Abstract
Recent studies have revealed that insertions and deletions (indels) are more different in their formation than previously assumed. What remains enigmatic is how the local DNA sequence context contributes to these differences. To investigate the relative impact of various molecular mechanisms to indel formation, we analyzed sequence contexts of indels in the non protein- or RNA-coding, nonrepetitive (NCNR) portion of the human genome. We considered small (≤30-bp) indels occurring in the human lineage since its divergence from chimpanzee and used wavelet techniques to study, simultaneously for multiple scales, the spatial patterns of short sequence motifs associated with indel mutagenesis. In particular, we focused on motifs associated with DNA polymerase activity, topoisomerase cleavage, double-strand breaks (DSBs), and their repair. We came to the following conclusions. First, many motifs are characterized by unique enrichment profiles in the vicinity of indels vs. indel-free portions of the genome, verifying the importance of sequence context in indel mutagenesis. Second, only limited similarity in motif frequency profiles is evident flanking insertions vs. deletions, confirming differences in their mutagenesis. Third, substantial similarity in frequency profiles exists between pairs of individual motifs flanking insertions (and separately deletions), suggesting “cooperation” among motifs, and thus molecular mechanisms, during indel formation. Fourth, the wavelet analyses demonstrate that all these patterns are highly dependent on scale (the size of an interval considered). Finally, our results depict a model of indel mutagenesis comprising both replication and recombination (via repair of paused replication forks and site-specific recombination).
Indels cause multiple human genetic diseases (Cooper et al. 2006) and are a source of natural intra- and interspecific genetic variation (e.g., Mills et al. 2006; Clark et al. 2007). Recent studies highlight the differences in rates of small (≤100-bp) insertions vs. deletions (e.g., Chen et al. 2007; Messer and Arndt 2007) and suggest that these two types of mutations might at least in part be caused by distinct molecular mechanisms (Kvikstad et al. 2007). In contrast to earlier reports attributing all indels to replication slippage errors (Ball et al. 2005; Messer and Arndt 2007), we have previously demonstrated the significance of recombination to the formation of small indels, in particular insertions (Kvikstad et al. 2007). Several analyses of protein coding regions have demonstrated sequence context dissimilarities between insertions and deletions, such as differences in local base composition and distinct hotspots (Halangoda et al. 2001; Kondrashov and Rogozin 2004; Ball et al. 2005), which again indicate differences in their mutagenesis. Recently, Tanay and Siggia (2008) analyzed the base composition of sequences flanking insertions and deletions and developed models of genome-wide indel propensity that differ between the two mutation types.
Despite these exciting advances, many questions still remain. For example, it is not known to what extent natural selection could have affected signatures of hotspots and motifs in the vicinity of indels identified in genic sequences (e.g., Kondrashov and Rogozin 2004; Ball et al. 2005). Heterogeneity in base composition and substitution rate along the genome may have confounded some results (e.g., Halangoda et al. 2001; Tanay and Siggia 2008). Additionally, just as combinations of specific binding sites act jointly to promote transcription (for review, see Ji and Wong 2006), the simultaneous presence of multiple motifs could provide important clues to indel mutagenesis and requires investigation. Finally, determining the scale at which a hotspot or a motif can be detected and implicated in a particular molecular process constitutes an active area of research (e.g., Berry et al. 2006). Thus, analyzing sequence contexts flanking a genome-wide set of indels located outside of genes with a multiscale methodology is likely to aid in discerning the biological mechanisms underlying these mutations.
Here we utilize wavelet techniques to study sequences flanking small indels in the NCNR portion of the human genome. Wavelet transformations, traditionally used in the study of time series, allow one to re-express a signal preserving both its global trends and its local fluctuations (Percival and Walden 2006). For wavelet analysis of DNA, the nucleotide sequence is regarded as “time,” and features mapped along the sequence represent the “signal.” A wavelet transformation is multiscale since a scale (or window size) is not chosen a priori; rather, a signal is analyzed across multiple scales simultaneously. Exploiting this fact, we expand on previous presence/absence enrichment tests (Abeysinghe et al. 2003; Kondrashov and Rogozin 2004; Ball et al. 2005) and use wavelet transformations for the detection of spatial patterns (i.e., local fluctuations in occurrence) among sequence motifs flanking small indels. To our knowledge, this represents the first application of wavelet techniques to the entire human genome.
Specifically, we address the following questions. First, are regions flanking indels similar or different in terms of their motif content when compared to the indel-free portion of the genome? Second, can we discern significance in the spatial patterns of motifs in proximity to indels? Third, do motifs show similar or different spatial patterns when flanking insertions vs. deletions? And finally, is it possible to detect colocation of different motifs, indicative of the simultaneous action of different mechanisms in the generation of indels?
Results
Small indels occurring in the human lineage since its divergence from chimpanzee were identified in the NCNR portion of the genome (see Methods; Supplemental material). A total of 65,353 and 135,989 human-specific insertions and deletions, respectively, were detected in the human–chimpanzee–macaque three-way MULTIZ alignments (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), as described by Kvikstad et al. (2007). An excess of deletions over insertions is consistent with other studies (Chen et al. 2007; Kvikstad et al. 2007; Messer and Arndt 2007; Tanay and Siggia 2008).
To implement our analyses, we created five nonoverlapping human “subgenomes” (sets of sequences): four subgenomes corresponded to the regions flanking insertion or deletion breakpoints, separately, and in 5′ or 3′ position along the chromosome (in the reference genome), separately. The 5′ and 3′ subgenomes were considered separately because asymmetries in processing of stalled replication forks and double-strand break (DSB) repair have been noted for motifs in specific strand orientations (Kosmider and Wells 2006; Pollard et al. 2007); both processes potentially contribute to indel formation (see below). Additionally, a fifth (“control”) subgenome was built from the NCNR indel- and microsatellite-free portion of the human genome (see Methods).
Each subgenome was screened for the presence of motifs known to promote recombination, induce replication pausing and/or frameshift, and affect DNA stability, as well as several insertion/deletion hotspots (Table 1; Abeysinghe et al. 2003; Ball et al. 2005). We detected matches to each motif, its complement, mirror image (reverse), and reverse complement. As a result, a total of 126 motifs grouped into eight classes were sought. For each motif, we computed a total frequency profile in each subgenome by summing motif counts across all insertion/deletion events in contiguous 10-bp increments (see Methods). Total frequency profiles thus “average out” potential genome landscape effects. Moreover, no significant base composition differences were detected between indel and control subgenomes (P-values of 0.701, 0.671, 0.134, 0.127 for deletions 5′, deletions 3′, insertions 5′, and insertions 3′, respectively; Wilcoxon two-sided nonparametric tests). Hence, genome-wide heterogeneity in base composition is unlikely to affect motif detection in each subgenome.
