Abstract
Regulation of chromatin structure through post-translational modifications of histones (e.g. acetylation) has emerged as an important mechanism to translate a variety of environmental stimuli, including drugs of abuse, into specific changes in gene expression. Since alterations in gene expression are thought to contribute to the development and maintenance of the addicted state, recent efforts are aimed at identifying how drugs of abuse alter chromatin structure and the enzymes which regulate it. This review discusses how drugs of abuse alter histone acetylation in brain reward regions, through which enzymes this occurs, and ultimately what role histone acetylation plays in addiction-related behaviors.
Keywords: drug abuse, ΔFosB, histone, deacetylase, epigenetics
Introduction
Drug addiction is a chronic psychiatric condition of compulsive drug seeking and taking despite severe social and physical repercussions [1–4]. One of the most clinically challenging aspects of addiction is its persistence even after long periods of drug abstinence, which is highlighted by high rates of dug relapse for most addictive drugs. All drugs of abuse in some way converge on the mesolimbic dopamine system, a key brain circuit involved in reward [1, 3]. This circuit normally helps determine how rewarding certain environmental stimuli are and functions to reinforce evolutionally favorable activities, such as eating fatty foods or sex [5]. The dopaminergic cells of the ventral tegmental area (VTA) project into the forebrain and bathe several key limbic structures with dopamine, including the nucleus accumbens (NAc). The NAc integrates inputs from the prefrontal cortex (PFC), amygdala, hippocampus, and several other limbic structures to synthesize the appropriate synaptic and behavioral responses to rewarding stimuli including drugs of abuse [1, 3]. Addictive drugs such as cocaine have been shown to induce long-lasting structural, electrophysiological, and transcriptional changes in the NAc, and some of these changes have been linked to addictive behaviors such as drug self-administration and relapse [1–4]. One of the best studied examples is the transcription factor ΔFosB, a splice product of the immediate early gene fosB, which accumulates several fold in the NAc after repeated drug exposure due to its protein stability [6–8]. The drug-induced accumulation of ΔFosB contributes to addiction-related behaviors, as its overexpression in the NAc sensitizes mice to the locomotor-activating and rewarding effects of cocaine and morphine [9, 10], as well as promotes the motivation to self-administer cocaine [11]. In addition to ΔFosB, gene expression microarrays have been used to identify other dysregulated genes in the NAc that may also contribute to the addicted state [12–18]. These genome-wide studies begin to illustrate the potent regulatory control drugs of abuse have on gene activity in the NAc, as numerous genes are up- or down-regulated in response to chronic drug exposure. While much is known about the upstream signaling mechanisms initiated by drug-induced increases in dopamine and other neurotransmitters in the NAc [1–4], far less is known about the downstream mechanisms which integrate neurotransmitter signaling into long-lasting genome-wide alterations in transcription.
A key cellular mechanism that integrates diverse environmental stimuli with changes in gene expression is chromatin remodeling [19, 20]. Signal-dependent enzymes can alter the structure of chromatin at specific gene loci to facilitate the activation or repression of specific transcriptional programs. Chromatin is made up of DNA and the histone proteins around which the DNA is wrapped. Histones are assembled into an octamer composed of two copies each of H2A, H2B, H3, and H4 [21]. Through a complex process not completely understood, chromatin is supercoiled into a highly condense structure that packages and organizes many meters of DNA into the nucleus of each cell. This highly condensed structure provides chromatin unique control over gene expression by gating access of transcriptional activators to DNA [22, 23]. Chromatin structure itself is regulated at specific gene loci by numerous mechanisms that serve to either physically relax (e.g. histone acetylation) or remodel (e.g. SWI-SNF-dependent nucleosome remodeling) chromatin structure, or provide docking sites to recruit additional transcriptional co-activators or repressors [24]. Such modifications include histone acetylation, phosphorylation, and methylation, among several others, which together determine the activity of the underlying gene [19, 24]. This review discusses the recent evidence that changes in histone acetylation, and the enzymes which control it, contribute to drug-induced alterations in gene expression and behavior. While histone acetylation is the best studied modification in brain, it should be emphasized that other chromatin modifications typically occur in parallel and also likely participate in the long-term plasticity underlying drug addiction.
