased from Integrated DNA Technologies, Inc. The sequences used in reporter gene constructs to mimic 5′-UTR and 3′-UTR sequences were 5′-GTGGTATTGTTGTTCTGTAAGCCACATAGGTTGTATTCTCTAGTTAACACATAGT-3′ and 5′-CACCTACCTCCTCAAATTGCACTCTCAGGGATTCTTTTTTTTTTCAAATAGAACT-3′, respectively. The corresponding mutated 5′-UTR sequence was 5′-GTGGTATTGTTGTTCTGTATATTTGATAGGTTGTATTCTCTAGTTAACACATAGT-3′, which contains the 5′-UTR sequences of lin-28. The expression reporter vector, pMIR-REPORT, was purchased from Ambion, Inc. (Cat. # AM5795). 5′-UTR sequences were cloned into the BamHI restriction site upstream of the luciferase coding sequence and the 3′-UTR sequences were cloned into the multiple cloning site using HindIII and SpeI. UTR sequences and their orientation in the constructs were confirmed by DNA sequencing (University of Michigan DNA sequencing core).
lin-4-like sequences
Strands that make up the lin-4-like duplex were purchased from Integrated DNA Technologies, Inc. The in-house designed sequences of the functional strand and opposing strand of lin-4-like were 5′-UCCCUGAGACCUGUGGCUUGA-3′ and 5′-AAGCCACAGGUCUCAGAAGUU-3′, respectively. The single-stranded molecules were later annealed using the manufacturer's protocol.
Cell culture and transfection for lin-28-like assay
HEK293 cells were grown to 80% confluence in Dulbecco Modified Eagle Medium (DMEM) with 10% fetal bovine serum. Cells were then trypsinized and plated in 12-well plates with ∼250,000–300,000 cells per well. Five hundred nanograms of each firefly reporter construct and 50 ng of internal control Renilla reporter pRL-tk (Promega, Cat. # E2241) were co-transfected with either 37 pmol of control miRNA (hsa-miR-16; Ambion, Inc., product ID PM10339) or 170 pmol of lin-4-like using Lipofectamine 2000 (Invitrogen). Owing to mismatches in the duplex we used, we increased the siRNA concentration to compensate for any inefficiency in annealing. Cells were lysed 24 h post-transfection and assayed for luciferase expression using the Dual-Luciferase Reporter Assay System (Promega, Cat. # E1910) and GloMax 96 Microplate Luminometer w/Dual injectors (Promega, Cat. # E6521) according to the manufacturer's protocol. Here, experiments were repeated two independent times in quadruplicate each time. Renilla-normalized luciferase values were normalized using values from a nonspecific miR-16 transfection. To determine if there was significant difference between 5ULuc3U and 5UmutLuc3U, we used the Wilcoxon rank-sum test to calculate P-values from the normalized luciferase values for each pair of constructs chosen.
5U3P site analysis using proteomics data
After downloading Supplemental Tables 2–5 of Baek et al.'s (2008) study, we totaled all 3′-UTR interaction sites for each transcript. We chose transcripts having at least one 3U target site, sorted them based on their degree of protein fold change, and selected the 10 transcripts with the most protein fold change. The accession numbers of the 10 transcripts related to each miRNA are as follows: hsa-miR-1: NM_019594, NM_001043352, NM_003769, NM_017444, NM_001111, NM_001083112, NM_001839, NM_012120, NM_172173, NM_001655; hsa-miR-124: NM_001084, NM_006289, NM_206855, NM_004099, NM_133452, NM_015493, NM_018719, NM_021961, NM_138473, NM_001753; hsa-miR-181a: NM_021629, NM_001037165, NM_006931, NM_002024, NM_014988, NM_004282, NM_015057, NM_000093, NM_002129, NM_006516, NM_170751; mmu-miR-223: NM_009984, NM_022880, NM_030253, NM_029564, NM_029364, NM_008633, NM_145452, G270135O13, NM_001033606, NM_011486. We searched the 5′-UTR of each transcript for interaction with the 3′-end of its corresponding miRNA using RNAhybrid and then consecutive sequence matches. Here we set the RNAhybrid energy cutoff at −13 kcal/mol and consecutive sequence matches at 7-mer or greater, in accordance with the characteristics of 3′-UTR interaction sites.