Table 1.
DNA sequence motifs
aMotif names in quotation marks were given for reference in this work only.
bIUPAC ambiguities: R = A/G, Y = C/T, K = G/T, M = A/C, S = G/C, W = A/T, N=A/C/G/T.
cMotifs longer than 10 nucleotides (unit for wavelet transform) were sought for simple enrichment test only, at scales of 20-bp and larger.
If the distinction between 5′ vs. 3′ position with respect to an indel breakpoint were unimportant, the total frequency profiles of any given motif should be symmetric 5′ vs. 3′, showing high correlation (e.g., as measured nonparametrically by Kendall tau). In contrast with this expectation, in our data set many motifs display asymmetrical frequency behaviors 5′ and 3′ to indel breakpoints (for one example motif, topoisomerase cleavage site 4; see Supplemental Fig. S1). Indeed, the 5′ vs. 3′ correlations of total frequency profiles are relatively weak for all motifs considered here (Kendall tau coefficients are all <0.5—computed separately for insertions' and deletions' flanks), suggesting differences in motif occurrences on the two sides of breakpoints. Similarly, if indel mutations were equally likely to occur on the two DNA strands, then any given motif involved in indel mutagenesis and its reverse complement should show high 5′ vs. 3′ correlation (i.e., similarity between a motif's occurrence and its reverse complement's occurrence on the opposite strand). Conversely, the 5′ vs. 3′ correlations of total frequency profiles for each motif (e.g., 5′) and its reverse complement (e.g., 3′) are relatively weak (Kendall tau coefficients are all <0.4—again, computed separately for insertions' and deletions' flanks).
We assessed the significance of 5′ vs. 3′ positional differences with a permutation scheme based on randomly reassigning 5′ and 3′ flank labels (Supplemental Fig. S1; Supplemental Table S1; see Methods). Interestingly, nearly twice as many motifs exhibit significant positional asymmetry when flanking deletions than insertions (Supplemental Table S1), and the flank closest to the indel (first 10 bp from the breakpoint) displays extreme positional asymmetry for as many as 25% of the motifs analyzed here (Supplemental Table S1; Supplemental Fig. S1)—no such bias was observed in the control subgenome. Significance of motif positional differences prompted us to consider frequency profiles separately for 5′ and 3′ subgenomes.
Simple (nonwavelet) motif overrepresentation
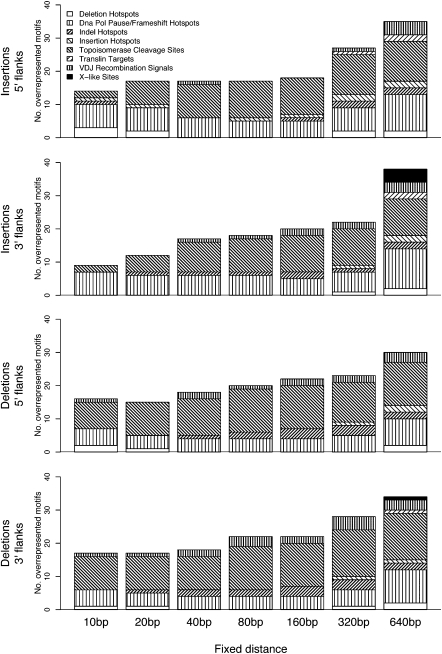
We commenced with an analysis of global trends using a simple (nonwavelet-based) procedure to detect motif overrepresentation in indel flanking sequences, because overrepresentation of particular motifs in the vicinity of indels suggests their importance to indel formation. For a given motif, its total occurrences were computed for each of the four indel-related subgenomes and contrasted with the control subgenome using a resampling scheme, separately at various fixed distances from indel breakpoint (Methods; Fig. 1; Supplemental Table S2). Importantly, a comparison of motif total occurrences aggregated across all distances (for each subgenome) reveals no striking differences (Supplemental Table S3), demonstrating the importance of scale for the discriminatory power of simple presence/absence overrepresentation testing.
Figure 1.
Distribution of overrepresented motifs (nonwavelet based) across motif classes (patterned areas) and distances from the breakpoint (bars), separately for the 5′ and 3′ flanks of insertions and deletions. Patterned areas are proportional to the number of significant motifs per class (significance is assessed resampling from the control subgenome, and subject to a false discovery rate [FDR] correction; FDR controlled at 5%).
DNA polymerase (pol) pause/frameshift hotspots, topoisomerase cleavage sites, and motifs promoting site-specific recombination are significantly overrepresented in sequences flanking insertions and deletions alike (Fig. 1). However, there are noticeable differences between insertions and deletions in the proportions of overrepresented motifs by class and by distance from the breakpoint. For instance, at all but the largest (320 and 640 bp) distances, DNA pol pause/frameshift hotspots represent a larger proportion of the overrepresented motifs proximal to insertions than to deletions. In contrast, at even the smallest (10 bp) distance, a higher proportion of topoisomerase cleavage sites are overrepresented in proximity to deletions than insertions. Among them, the example motif topoisomerase cleavage site 4 displays overrepresentation near deletions in 5′ orientation at all distances (Supplemental Table S2).
Our analysis of the NCNR portion of the genome indicates overrepresentation of V(D)J recombination signals, X-like sites, and translin binding targets in the vicinity of indels (Fig. 1), while no such trends were observed in previous reports focusing on indels located in genes (Ball et al. 2005)—confirming ascertainment bias when considering genic sequences. Surprisingly, both insertion hotspots and deletion hotspots (Steinmetz et al. 1987; Krawczak and Cooper 1991; Chuzhanova et al. 2002; Kondrashov and Rogozin 2004) display overrepresentation around both insertions and deletions, depending on the distance analyzed (Fig. 1). Deletion hotspots in particular are overrepresented flanking insertions and deletions at the smallest distance considered (10 bp).
Multiscale wavelet analyses
In addition to overrepresentation, the spatial pattern in a motif's occurrence can also convey information suggestive of a role in indel mutagenesis. For a given scale, the motif could have similar global trends in indel flanks and controls (i.e., no overrepresentation), but a very different profile of local fluctuations (i.e., spatial pattern), or vice versa. In the following, we utilize wavelet transformations to investigate motifs' spatial patterns, at multiple scales simultaneously.