Histone Acetylation
Histone acetylation occurs on histones H2A, H2B, H3, and H4, but it is best described in brain on H3 where it can occur on the N-terminal tails at lysines 9, 14, 18, and 23 and on H4 at lysines 5, 8, 12, and 16 [24]. Histone acetylation is thought to affect the activity of a gene through two main mechanisms. First, acetylation of lysine residues reduces their positive charge and thus their electrostatic attraction to the negatively charged backbone of DNA. This reduced electrostatic attraction is thought to result in a “looser” chromatin structure allowing greater access of transcriptional activators to the underlying gene. Second, acetylated histones serve as a substrate for bromodomain-containing proteins [25]. An example of such a protein is TAFII250, which contains two bromodomains that recognize polyacetylated histone H4 [26]. TAFII250 along with the TATA-binding protein (TBP), recruit other components of the transcriptional machinery, including RNA polymerase, which ultimately transcribes the gene [27]. As would be predicted from its biochemical role, genome-wide promoter analyses in a variety of cells, tissues, and species have identified strong correlations between histone H3 and H4 acetylation and highly transcribed genes [28, 29]. Thus, histone acetylation is almost universally thought of as a “mark” of activated chromatin. In addition, histone H3 acetylation at certain genes is coupled to phosphorylation of a neighboring serine residue, serine 10, on histone H3 (H3S10), a process known as phospho-acetylation [24]. Histone phospho-acetylation occurs in concert because H3S10 phosphorylation provides a docking site for the histone acetyltransferase (HAT), GCN5, on chromatin [30]. GCN5 then maintains a hyperacetylated state, which promotes gene activation as described above. Moreover, acetylation of lysine K9 on histone H3 prevents the methylation of this same residue, which is a mark of repressed chromatin, and the subsequent recruitment of HP1 (heterochromatin protein 1). These effects of lysine K9 acetylation further promote active transcription [24].
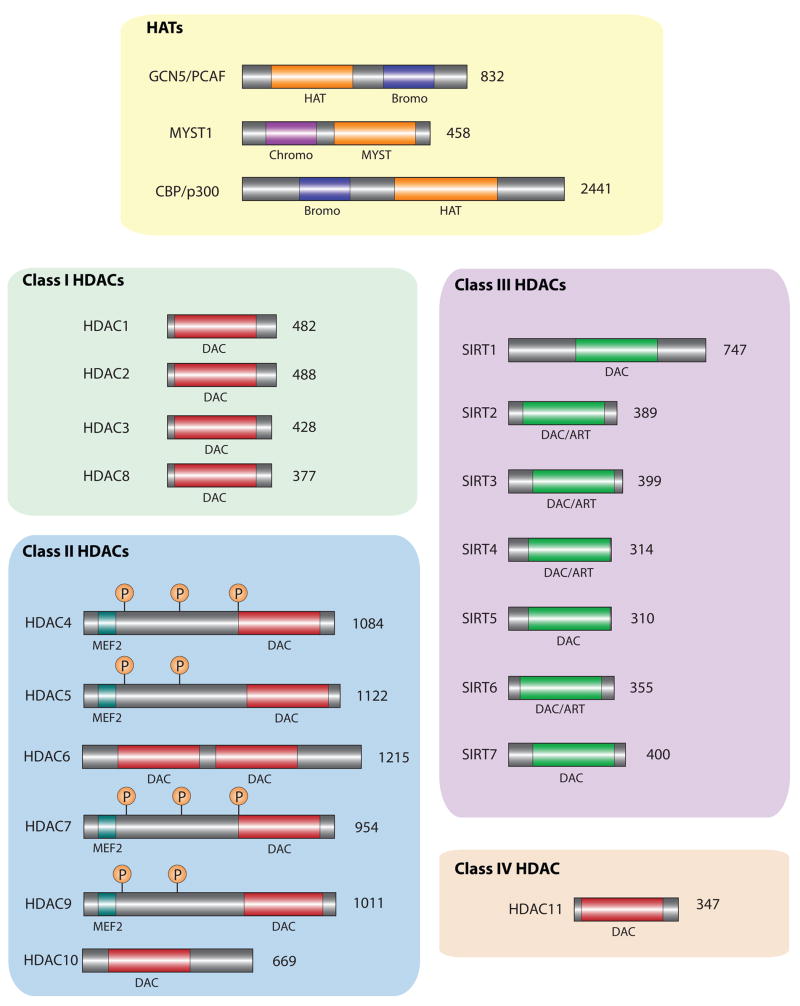
Histone acetylation is highly regulated by two families of enzymes: HATs, as mentioned earlier, and histone deacetylases (HDACs). HATs catalyze the transfer of acetyl groups from acetylCoA to a specific lysine residue, while HDACs catalyze the removal of acetyl groups from histones to yield free acetic acid (HDACs 1-11) or nicotinamide and 2′-O-acetyl-ADP-ribose (SIRT1-7) (Figure 1).
Figure 1. Histone acetyltransferases and deacetylases.
The domain organization of three major families of histone acetyltransferases (HATs) is shown, GCN5/PCAF, MYST, and CBP/p300. Conserved members of these families are found in yeast. The GCN5/PCAF and CBP/p300 families share a similar catalytic domain (HAT) while the MYST family uses the distinct but functionally similar MYST catalytic domain. The MYST family is much more diverse and the example depicted does not represent all family members. The GCN5/PCAF and CBP/p300 families contain bromodomains, which can bind to acetylated lysine residues on histone proteins. The function of chromodomains in MYST family members is still unclear, but, in other proteins, these domains can recognize methylated histones. Histone deacetylases (HDACs) are divided into four major classes, Class I-IV, based on structural homology to yeast proteins. Class I and II use zinc as a cofactor, and have similar catalytic domains (DAC). Class II HDACs are distinguished from Class I by their large N-terminal regulatory region, which recognizes transcription factors (e.g., MEF2) and are phosphorylated to control subcellular localization. The Class IV HDAC, HDAC11, is structurally similar to both Class I and Class II but has not been well studied. The catalytic deacetylase domain on Class III HDACs requires NAD+ as a cofactor and, in addition to deacetylating proteins, this domain has also been reported to have ADP ribosyltransferase activity (ART).