miBridge targets
All mature miRNAs were downloaded from miRBase (Release 11.0) (Griffiths-Jones et al. 2008) and split into their respective 5′- and 3′-ends, making miRNA halves. All mRNA coordinate data and genome sequences of all human chromosomes were downloaded from the UCSC genome browser (Karolchik et al. 2008). An in-house program extracted 5′-UTR and 3′-UTR sequences from chromosome sequences using RefSeq coordinate data (thus only RefSeq data are included in the database). We first searched the 5′-end of each miRNA for entire 3′-UTRs using RNAhybrid parameters of –e –13 –f 2,7, and the 3′-end of miRNA for entire 5′-UTRs using a parameter of –e –13. RNAhybrid outputs were filtered for consecutive 7-mer without GU matches. Only those miRNAs and targets commonly identified in both searches were retained as miBridge targets.
hsa-miR-605 transfection for SEC24D assay
For transient transfection, HeLa cells were seeded at 60% confluency, and transfections carried out using Pre-miR miRNA Precursor Starter Kit (Ambion, AM1540). HeLa cells were transfected with Negative Control-1 Precursor, Pre-mir-605, or anti-miR-605 miRNAs in 12-well plates using siPORT NeoFX according to the manufacturer's protocol.
Western blot analysis
Cells were harvested three days post-transfection for Western blot analysis. Protein samples were extracted from cells using PRO-PREP protein extraction solution (Intron Biotechnology, 17081), and protein concentration was measured by BCA protein assay kit (Pierce, 23227). Forty micrograms of proteins were separated by SDS-PAGE and then transferred to nitrocellulose membranes (MILLIPORE, HAHY00010). The membranes were blocked for 1 h in blocking buffer (1×-Tris-bufered saline, 5% nonfat dry milk, and 0.1% Tween 20), which was replaced by anti-SEC24D monoclonal antibody (Abnova, H00009871-M04) in blocking buffer, overnight at 4°C. Primary antibody was detected using horseradish peroxidase-linked goat anti-mouse antibody (Amersham Biosciences, NA931V) and visualized with the SUPEX ECL reaction kit (Neurotics, MNPS-200401). The blots were reblotted with HRP-conjugated-β-actin (ACTB) antibodies (Santa Cruz Biotechnology, Inc., sc-47778) without stripping. The bands were scanned with HP Scanjet 3570c (Hewlett-Packard) and quantified using NIH image software.
Quantitation of SEC24D mRNA with real-time PCR
Cells were harvested two days post-transfection, and the total RNA was extracted with the mirVana miRNA isolation kit (Ambion, AM1560) according to the manufacturer's instructions. To validate SEC24D mRNA expression, qRT-PCRs were performed using TaqMan Gene Expression Master Mix (Ambion, 4369016) and TaqMan probe assays for SEC24D (Assays-on-Demand, Hs00207926_m; Applied Biosystems) and GAPDH (Assays-on-Demand, Hs99999905_m; Applied Biosystems) following the manufacturer's instructions. All the primers and probes are cDNA-specific. We quantified transcripts of GAPDH as the endogenous RNA control, normalizing each sample on the basis of its GAPDH content. Each sample was tested in triplicate for each gene. Real-time PCR was carried out according to the manufacturer's instructions (Applied Biosystems 7500).
Acknowledgments
We thank X. Xie for providing the UTR motif data set; Anders H. Lund and Ulf A. Ørom for discussion and preliminary experiments; Jamal Kasham and Heekon Cha for miBridge programming assistance; Jeff de Wet, John Kim, and Matthias Kretzler for discussion; YongSun Lee, Neil Smalheiser, and Nils Walters for manuscript review; and Paul D. Trombley for artwork. B.D.A and A.M.C. are supported by the National Institutes of Health to the National Center for Integrative Biomedical Informatics NCIBI (U54-DA021519); J.I.Y. is supported by National Research and Development Program for Cancer Control (0720270), Basic Research Program of Korea Science and Engineering Foundation KOSEF (R01-2006-10203, R15-2008-01565), and Korea Research Foundation Grant (KRF-2007-314-C00199); and S.H.H. was supported by the Center for Biological Modulators of the 21st Century Frontier Research and Development Program (CBM31-B3000-01-00-01), the Ministry of Science and Technology, Korea.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.089367.108.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes & Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Pillai RS. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions. BMC Bioinformatics. 2004;5:139. doi: 10.1186/1471-2105-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes & Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc Natl Acad Sci. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Post-transcriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]