Motif enrichment profiles
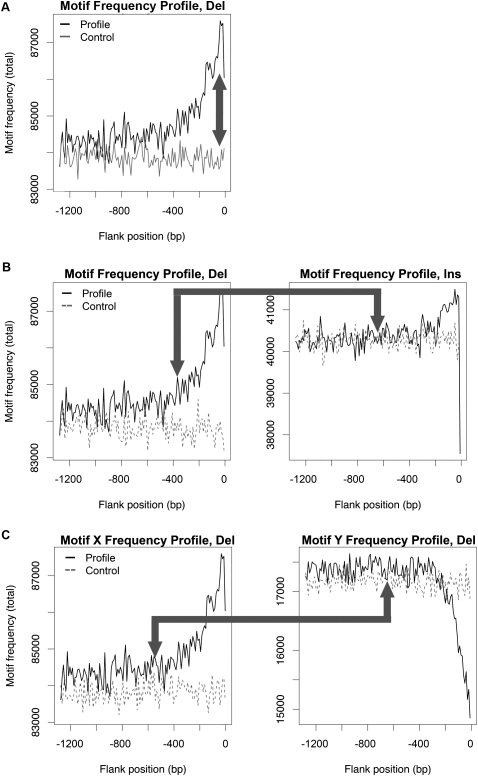
For a given motif, we created four enrichment profiles, each computed as the difference between its total frequency profile (the sum of motif counts across all insertion/deletion events in contiguous 10-bp increments, see above) from an indel-related subgenome and its corresponding profile from the control subgenome (Fig. 2A; see Methods). A multiscale analysis using wavelet-transformed enrichment profiles and their second moments was conducted to assess significance of the magnitude of each enrichment profile (see Methods). This approach is illustrated for the example motif topoisomerase cleavage site 4, which displays a significant enrichment profile flanking deletions in 5′ position (Fig. 2A; Supplemental S2).
Figure 2.
Multiscale wavelet analyses (for details, see text; Supplemental Figs. S2–S4): total frequency profiles illustrating three types of comparisons (arrows) using topoisomerase cleavage site 4 as an example motif. (A) Indel vs. control: enrichment profiles. The spatial occurrence pattern characterizing a motif is investigated forming an enrichment profile, i.e., the difference between the motif's total frequency profile in an indel-related subgenome (e.g., the 5′ flank of deletions; black line) and that in the corresponding control subgenome (gray line). (B) Insertions vs. deletions: similarity between profiles. This is investigated by comparing the motif's total frequency profile in a deletion-related subgenome (e.g., the 5′ flank; black line; left) with that in the corresponding insertion-related subgenome (black line; right). Control profiles (dashed lines) are provided for visual reference only. (C) Motif X vs. motif Y: colocation in profiles. Along with topoisomerase cleavage site 4 (X), here we consider DNA pol pause/frameshift hotspot 1 (Y). Colocation is investigated by comparing the total frequency profiles of X vs. Y in an indel-related subgenome (e.g., the 5′ flank of deletions; black lines for X on the left, and Y on the right). Control profiles (dashed lines) are provided for visual reference only.
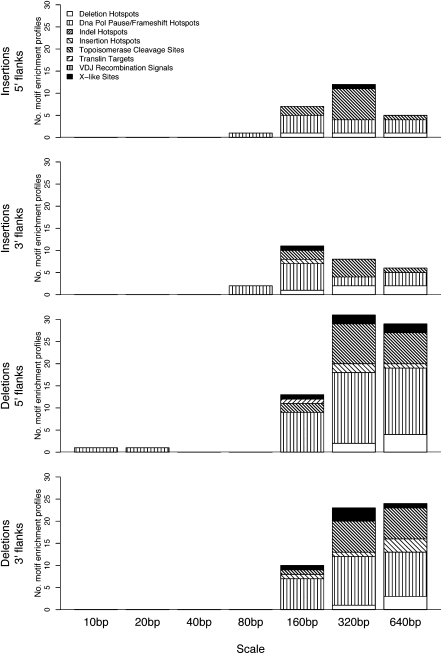
Results summarized by motif class (Fig. 3; for individual motifs, see Supplemental Table S4) reveal that a greater number of motifs have significant enrichment profiles flanking indels at intermediate to large (160–640 bp) than at small scales. Nevertheless, DNA pol pause/frameshift motifs exhibit significant enrichment profiles flanking deletions at the finest (10–20 bp) scales (only 5′ with respect to breakpoint); for insertions, they become significant at intermediate (80 bp) scales.
Figure 3.
Distribution of motifs with significant enrichment profiles (wavelet-based) across motif classes (patterned areas) and scales (bars), separately for the 5′ and 3′ flanks of insertions and deletions. Patterned areas are proportional to the number of significant motifs per class (magnitude is measured taking second moments after wavelet transform of each enrichment profile, and significance is assessed through a random permutation scheme, and subject to a FDR correction; FDR controlled at 5%).
Interestingly, there are motif classes (e.g., insertion and deletion hotspots, topoisomerase cleavage sites) that display both significant global trends as measured by nonwavelet overrepresentation (Fig. 1) and significant local fluctuations captured by enrichment profiles (Fig. 3). However, some motif classes display significance in only one of these two measures. For instance, despite lacking significant overrepresentation 5′ to deletions (Fig. 1), translin target binding sites exhibit significant enrichment profiles in this position flanking deletions at a 160-bp scale (and never for insertions) (Fig. 3). Conversely, neither indel hotspots nor VDJ recombination signals exhibit significant enrichment profiles (Fig. 3), although they are significantly overrepresented proximal to both insertions and deletions (Fig. 1). These results corroborate differences in the information carried by total occurrences versus spatial patterns.
Potentially, indels of different sizes (e.g., 1-bp events) or evolutionary “ages” (times of occurrence, e.g., events polymorphic in human populations) could show distinct motif enrichment profiles (see Supplemental material; Supplemental Tables S5–S8) suggestive of distinct mutagenic mechanisms. However, preliminary investigation of data from chromosome 1 indicates that the majority of motifs with significant enrichment profiles around 1 bp or polymorphic indels represent subsets of the motifs identified considering all events. Thus, variation due to indel size and/or evolutionary “age” is unlikely to significantly affect our conclusions.
Similarity between motif frequency profiles flanking insertions and deletions
Next, we investigated whether a motif presents similar spatial patterns flanking both insertions and deletions, suggesting shared mechanisms of origin for these two mutation types. To measure similarity, we used the coefficients of wavelet-transformed frequency profiles to compute multiscale correlations between insertion and deletion behavior (a motif with itself) in the 5′ flank, and separately, in the 3′ flank (Fig. 2B; Supplemental Fig. S3; Methods).