HATs are divided into three main families, GNAT (Gcn5-related N-acetyltransferase), MYST (MOZ/Ybf2/Sas2,3/Tip60), and CBP/p300 (CREB-binding protein/protein acetyltransferase of 300 kD). The most prominent members of the GNAT family are GCN5 and PCAF (p300/CBP-Associated Factor), both of which acetylate histone H3 and H4. While both enzymes are expressed ubiquitously, levels of GCN5 are highest in brain and testes, and levels of PCAF are highest in liver [31]. The MYST family of HATs is related by sequence similarity but has many diverse functions, from regulating HIV gene expression (Tip60) to leukemia (MOZ, MORF). Most MYST HATs have been shown to acetylate histone H3, H4, and H2A. The CBP/p300 family of HATs is the best studied in brain and has been implicated in learning and memory and drug addiction [32–35]. Mutations in CBP cause Rubinstein–Taybi Syndrome, an autosomal dominant condition characterized by mental retardation and other abnormalities [20]. CBP and p300 are distinct proteins but often referred to interchangeably due to their high degree of similarity. CBP/p300 can acetylate H2A, H2B, H3, and H4 and are recruited to specific genes through their interactions with several transcription factors (e.g. CREB, Fos/Jun). Importantly, each of these classes of HATs can acetylate non-histone proteins as well, which is known to play an important role in a variety of cell signaling pathways [36]. Interestingly, several transcription factors, such as Clock, have been reported to possess HAT activity themselves. Studies are underway to determine the contribution of such acetylation to the regulation of gene activity [37, 38].
HDACs are divided into four distinct classes based on their homology to the yeast proteins rpd3 (Class I), hda1 (Class II), sir2, (Class III), and a unique Class IV deacetylase that shares features with both rpd3 and hda1. In mammals, Class I HDACs (HDAC1, 2, 3, 8) are ubiquitously expressed and are far more active against histone substrates than Class II HDACs (HDAC4, 5, 6, 7, 9) [39]. Both Class I and Class II HDACs share a conserved catalytic domain, which requires zinc for activity, but Class II HDACs are much larger proteins which contain an N-terminal regulatory region that controls subcellular localization and interaction partners [40]. Class III HDACs, the sirtuins (SIRT1-7), are distinct from the other classes structurally and mechanistically; they require NAD+ as a cofactor instead of zinc to catalyze histone deacetylation (Figure 1) [41]. HDACs are highly regulated through post-translational modifications that control their activity, subcellular localization, or stability. The catalytic activity of Class I HDACs is regulated by phosphorylation by casein kinase II for HDACs 1–3 [42], and by PKA for HDAC8 [43, 44]. Class II HDACs respond to numerous environmental stimuli, from neural signaling to DNA damage. With neural signaling, for example, Ca2+ influx after depolarization results in activation of CaMKII, which in turns phosphorylates the class II HDACs HDAC4 and HDAC5 [45]. Phosphorylated HDAC4/5 recruit 14-3-3 proteins, which facilitate their nuclear export and reduce their activity in the nucleus [46]. Little is known about cytoplasmic functions of Class II HDACs with an exception of HDAC6, which is exclusively cytoplasmic and involved in deacetylating tubulin and shuttling misfolded proteins to the proteasome [47, 48]. Since Class III HDACs require NAD+ as a cofactor, their activity is traditionally thought to be dependent on the metabolic state of the cell (e.g. NAD+/NADH ratio). However, a recent study identified DBC1 (deleted in breast cancer 1) as a potent inhibitor of SIRT1 activity [49, 50], suggesting other upstream regulatory mechanisms also exist.
Together, the large families of HATs and HDACs integrate a wide range of extracellular signals to determine the appropriate balance of acetylation/deacetylation on diverse protein substrates. In the case of histone substrates, the balance of acetylation plays a critical role in the activation or repression of specific gene programs that ultimately mediate long-term adaptations in neural function and behavior.
Drug-Induced Changes in Histone Acetylation
Cocaine/Amphetamine
Cocaine and amphetamine substantially elevate dopamine levels in several forebrain regions involved in reward processing such as the NAc and PFC [1, 51]. The molecular changes induced by cocaine and amphetamine in brain reward regions are typically studied in rodents using investigator-administered or self-administered drug paradigms. Investigator-administration is a relatively high throughput method used to examine differences caused by acute or chronic drug exposure at varying time points after treatment. Self-administration paradigms, while labor-intensive, are better models of addiction and permit the study of rodents that, like humans, choose to take cocaine or another drug of abuse. Additionally, self-administration permits the study of molecular mechanisms involved in drug relapse. Both of these paradigms have been used to identify transient and long-lasting changes in histone acetylation after cocaine or amphetamine exposure [52–54].