Unexpectedly, we discover significant similarity in spatial patterns (significantly positive Kendall tau correlation) around insertions and deletions for only a limited number of motifs at small scales (≤80-bp): 13 and 12 in 5′ and 3′ orientations, respectively (Fig. 4, two motifs are significantly correlated in both orientations while only six motifs are among the top 25% most “abundant” motifs identified; Supplemental Table S3). DNA pol pause/frameshift hotspots and topoisomerase cleavage sites possess significant similarity between insertions and deletions, as do several of the insertion and deletion hotspots (Fig. 4), confirming that the latter motifs fail to discriminate the two mutation types (see above). V(D)J recombination signals and X-like sites also have significant similarity between insertions and deletions, but only 5′ from the breakpoint (Fig. 4A). Such a small number of motifs with significant similarity in spatial patterns around insertions and deletions is surprising given that many motifs were previously thought to play similar roles during generation of both mutation types (Ball et al. 2005). Moreover, three motifs show significant negative correlations in spatial patterns around insertions vs. deletions, including the example topoisomerase cleavage site 4 (Fig. 2B; Supplemental Fig. S3), suggesting that these motifs may differentiate between the two mutation types (Fig. 4).
Figure 4.
Significant Kendall tau correlations of total motif frequency profiles around insertions vs. deletions, separately for various scales, and for 5′ (A) and 3′ (B) flanks. Positive and negative correlations are indicated by sign, with bolded cells corresponding to those significant after FDR correction (FDR controlled at 5%; −log10 of adjusted P-values are reported in parentheses, with “>16” signifying a P-value of <10−16). Motif names are given suffixes to indicate reverse (r), complement (c), and reverse complement (rc). Scales below 20 and above 80 bp are not reported because no motifs had significant correlations at such scales after FDR correction.
Similarity in frequency profiles between pairs of motifs (colocation)
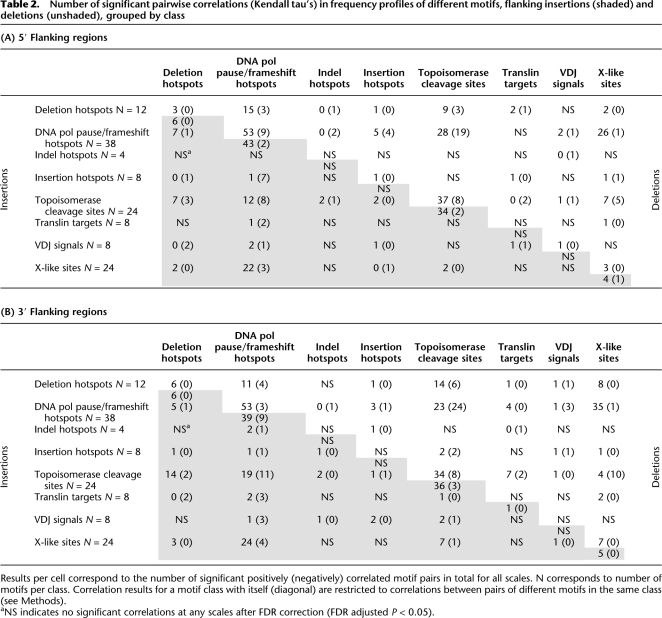
Last, we screened for potential signatures of simultaneous mutagenic action for pairs of motifs, as indicated by colocation around indels. The coefficients of wavelet-transformed frequency profiles for two motifs, say X and Y (Fig. 2C; Supplemental Fig. S4), were used to compute multiscale Kendall tau correlations between their spatial occurrences, separately for 5′ and 3′ flanks, and for insertions and deletions. Significantly positive correlations suggest “cooperation.” Figure 2C (for details, see Supplemental Fig. S4) compares the frequency profiles for our example motif topoisomerase cleavage site 4 and a DNA pol pause/frameshift hotspot, both considered in the 5′ flank of deletions. At small scales, these two motifs show significantly positive correlation in spatial occurrence patterns, whereas at larger scales their occurrences are anti-correlated (although this is not significant) (Supplemental Fig. S4). This fine-scale colocation would remain undetected if correlation were assessed at larger scales only. As there are 7875 pairs of motifs (and thus 7875 tests for each of the four subgenomes; see Methods), we present summary results for motifs grouped by class and across all scales (Table 2; for scale-specific results, see Supplemental Table S9).
Table 2.
Number of significant pairwise correlations (Kendall tau's) in frequency profiles of different motifs, flanking insertions (shaded) and deletions (unshaded), grouped by class
Results per cell correspond to the number of significant positively (negatively) correlated motif pairs in total for all scales. N corresponds to number of motifs per class. Correlation results for a motif class with itself (diagonal) are restricted to correlations between pairs of different motifs in the same class (see Methods).
aNS indicates no significant correlations at any scales after FDR correction (FDR adjusted P < 0.05).
We detected 185 (261) and 220 (289) significantly correlated pairs of motifs in 5′ and 3′ positions from insertion (deletion) breakpoint. Despite similar overall numbers, insertions and deletions display noticeably different trends (Table 2). For instance, translin targets positively correlate with more motifs when flanking deletions than insertions in 3′ positions, while these motifs are more frequently observed to anti-correlate in pairwise comparisons flanking insertions (translin targets are found in relatively similar abundance flanking both mutation types) (Supplemental Table S3). Interestingly, indel hotspots positively correlate with a greater number of motif classes (including V(D)J recombination signals and DNA pol pause/frameshift hotspots) when proximal to insertions than deletions.
In addition to the differences described above, there are colocation behaviors that are shared between insertions and deletions. Among them, the deletion hotspots positively correlate with similar motif classes 5′ and 3′ to both insertions and deletions (Table 2), confirming that these motifs do not distinguish the two types of mutations (see also Figs. 1, 3, 4). Similarly, different topoisomerase cleavage sites, DNA pol pause/frameshift hotspots, and X-like sites (that are among the most abundant motifs) (Supplemental Table S3) positively correlate within and among these motif classes at multiple scales when flanking both insertions and deletions (Table 2; Supplemental Table S9).
Discussion
Our novel wavelet-based analyses of motifs associated with DNA pol activity, topoisomerase cleavage, DSBs, and their repair emphasize differences between insertions and deletions in NCNR DNA. Additionally, we find motifs possessing patterns of overrepresentation, spatial occurrence, and colocation—analyzed here for the first time—that frequently differ in the proximity of insertions versus deletions, and at different scales. Taken together, these results provide clues to the relative contributions of different mechanisms associated with indel mutagenesis.