Acute exposure to cocaine or amphetamine, for example, which are known to rapidly induce the immediate early genes c-fos and fosb in the NAc, increases histone H4 acetylation and phospho-acetylation of H3 on their proximal gene promoters (Figure 1) [53, 54]. Time course analysis after cocaine revealed that this modification occurs within 30 minutes and disappears by 3 hours, consistent with the induction kinetics of these immediate early genes [53]. At least for fosb, this increase in histone acetylation is dependent on the HAT, CBP [34]. While the specific HAT responsible for acetylating c-fos after cocaine remains to be identified, there is evidence that the downstream mitogen activated protein kinase, MSK1 is responsible for cocaine-induced increases in H3S10 phosphorylation (Figure 1) [55]. A recent study has also implicated the nuclear accumulation of DARPP32, a protein phosphatase 1 (PP1) inhibitor, in mediating cocaine induction of H3S10 phosphorylation [56]. Interestingly, despite several control gene promoters where acute cocaine does not affect histone acetylation (β-tubulin, tyrosine hydroxylase, histone H4) [53], acute cocaine does increase global levels of histone H4 acetylation and histone H3 phospho-acetylation, but not H3 acetylation alone within 30 minutes [53, 55]. Thus, global changes in histone modifications, which have been observed in learning models and in response to environmental enrichment [57, 58], may be accounted for by altered acetylation at a specific subset of genes.
The induction of immediate early genes such as c-fos by cocaine or amphetamine after repeated drug exposure is strongly attenuated, a point at which histone H4 on c-fos promoter is hypoacetylated [53, 59]. In addition to desensitizing immediate early genes, repeated cocaine exposure (either forced- or self-administered) is also known to induce a distinct set of genes in the NAc (e.g. cdk5 and bdnf), some of which remain elevated for days to weeks [13, 16, 60, 61]. Consistent with such stable changes in gene expression, increased histone H3 acetylation was observed on the gene promoters of both cdk5 and bdnf for 1–7 days following the final dose of cocaine [53]. Stable changes in histone acetylation and gene expression have been observed for nearly two weeks following withdrawal from cocaine self-administration in the prefrontal cortex as well [52]. For example, npy (neuropeptide Y) expression was found to be upregulated and its gene promoter hyperacetylated, while egr-1 (early growth response 1) was found to be downregulated and hypoacetylated after cocaine withdrawal [52]. Together, these findings tightly correlate gene activity in the brain in vivo to levels of histone acetylation, a finding which is consistent with the role of histone acetylation in cell culture and yeast.
Ethanol and other drugs of abuse
Ethanol, a central nervous system depressant, is one of the most widely available and commonly abused drugs. The precise signaling mechanisms by which ethanol alters cognition are still under investigation, but its binding to and modulation of GABA receptors is thought to contribute [62]. The upstream signaling pathways activated by ethanol appear to converge on chromatin in the nucleus, as circular dichronism of brain lysates from ethanol-treated rats suggest a more relaxed chromatin structure than their controls [63]. More recent studies have focused on specific brain regions involved in ethanol-regulated behavioral responses in rats. Anxiety induced by ethanol withdrawal is an important negative reinforcing factor in addicts, and is thought to be mediated in part by the amygdala. Indeed, a recent study has found that ethanol withdrawal alters HDAC activity, gene expression, and chromatin structure in this brain region, and that these changes contribute to increases in anxiety [64]. Specifically, withdrawal from chronic ethanol exposure increased HDAC activity and reduced histone acetylation in the amygdala, and when this increased HDAC activity was blocked with an HDAC inhibitor, rats failed to develop withdrawal-induced anxiety (Figure 1) [64].
In Drosophila, changes in histone acetylation have also been implicated in the molecular and behavioral responses to the volatile organic solvent, benzyl alcohol [65]. Many organic solvents are abused as inhalants, which often induce rapid drug tolerance, thereby increasing the amount of drug needed to attain an equivalent behavioral effect. Ultimately, tolerance to inhalants can result in addiction and severe neurotoxicity. In Drosophila, a key mechanism involved in benzyl alcohol tolerance is the upregulation of slo, a Ca2+-activated K+ channel [66]. Induction of slo is associated with increases in histone H4 acetylation on its promoter, which is likely important for gene activation because pharmacological inhibition of HDAC activity increased both slo gene acetylation and mRNA expression [65]. Moreover, HDAC inhibition potentiated tolerance to benzyl alcohol, which further implicates histone acetylation in the behavioral responses to drugs of abuse.
Genome-wide analysis of histone acetylation in the NAc
Analyzing changes in histone acetylation at specific gene promoters via quantitative ChIP (qChIP) has demonstrated a key role of this modification in regulating gene expression in response to drugs of abuse [34, 52–54]. However, qChIP is relatively low throughput and is generally used to study carefully chosen candidate genes. Therefore, coupling ChIP to genome-wide microarrays (ChIP-chip) or massively parallel sequencing (ChIP-seq) provides the technology necessary to simultaneously identify drug-induced changes in histone acetylation throughout the genome, and such analyses have now been performed to assess cocaine action on chromatin structure in the NAc [67].
ChIP-chip analysis of histone acetylation in the NAc of cocaine-treated mice has provided new insight into both basic transcriptional mechanisms occurring in vivo as well as novel pathways involved in cocaine action. One such insight directly follows from a previous report using qChIP to analyze histone acetylation on a few genes already known to be involved in cocaine responses. On the small number genes studied, a pattern emerged whereby genes which are induced by acute cocaine (e.g. c-fos, fosb) were also hyperacetylated at histone H4, while genes which are induced by chronic cocaine (e.g. cdk5, bdnf) were hyperacetylated at histone H3 [53]. Similar findings were observed in acute and chronic seizure models [68]. Moreover, a switch between H4 and H3 acetylation was observed on the promoter of fosb, an immediate early gene which is induced after both acute and chronic cocaine [53]. Genome-wide ChIP-chip analysis reveals that while there are more H3 acetylated genes in the NAc of mice exposed to chronic cocaine, there is also a significant set of previously unrecognized, chronically-induced genes that are hyperacetylated only on H4 (Figure 2D) [67]. There was also an even smaller subset of genes which were acetylated on both histones H3 and H4. The mechanisms which determine whether a gene is hyperacetylated at H3 or H4 remain unclear, but these new data suggest that after chronic cocaine, there may be two distinct pathways to activate gene expression, one through acetylation of H3 and the other through H4. Additionally, these data also suggest that the H4 to H3 switch observed in both cocaine and seizure models may be reserved for certain types of genes, such as immediate early genes.
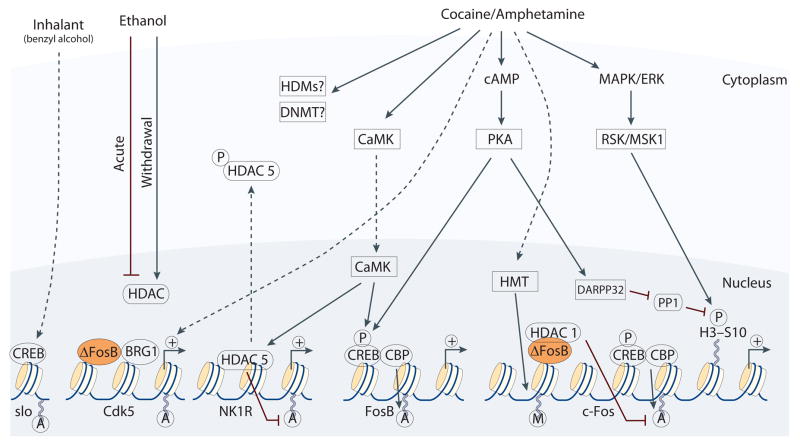
Figure 2. Regulation of chromatin structure by drugs of abuse.
Drug-induced signaling events are depicted for cocaine/amphetamine, ethanol, and an inhalant, benzyl alcohol. Cocaine and amphetamine can increase cAMP levels in striatum, which activates protein kinase A (PKA) and leads to phosphorylation of its targets. This includes the cAMP response element binding protein (CREB), the phosphorylation of which induces its association with the histone acetyltransferase, CREB binding protein (CBP) to acetylate histones and facilitate gene activation. This is known to occur on many genes including fosB and c-fos in response to psychostimulant exposure. ΔFosB is also upregulated by chronic psychostimulant treatments, and is known to activate certain genes (e.g., cdk5) where it recruits the SWI-SNF chromatin remodeling enzyme, BRG1, and represses others (e.g., c-fos) where it recruits HDAC1. This repression of c-fos also involves increased repressive histone methylation, which is thought to occur via the induction of specific histone methyltransferases. It is not yet known how cocaine regulates histone demethylases (HDM) or DNA methyltransferases (DNMTs). Cocaine also activates the mitogen activated protein kinase (MAPK) cascade, which through MSK1 can phosphorylate CREB and histone H3 at serine 10. Cocaine promotes H3 phosphorylation via a distinct pathway, whereby PKA activates protein phosphatase 2A, leading to the dephosphorylation of serine 97 of DARPP32. This causes DARPP32 to accumulate in the nucleus and inhibit protein phosphatase 1 (PP1) which normally dephosphorylates H3. Chronic exposure to psychostimulants is also known to increase glutamatergic stignaling from the prefrontal cortex to the NAc. Glutamatergic signaling elevates Ca2+ levels in NAc synapses and activates CaMK (calcium/calmodulin protein kinases) signaling, which, in addition to phosphorylating CREB, also phosphorylates HDAC5. This results in nuclear export of HDAC5 and increased histone acetylation on its target genes (e.g., NK1R [NK1 or substance P receptor). Acute ethanol has been shown to reduce histone acetylation by increasing HDAC activity, while withdrawal from chronic ethanol increases histone acetylation by reducing HDAC activity. The organic solvent, benzyl alcohol, which is used to study the tolerance effects of inhalants, rapidly induces histone acetylation on the Ca2+-activated K+ channel, slo, to facilitate CREB-dependent transcription. Figure was modified with permission from [20].