Importance of replication
Consistent with previous reports (Ball et al. 2005), we observe that DNA pol pausing and/or frameshift motifs are important to indel formation. The analyses of overrepresentation, enrichment profiles, and similarity in spatial patterns of these motifs flanking insertions and deletions (Figs. 1, 3, 4) all suggest that replication is critical for indel mutagenesis (Fig. 5). Note that the DNA pol hotspots analyzed here correspond to eukaryotic polymerases alpha and beta (Table 1; Moon et al. 2007; McCulloch and Kunkel 2008). In particular, the former polymerase primarily synthesizes primer extension for Okazaki fragments on the lagging strand (McCulloch and Kunkel 2008), whereas the latter functions in DNA synthesis during base excision repair (Sweasy et al. 2006; Moon et al. 2007). The significance of DNA pol alpha motifs at small scales (e.g., Fig. 4; Supplemental Tables S2–S4) indicates that lesions might be more likely to escape repair on the lagging strand (Fig. 5) by bypassing replication checkpoint response (Tourriere and Pasero 2007; McCulloch and Kunkel 2008). Notably, we demonstrate that frequency profiles of motifs involved in pausing/frameshift are highly correlated with those of topoisomerase cleavage sites and other motif classes (translin binding targets, X-like sites and V(D)J recombination signals; see discussion below) (Table 2), suggesting that indel formation can be affected by the combined action of replication and other processes.
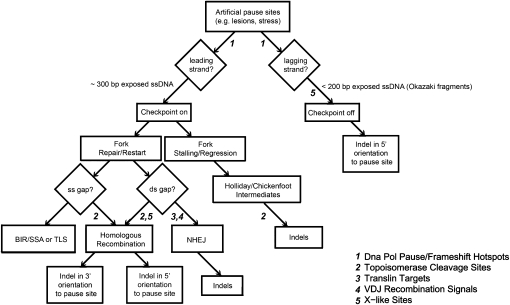
Figure 5.
Replication and recombination pathways proposed to contribute to insertion and deletion mutagenesis (numbers correspond to motif classes indicating involvement as evidenced by results; for details, see Discussion). DNA replication can pause when encountering natural pause sites or artificially due to lesions, stress, and/or specific sequence cues. Depending on the strand, single-stranded DNA (ssDNA) is exposed and replication checkpoint can be triggered or bypassed. Cells respond to replication pausing by repair or restart of the forks, depending on the type of lesion created at the paused site. Double-strand gaps are repaired via homologous recombination or nonhomologous end joining (NHEJ), whereas single-stranded gaps can be repaired via homologous recombination, break-induced replication (BIR) followed by single-strand annealing (SSA), or translesion synthesis (TLS). Our results confirm that NHEJ repair of double-stranded gaps and homologous recombination (of double- and/or single-stranded gaps) could contribute to indel mutagenesis. Stalling of the fork can result in regression and Holliday junction/chickenfoot intermediates, which can be repaired and thus cause indel formation. Checkpoint bypass can also lead to indels.
Interplay of recombination and replication in indel formation
The significance of topoisomerase cleavage sites in our analysis suggests that recombination-mediated repair of paused replication forks can contribute to indel mutagenesis (Fig. 5). We found that topoisomerase cleavage sites display overrepresentation, enrichment profiles, and positive profile correlations (Figs. 1, 3, 4) flanking both insertions and deletions. Since the type I class of topoisomerases (represented by the motifs analyzed here) break and rejoin gapped single-stranded DNA (ssDNA) during replication (Hyrien 2000; Wang 2002; Tourriere and Pasero 2007), these results again support the importance of replication for indel formation (Fig. 5). Moreover, colocation detected between frequency profiles of DNA pol pause/frameshift hotspots and topoisomerase cleavage sites at intermediate (80–160 bp) scales (Supplemental Table S4) may reflect checkpoint mediated restart of paused forks contributing to indel mutations (Fig. 5; Tourriere and Pasero 2007).
Site-specific recombination and indel formation
Our analyses of the NCNR genome indicate that translin targets, V(D)J recombination signals, and X-like sites (the motifs associated with site-specific recombination and genome instability) might be significant for small indel mutagenesis. First, translin may be involved in the formation of deletions. Enrichment profiles flanking deletions, but not insertions, are observed for translin targets (Fig. 3). These are binding sites for recognition of the translin protein to ssDNA at staggered breaks in the first steps of their repair (Kasai et al. 1997; Erdemir et al. 2002a,b; Sengupta and Rao 2002). Previously, overrepresentation of translin targets implicated NHEJ in mediating chromosomal rearrangements such as translocations and large deletions (Abeysinghe et al. 2003; Gajecka et al. 2006a,b) and microsatellite/telomere repeat expansions/contractions (Jacob et al. 2004). Our results suggest that translin may mediate small deletion formation as well. Additionally, we demonstrate for the first time that translin targets have frequency profiles positively correlated with other motifs (e.g., DNA pol pause/frameshift hotspots and topoisomerase cleavage sites) 3′ to deletion breakpoints (Table 2). Translin is thought to act in cellular response to DNA damage via the translin-associated factor X (TRAX) that interacts with the nuclear matrix protein C1D (Erdemir et al. 2002a,b; Cho et al. 2004). Since C1D regulates a number of cellular functions, including replication, NHEJ, homologous recombination, and topoisomerase activity (Erdemir et al. 2002a,b; Cho et al. 2004; Yang et al. 2004), our results suggest that recruitment of DSB repair, possibly mediated by TRAX and C1D, can contribute to indel (particularly deletion) formation (Fig. 5).
Second, V(D)J recombination may lead to both insertions and deletions. V(D)J recombination signals are overrepresented flanking insertions and deletions at almost all distances from the breakpoint (Fig. 1), and have similar frequency profiles 5′ to both mutation types (Fig. 4A). As these motifs correspond to the heptamer and nonamer signals required for creating the DSBs and NHEJ of immune receptor molecules (Lu et al. 2007), our results suggest illegitimate recruitment of V(D)J recombination or NHEJ leading to indel mutation (Fig. 5).
Finally, our results indicate that X-like sites may promote insertions and deletions, but in different ways (Fig. 5). We observe that X-like sites are enriched in the vicinity of indels (Fig. 2), particularly in the 3′ position to insertion breakpoints. X-like sites include the X element and other conserved sequences known to induce recombination (Table 1; Seitz and Kowalczykowski 2006). In agreement with this, an X-like consensus sequence previously hypothesized to promote insertions and deletions via V(D)J recombination (Wyatt et al. 1992) is significant to insertion formation (for the X-like motif 4, see Fig. 4A; Supplemental Tables S2, S4). Interestingly, the Fragile X breakpoint cluster motif displays significant enrichment profiles at multiple scales flanking deletions but not insertions (Supplemental Table S4). The Fragile X breakpoint cluster motif is known to adopt many non-B DNA structures (Bacolla et al. 2006). Indeed, the recombination-mediated repair of DSBs at this cluster has been reported to differ in induced mutations when present on leading vs. lagging strands (Kosmider and Wells 2006). This motif also displayed significant positional asymmetry (Supplemental Table S1). Thus, recombination-mediated repair of secondary structures at this X-site may lead to more deletions than insertions and in an orientation-dependent manner.