Another important result from these first genome-wide studies of histone acetylation in cocaine responses is the new set of gene targets and signaling pathways revealed. In addition to identifying several genes previously implicated in cocaine responses (e.g. the genes for CDK5 [Cyclin-dependent kinase 5], AGS3 [Activator of G-protein signaling 3], PER2 [Period 2], DYN [Dynorphin], etc.), a new class of histone deacetylases, the sirtuins, was also found to be regulated by cocaine in the NAc [67]. Both sirt1 and sirt2 are hyperacetylated at histone H3 and have higher levels of mRNA in response to chronic cocaine exposure. ΔFosB appears to contribute to these transcriptional changes. Moreover, pharmacological manipulation of these enzymes potently regulates cocaine reward, illustrating how genome-wide ChIP-chip analyses can ultimately reveal novel gene targets involved in behavioral responses to drugs of abuse.
Role of HATs and HDACs in drug abuse
Histone acetyltransferases
As discussed above, HATs are enzymes which catalyze the addition of acetyl groups onto histone proteins. One of the best studied HATs is CBP, which is known to associate with phosphorylated CREB and assist in target gene activation [36, 69]. While cocaine increases H4 acetylation on the fosb promoter in striatum of wild type mice, this does not occur in CBP-deficient mice [34]. Moreover, this cocaine-induced acetylation event on fosb and perhaps other genes may be behaviorally important, as CBP-deficient mice are also less sensitive to the locomotor-activating effects of cocaine. These findings suggest that CBP is an important HAT in mediating at least some of the cocaine-induced increases in histone acetylation and ultimately the downstream behavioral responses [34]. The reduced sensitivity of CBP-deficient mice to cocaine may be independent of CREB, since other manipulations which reduce CREB activity in striatum typically enhance behavioral responses to cocaine [70, 71]. However, since CBP-deficient mice have reduced CBP levels in all brain regions throughout development, it will be important to determine which specific regions in the adult brain are important for CBP’s behavioral effects. Moreover, these studies may also provide insight into what role CREB plays in mediating the downstream functions of CBP in cocaine responses. Mice expressing mutant forms of CREB that cannot interact with CBP [72] may also help address this question.
Histone deacetylases
Small molecule inhibitors of HDACs have been used in many systems since they were first identified in 1978 [73], but have only recently been used to study behavior. Sodium butyrate, a highly non-specific HDAC inhibitor, was the first discovered [73], followed by more specific and potent inhibitors trichostatin A (TSA) [74], a fungal antibiotic, and suberoylanilide hydroxamic acid (SAHA) [75], which is currently FDA-approved for treatment of cutaneous T-cell lymophoma. Recent studies have shown that each of these HDAC inhibitors potentiates the locomotor-activating effects of psychostimulants, as measured by the number of horizontal infrared beam breaks, as well as the rewarding effects psychostimulants, as measured by conditioned place preference [53, 54, 76]. Importantly, systemic administration of sodium butyrate or TSA, or the direct infusion of SAHA into the NAc, is sufficient to potentiate the locomotor activating or rewarding effects of cocaine, amphetamine, and D1 agonists [53, 54, 76, 77]. Conversely, overexpression of the Class II HDACs, HDAC4 or HDAC5 (but not HDAC9), in the NAc significantly attenuates cocaine reward [53, 54], but it is not yet known if HDAC4/5 overexpression also reduces the locomotor-activating effects of cocaine. The behavioral effects of HDAC overexpression in the NAc likely require histone deacetylation, since deletion of the catalytic deacetylase domain of HDAC5 prevents its inhibitory effects on cocaine reward [54]. However, since HDAC5 also interacts with HDAC3 at this same domain [78], it is unclear whether HDAC5’s effects on cocaine reward are due to its own catalytic activity or its interaction with HDAC3.
In the NAc, endogenous HDAC5 also plays a key role in regulating gene expression in response to chronic, but not acute, cocaine exposure. Only repeated exposure to cocaine induces phosphorylation and nuclear export of HDAC5, thus reducing histone deacetylation and increasing gene expression of certain HDAC5 target genes [54]. Consistent with its regulation only by chronic cocaine exposure, drug-naïve HDAC5 knockout mice display normal cocaine reward. After a prior cocaine exposure, however, HDAC5 knockout mice hypersensitize to the rewarding effects of cocaine [54]. These findings raise the possibility that HDAC5 is involved in the behavioral transitions which occur between acute and chronic cocaine exposure (e.g., experimental drug use to compulsive drug use). The receptor for substance P, NK1, is one relevant target gene for HDAC5: its expression is induced to a greater extent by chronic cocaine in HDAC5 knockout mice and this induction appears to contribute to enhanced behavioral responses to the drug [45].