Chromatin organization and indel formation
From our wavelet-based analyses, the 80-bp scale emerges as particularly relevant given the significance of spatial occurrence patterns at this scale for many motif classes, including DNA pol pause/frameshift hotspots, topoisomerase cleavage sites, V(D)J recombination signals, and X-like sites (e.g., Fig. 4; Supplemental Table S4). Notably, 80 bp corresponds to approximately half the length of DNA involved in nucleosome positioning (∼150 bp) (Luger et al. 1997; Kornberg and Lorch 1999; Segal et al. 2006; Gupta et al. 2008). Since wavelets capture fluctuations in signals representing changes from “peaks” (e.g., nucleosome spacer regions) to “valleys” (e.g., nucleosome-occupied regions), our results corroborate the importance of chromatin structure in the regulation of molecular processes (e.g., Wang 2002; Nightingale et al. 2007) and, as recently noted in medaka, the potential formation of indels (Sasaki et al. 2009).
Hotspots: Lessons from noncoding DNA
Our results for hotspots reveal similarities in the frequencies of these motifs in insertion- and deletion-flanking regions. Both insertion and deletion hotspots show overrepresentation (Fig. 1), enrichment profiles (Fig. 3), and interesting correlations in frequency profiles (Fig. 4) around both mutation types. Therefore, the classification of hotspots as insertion- or deletion-specific does not hold when analyzing the NCNR genome. Indel (as opposed to insertion or deletion) hotspots behave differently: Their frequency profiles correlate with those of several biological classes of motifs (topoisomerase cleavage sites, V(D)J recombination signals, and DNA pol pause/frameshift hotspots) around insertions, but not deletions (Table 2). Thus, indel hotspots might act in conjunction with other sequences to facilitate the formation of more insertions than deletions, during the processes noted above.
5′ vs. 3′ positional asymmetry
We noted differences in motif occurrence patterns between the 5′ and 3′ indel-related subgenomes, suggesting the possibility for asymmetrical interactions between DNA flanking sequences and processes leading to indel formation. In contrast to the control subgenome, several motifs exhibit significantly different frequency profiles 5′ and 3′ of indels (particularly deletions) (Supplemental Fig. S1; Supplemental Table S1). The extreme discrepancy in positional behavior for the flanks closest to the event implies direct involvement of the motif in promoting mutagenesis, particularly for deletions (∼85% of small insertions are tandem duplications) (Messer and Arndt 2007). Uncoupling of leading vs. lagging strand synthesis during the processing of stalled replication forks could potentially account for such asymmetry, since as many as a third of human replication forks experience arrest (Conti et al. 2007). Furthermore, the processes of transcription-mediated mutation (Green et al. 2003), homologous recombination, and chromatin assembly have inferred strand asymmetries based on footprints of their interactions with DNA sequences (Touchon and Rocha 2008). Indeed, processing of stalled replication forks and repair of DSBs at motifs involved in Fragile X Syndrome and Friedreich ataxia, respectively, exhibit specific leading versus lagging strand orientation effects on the subsequent mutational spectra (Kosmider and Wells 2006; Pollard et al. 2007); the same may hold true for motifs analyzed here that likely promote indels.
Methods
Indel identification and motif detection
Small (≤30-bp) indels occurring in the human lineage since its divergence from chimpanzee were identified from the human–chimpanzee–macaque (hg18-panTro2-rheMac2) three-way MULTIZ alignments (Blanchette et al. 2004), following the methods described by Kvikstad et al. (2007), which include rigorous filtering to remove false positives likely attributable to issues of sequence, assembly and alignment accuracy (for details, see Supplemental material). Indels were further excluded if they intersected with human microsatellites, as annotated by Kelkar et al. (2008). To reduce potential effects of natural selection (see also Supplemental material), we restricted our analysis to indels occurring in the “noncoding” portion of the human genome by excluding “known genes” plus their 5 kb upstream and downstream regions. We determined that only 0.02% of the resulting sequences overlapped with annotated RNA-coding genes (Blankenberg et al. 2007), less than the genome-wide annotation (Karolchik et al. 2008). Thus, to the best of our ability, we isolated the nongenic portion of the genome. Furthermore, we excluded repetitive elements annotated by RepeatMasker (http://www.repeatmasker.org/) in any of the three species to produce the resulting NCNR genome.
Experimentally determined consensus hotspots or recognition sites for various classes of motifs were obtained from Abeysinghe et al. (2003) and Ball et al. (2005) (Table 1). Computationally predicted indel, insertion, and deletion mutation hotspots, separately, were defined as according to the method of Steinmetz et al. (1987), Krawczak and Cooper (1991), Chuzhanova et al. (2002), and Kondrashov and Rogozin (2004). Perl scripts were developed to screen nucleotide sequences for perfect matches to a given set of input motifs, including match to each motif, its complement, reverse (i.e., mirror image), and reverse complement, separately.
Motif total frequency profiles
An ordered series was created for each of the four indel-related subgenomes by breaking the sequence upstream (downstream) of each insertion (deletion) into 128 consecutive, nonoverlapping increments of 10 bp. Each 10-bp increment was screened for motif occurrences as described above (motifs overlapping increment endpoints were not scored), and increments were then pooled across all insertions (deletions) for each position relative to the insertion (deletion) breakpoint. Total numbers of occurrences for each motif in each increment were obtained to construct each total frequency profile.
A subset of motifs (Supplemental Table S1) displayed different frequency profiles upstream vs. downstream of indel breakpoints (e.g., Supplemental Fig. S1). To assess the significance of these asymmetries, we reconstructed each motif's total frequency profile after randomly permuting 5′ and 3′ position labels—this was repeated 100,000 times to create “null” distributions for the profiles, which allowed us to detect significant asymmetries, especially in the first 10-bp increments (the ones closest to the indel event) (Supplemental Table S1).
The control subgenome corresponding to the NCNR indel-free (i.e., excluding 1280 bp 5′ and 3′ to all indels) and microsatellite-free portion of the human genome was obtained using Galaxy (Blankenberg et al. 2007). To construct control (total) frequency profiles for each motif, we randomly sampled segments of 1280 bp from the control subgenome in equal number to the sequences comprising the insertion (deletion) subgenomes, respectively. These segments were then broken into 128 contiguous 10-bp increments and screened for motif occurrences, applying the procedure used for indel-related subgenomes.