The Class I HDAC, HDAC1 is also involved in the cellular responses to chronic psychostimulant exposure: it binds to Δ FosB and facilitates repression of downstream target genes [59]. An example of this occurs at the c-fos promoter, where ΔFosB accumulates after a chronic course of psychostimulants to desensitize its activation to subsequent drug exposure. Prior administration of the HDAC inhibitor, sodium butyrate, or local genetic deletion of the HDAC1 gene from the NAc, increases c-fos induction despite previous treatment with chronic psychostimulants [59]. This ΔFosB/HDAC1-mediated desensitization of c-fos is part of the “molecular switch” whereby acute Fos family proteins are gradually replaced by ΔFosB during a course of chronic drug exposure. Further studies are needed to identify other genes that are repressed via this unique mechanism.
Functions of histone acetylation in vivo
These studies in drug abuse, together with findings in models of leaning and memory, depression, and chronic pain, suggest that histone acetylation controls the saliency of a wide variety of environmental stimuli [20, 34, 53, 54, 57, 58, 79, 80]. In fear conditioning, a commonly used behavioral test of learning and memory, HDAC inhibitors significantly improve formation of long-term memory [57, 58]. In social defeat stress, a behavioral model which elicits a depression-like syndrome in mice [81, 82], mice lacking HDAC5 develop a stronger depressive-like syndrome than wild type mice [54]. After spinal nerve injury, which elicits a chronic pain syndrome, HDAC5 knockout mice also become hyperalgesic to mechanical stimuli [79]. While reduced HDAC activity hypersensitizes mice to stress or pain, it also significantly potentiates the efficacy of several antidepressants [79, 83, 84]. Finally, as discussed above, inhibition of HDACs in the NAc potentiates the rewarding effect of psychostimulants while overexpressing HDACs in this brain region attenuates this behavior [53, 54]. Thus, pharmacological and genetic manipulations that result in elevated histone acetylation appear to potentiate the saliency of many types of environmental stimuli.
Despite the substantial progress in identifying the genes at which drugs of abuse alter histone acetylation, many questions remain. For example, we still do not know if histone acetylation in brain is a primary cause of changes in gene expression or if it is simply a reflection of such changes. This is complicated by the fact that at most genes identified to date, gene expression and histone acetylation are both elevated and return to baseline with a similar time frame. However, the cocaine-induced regulation of bdnf provides unique insight into the possible interplay between acetylation and transcription in vivo. Rats in withdrawal from cocaine self-administration show a marked increase in histone acetylation in the NAc and dorsal striatum on the bdnf promoter within 24 hrs after the final drug exposure [53]. However, no detectable increases in BDNF protein are observed for nearly a week of drug withdrawal [61]. This is an example where histone acetylation on a gene promoter precedes the induction of gene expression, and suggests that, in addition to marking an actively transcribed gene, histone acetylation may also prime a gene for more rapid or robust transcription in response to a subsequent stimulus without altering its basal level of gene activity. Better knowledge of the genes at which cocaine regulates histone acetylation may result in the identification of novel gene targets and signaling pathways involved in addiction-related behaviors that were previously not appreciated by studies of steady-state mRNA levels.
Future studies and challenges
While drug-induced alterations in histone acetylation have to date been implicated in the behavioral responses to drugs of abuse in simple models such as conditioned place preference and locomotor assays, future research is needed to translate these findings to more sophisticated models of human addiction, such as self-administration and relapse paradigms. These pre-clinical behavioral models can directly assess the role of histone acetylation in the pathogenesis and maintenance of the addicted state by studying how animals acquire drug self-administration and how they relapse after periods of drug withdrawal.
The study of gene expression and chromatin remodeling in adult brain is made difficult by two major technical challenges. The first challenge is inherent to the structure of the brain: it is highly heterogeneous and contains many subtypes of neurons and glia, each of which are further distinguished based on connectivity and function. Thus, the cellular and molecular responses to cocaine can vary tremendously even within a small subregion of the NAc. In response to cocaine, for example, neurons which express the Gs-coupled dopamine D1 receptors in the NAc show higher levels of cAMP and its downstream consequences, while neurons expressing the Gi-coupled D2 receptors respond oppositely. There is even a small population of striatal neurons which express both D1 and D2 receptors, where they are reported to form heterooligomers that couple to Gq signaling [85], even further complicating the cellular responses to cocaine. The end result of studying tissue lysates from such a heterogeneous structure is that most of the observed effects will be very small since they are averaged together with several other, differently responding, cell types. This is particularly problematic for microarray analyses, which depend on strong effects for statistical significance. One solution that is being currently developed is fluorescence activated cell sorting (FACS) in conjunction with BAC-transgenic mice that express GFP in specific cell types (e.g., dopamine D1 vs. D2 neurons) [86]. This new technology will permit the study of how cocaine alters gene expression and histone modifications in isolated cell populations.