Simple motif overrepresentation
Overrepresentation of motifs upstream (5′) and downstream (3′) of insertions and deletions, separately, was assessed in comparison to their occurrence in the control subgenome. This was performed at various fixed distances (10, 20, 40, 80, 160, 320, and 640 bp) from the indel breakpoints. For each distance, a number of sequences equal to that in the corresponding indel-related subgenome was randomly sampled from the control subgenome and scanned to obtain frequencies for each motif. This was repeated 1000 times to create empirical P-values for each motif, each event type (insertion, deletion), each flank (5′, 3′), and each scale (Supplemental Table S2). These P-values were then adjusted for multiple testing according to the method of Benjamini and Hochberg (1995), as to control the FDR. Significance was reported in all cases with an adjusted P-value < 0.05 (i.e., capping the FDR at 5%).
Multiscale analyses using wavelets
Three multiscale analyses were conducted using wavelet transforms (see Supplemental material). In each, the relevant input signal was decomposed using the Haar wavelet basis function to construct a discrete wavelet transformation (similar results were obtained using other wavelet filters; data not shown). All analyses were performed using the wavelet libraries (“waveslim,” “wavethresh”) available in the R statistical package (R Development Core Team 2005).
Motif enrichment profiles
To compare a motif's spatial patterns in 5′ and 3′ indel-flanking regions to those in control regions, we formed four enrichment profiles for each motif as differences between its total frequency profiles in the four indel-related subgenomes and the corresponding frequency profiles derived from the control subgenome (Fig. 2A). Next, we obtained wavelet decompositions of the enrichment profiles and analyzed the resulting coefficients' second raw moments (squared deviations from zero) on a multiscale basis (Supplemental Fig. S2). These second moments measured the size of the difference between motifs occurrence patterns in indel flanks vs. control regions. Using a random permutation scheme (see Supplemental material), we tested the null hypothesis that this difference is equal to zero (no difference between indel-related subgenome and control), at various scales. This involved 1000 permutations of the frequency profiles' ordered series before computing differences (enrichment profile) and their wavelet decomposition, each followed by the multiscale analysis of second raw moments (Supplemental Fig. S2; Supplemental Table S4). Multiple testing correction was performed as described above for simple motif overrepresentation.
Shared profiles: Similarity between motif frequency profiles flanking insertions and deletions
To investigate the similarity of a motif occurrence patterns around insertions and deletions, we sought positive correlations between its total frequency profiles in the 5′ and 3′ flanks (separately) of the two types of events (Fig. 2B). For each comparison, we obtained the wavelet decompositions of the two frequency profiles in question, and analyzed their Kendall tau (Kendall 1938) rank-based correlations on a multiscale basis (Supplemental Fig. S3). These provided a robust measure of association between motif occurrence patterns in indel flanks and one that was more conservative than either the Pearson correlation or the Spearman's Rho (Colwell and Gillett 1982), resulting in higher P-values and broader null bands when assessing significance through permutations (data not shown). Similar to above, we used a random permutation scheme to test the null hypothesis of no association, at various scales. Here, permutations of the frequency profiles' ordered series were performed, and wavelet decompositions were computed, followed by multiscale analyses of Kendall tau correlations (Supplemental Fig. S3). Multiple testing correction was performed as described above for simple motif overrepresentation.
Colocation: Similarity in frequency profiles between pairs of motifs
To investigate the similarity of occurrence patterns between two different motifs (colocation), we sought positive correlations between their total frequency profiles in the flanks of insertions and (separately) deletions, 5′ and (separately) 3′ (Fig. 2C). For each comparison (7875 in total), we obtained the wavelet decompositions of the frequency profiles in question, and once again analyzed the Kendall tau rank-based correlations on a multiscale basis. Once more, testing for no association was performed with 1000 permutations of the frequency profiles ordered series before computing wavelet decompositions, each followed by the multiscale analysis of Kendall tau correlations (Supplemental Fig. S4; Supplemental Table S9). Multiple testing correction was performed as described above for simple motif overrepresentation.
Acknowledgments
We thank Kristin Eckert and three anonymous reviewers for helpful comments; Yogeshwar Kelkar for providing human microsatellite data; and Jeffrey Sorley for graphic design. This study was supported by NIH grant R01-GM072264 (to K.D.M.) and by Penn State Academic Computing Fellowship (to E.M.K.).
Footnotes
[Supplemental material is available online at www.genome.org and at http://bx.psu.edu/makova_lab/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.088922.108.
References
- Abeysinghe S, Chuzhanova N, Krawczak M, Ball E, Cooper D. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Wojciechowska M, Kosmider B, Larson JE, Wells RD. Gross rearrangements caused by long triplet and other repeat sequences. In: Wells RD, Ashizawa T, editors. Genetic instabilities and neurological diseases. Academic Press; San Diego, CA: 2006. pp. 717–733. [Google Scholar]
- Ball E, Stenson P, Abeysinghe S, Krawczak M, Cooper D, Chuzhanova N. Microdeletions and microinsertions causing human genetic disease: Common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat. 2005;26:205–213. doi: 10.1002/humu.20212. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova K, et al. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Chen C, Li W, Chuang T. Human-specific insertions and deletions inferred from mammalian genome sequences. Genome Res. 2007;17:16–22. doi: 10.1101/gr.5429606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Chennathukuzhi VM, Handel MA, Eppig J, Hecht NB. The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleoplasmic distribution in male germ cells. J Biol Chem. 2004;279:31514–31523. doi: 10.1074/jbc.M401442200. [DOI] [PubMed] [Google Scholar]
- Chuzhanova N, Anassis EJ, Ball EV, Krawczak M, Cooper DN. Meta-analysis of indels causing human genetic disease: Mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat. 2002;21:28–44. doi: 10.1002/humu.10146. [DOI] [PubMed] [Google Scholar]
- Clark T, Andrew T, Cooper G, Margulies E, Mullikin J, Balding D. Functional constraint and small insertions and deletions in the ENCODE regions of the human genome. Genome Biol. 2007;8:R180. doi: 10.1186/gb-2007-8-9-r180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell D, Gillett J. Spearman versus Kendall. The Mathematical Gazette. 1982;66:307–309. [Google Scholar]
- Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Human gene mutation. BIOS Scientific; Oxford, UK: 1993. [Google Scholar]
- Cooper D, Stenson P, Chuzhanova N. Current protocols in bioinformatics. John Wiley & Sons; New York: 2006. The human gene mutation database (HGMD) and its exploitation in the study of mutational mechanisms. [DOI] [PubMed] [Google Scholar]
- Erdemir T, Bilican B, Cagatay T, Goding CR, Yavuzer U. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol Microbiol. 2002a;46:947–957. doi: 10.1046/j.1365-2958.2002.03224.x. [DOI] [PubMed] [Google Scholar]
- Erdemir T, Bilican B, Oncel D, Goding CR, Yavuzer U. DNA damage-dependent interaction of the nuclear matrix protein C1D with translin-associated factor X (TRAX) J Cell Sci. 2002b;115:201–216. doi: 10.1242/jcs.115.1.207. [DOI] [PubMed] [Google Scholar]
- Gajecka M, Glotzbach CD, Shaffer LG. Characterization of a complex rearrangement with interstitial deletions and inversion on human chromosome 1. Chromosome Res. 2006a;14:277–282. doi: 10.1007/s10577-006-1044-7. [DOI] [PubMed] [Google Scholar]
- Gajecka M, Pavlicek A, Glotzbach CD, Ballif BC, Jarmuz M, Jurka J, Shaffer LG. Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum Genet. 2006b;120:519–526. doi: 10.1007/s00439-006-0222-1. [DOI] [PubMed] [Google Scholar]
- Green P, Ewing B, Miller W, Thomas PJ, Program NCS, Green ED. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- Gupta S, Dennis J, Thurman RE, Kingston R, Stamatoyannopoulos JA, Noble WS. Predicting human nucleosome occupancy from primary sequence. PLoS Comput Biol. 2008;4:e1000134. doi: 10.1371/journal.pcbi.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halangoda A, Still JG, Hill KA, Sommer SS. Spontaneous microdeletions and microinsertions in a transgenic mouse mutation detection system: Analysis of age, tissue, and sequence specificity. Environ Mol Mutagen. 2001;37:311–323. doi: 10.1002/em.1038. [DOI] [PubMed] [Google Scholar]
- Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- Jacob E, Pucshansky L, Zeruya E, Baran N, Manor H. The human protein translin specifically binds single-stranded microsatellite repeats, d(GT)n, and G-strand telomeric repeats, d(TTAGGG)n: A study of the binding parameters. J Mol Biol. 2004;344:939–950. doi: 10.1016/j.jmb.2004.09.095. [DOI] [PubMed] [Google Scholar]
- Ji H, Wong W. Computational biology: Toward deciphering gene regulatory information in mammalian genomes. Biometrics. 2006;62:645–663. doi: 10.1111/j.1541-0420.2006.00625.x. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn R, Baertsch R, Barber G, Clawson H, Diekhans M, Giardine B, Harte R, Hinrichs A, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M, Matsuzaki T, Katayanagi K, Omori A, Maziarz RT, Strominger JL, Aoki K, Suzuki K. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J Biol Chem. 1997;272:11402–11407. doi: 10.1074/jbc.272.17.11402. [DOI] [PubMed] [Google Scholar]
- Kelkar Y, Tyekucheva S, Chiaromonte F, Makova K. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. A new measure of rank correlation. Biometrika. 1938;30:81–89. [Google Scholar]
- Kondrashov A, Rogozin I. Context of deletions and insertions in human coding sequences. Hum Mutat. 2004;23:177–185. doi: 10.1002/humu.10312. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell Res. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kosmider B, Wells RD. Double-strand breaks in the myotonic dystrophy type 1 and the fragile X syndrome triplet repeat sequences induce different types of mutations in DNA flanking sequences in Escherichia coli. Nucleic Acids Res. 2006;34:5369–5382. doi: 10.1093/nar/gkl612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Cooper D. Gene deletions causing human genetic disease: Mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- Kvikstad E, Tyekucheva S, Chiaromonte F, Makova K. A macaque's-eye view of human insertions and deletions: Differences in mechanisms. PLoS Comput Biol. 2007;3:e176. doi: 10.1371/journal.pcbi.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Schwartz K, Lieber MR. Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination. Nucleic Acids Res. 2007;35:6917–6923. doi: 10.1093/nar/gkm823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Arndt PF. The majority of recent short DNA insertions in the human genome are tandem duplications. Mol Biol Evol. 2007;24:1190–1197. doi: 10.1093/molbev/msm035. [DOI] [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, Wilson SH, Pederson LC. The X family portrait: Structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale KP, Baumann M, Eberharter A, Mamais A, Becker PB, Boyes J. Acetylation increases access of remodeling complexes to their nucleosome targets to enhance initiation of V(D)J recombination. Nucleic Acids Res. 2007;35:6311–6321. doi: 10.1093/nar/gkm650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival DB, Walden AT. Wavelet methods for time series analysis. Cambridge University Press; New York: 2006. [Google Scholar]
- Pollard LM, Chutake YK, Rindler PM, Bidichandani SI. Deficiency of the RecA-dependent RecFOR and RecBCD pathways causes increased instability of the (GAA/TTC)n sequence when GAA is the lagging strand template. Nucleic Acids Res. 2007;35:6884–6894. doi: 10.1093/nar/gkm810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2005. [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;13:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Mello CC, Shimada A, Nakatani Y, Hashimoto S-i, Ogawa M, Sasaki A, Saito T, Suzuki Y, Sugano S, et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang J-PZ, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz EM, Kowalczykowski SC. Human Rad51 protein displays enhanced homologous pairing of DNA sequences resembling those at genetically unstable loci. Nucleic Acids Res. 2006;34:2847–2852. doi: 10.1093/nar/gkl355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta K, Rao BJ. Translin binding to DNA: Recruitment through DNA ends and consequent conformational transitions. Biochemistry. 2002;41:15315–15326. doi: 10.1021/bi026378m. [DOI] [PubMed] [Google Scholar]
- Steinmetz M, Vematsu Y, Lindahl K. Hotspots of homologous recombination in mammalian genomes. Trends Genet. 1987;3:7–10. [Google Scholar]
- Sweasy J, Lauper J, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- Tanay A, Siggia E. Sequence context affects the rate of short insertions and deletions in flies and primates. Genome Biol. 2008;9:R37. doi: 10.1186/gb-2008-9-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Rocha EPC. From GC skews to wavelets: A gentle guide to the analysis of compositional asymmetries in genomic data. Biochimie. 2008;90:648–659. doi: 10.1016/j.biochi.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair. 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Wang J. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Wyatt RT, Rudders RA, Zelenetz A, Delellis RA, Krontiris TG. BCL2 Oncogene translocation is mediated by a χ-like consensus. J Exp Med. 1992;175:1575–1588. doi: 10.1084/jem.175.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Cho YS, Chennathukuzhi VM, Underkoffler LA, Loomes K, Hecht NB. Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J Biol Chem. 2004;279:12605–12614. doi: 10.1074/jbc.M313133200. [DOI] [PubMed] [Google Scholar]