The second major challenge is determining causality from correlation. How does one determine whether histone acetylation at the bdnf promoter causes a transcriptional or behavioral response in vivo? To address this question, one must induce or prevent histone acetylation on the bdnf gene specifically. Simply overexpressing a HAT or inhibiting HDACs would likely hyperacetylate numerous other target genes in addition to bndf, thus confounding the transcriptional and behavioral interpretations. Recently, an exciting breakthrough using zinc finger proteins may enable us to directly address this very challenging question. Zinc finger peptides can be designed or screened for highly sequence-specific DNA binding properties. These zinc finger peptides can then be fused to a chromatin remodeling enzyme, which would effectively target that enzyme to the promoter of a specific gene. This was accomplished for the first time using a DNA methyltransferase in cell culture [87, 88], and may now permit behavioral neuroscientists, using viral-mediated gene transfer, to ask whether histone modifications at specific genes are indeed causally linked to the transcriptional and behavioral phenotypes observed.
Conclusions
Histone acetylation has been shown to be involved in modulating the saliency of many environmental stimuli. Conditions which increase levels of histone acetylation appear to sensitize mice to cocaine, stress, pain, etc. while conditions that reduce histone acetylation diminish sensitivity to these stimuli. This has important implications in the pathogenesis of drug addiction, depression, chronic pain, and memory disorders. Ultimately, the key function of histone acetylation is to increase the transcription or the transcriptional potential of genes which eventually alter neural function [53]. Thus, any study of chromatin modifications is, in theory, inexorably linked with the study of the underlying gene activity, and may reveal novel signaling pathways not previously identified from studies of steady-state mRNA and protein levels. Regulation of chromatin structure also offers a fundamentally new approach for the development of more effective treatments for drug addiction and other neuropsychiatric disorders.
Acknowledgments
Preparation of this review was supported by grants from NIDA (EJN) and the UT Southwestern Medical Scientist Training Program (WR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–98. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 2.Kalivas PW, Volkow N, Seamans J. Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron. 2005;45:647–50. doi: 10.1016/j.neuron.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Koob G, Kreek MJ. Stress, dysregulation of drug reward pathways, and the transition to drug dependence. Am J Psychiatry. 2007;164:1149–59. doi: 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–28. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 5.Kelley AE, Berridge KC. The neuroscience of natural rewards: relevance to addictive drugs. J Neurosci. 2002;22:3306–11. doi: 10.1523/JNEUROSCI.22-09-03306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carle TL, Ohnishi YN, Ohnishi YH, Alibhai IN, Wilkinson MB, Kumar A, et al. Proteasome-dependent and -independent mechanisms for FosB destabilization: identification of FosB degron domains and implications for DeltaFosB stability. Eur J Neurosci. 2007;25:3009–19. doi: 10.1111/j.1460-9568.2007.05575.x. [DOI] [PubMed] [Google Scholar]
- 7.McClung C, Ulery P, Perrotti L, Zachariou V, Berton O, Nestler E. DeltaFosB: a molecular switch for long-term adaptation in the brain. Mol Brain Res. 2003;132:146–54. doi: 10.1016/j.molbrainres.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Ulery PG, Rudenko G, Nestler EJ. Regulation of DeltaFosB stability by phosphorylation. J Neurosci. 2006;26:5131–42. doi: 10.1523/JNEUROSCI.4970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelz MB, Chen J, Carlezon WA, Jr, Whisler K, Gilden L, Beckmann AM, et al. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–6. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- 10.Zachariou V, Bolanos CA, Selley DE, Theobald D, Cassidy MP, Kelz MB, et al. An essential role for DeltaFosB in the nucleus accumbens in morphine action. Nat Neurosci. 2006;9:205–11. doi: 10.1038/nn1636. [DOI] [PubMed] [Google Scholar]
- 11.Colby CR, Whisler K, Steffen C, Nestler EJ, Self DW. Striatal cell type-specific overexpression of DeltaFosB enhances incentive for cocaine. J Neurosci. 2003;23:2488–93. doi: 10.1523/JNEUROSCI.23-06-02488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman WM, Nader MA, Nader SH, Robertson DJ, Gioia L, Mitchell SM, et al. Chronic cocaine-mediated changes in non-human primate nucleus accumbens gene expression. J Neurochem. 2001;77:542–9. doi: 10.1046/j.1471-4159.2001.00252.x. [DOI] [PubMed] [Google Scholar]
- 13.McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–15. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 14.McClung CA, Nestler EJ, Zachariou V. Regulation of gene expression by chronic morphine and morphine withdrawal in the locus ceruleus and ventral tegmental area. J Neurosci. 2005;25:6005–15. doi: 10.1523/JNEUROSCI.0062-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winstanley CA, LaPlant Q, Theobald DE, Green TA, Bachtell RK, Perrotti LI, et al. DeltaFosB induction in orbitofrontal cortex mediates tolerance to cocaine-induced cognitive dysfunction. J Neurosci. 2007;27:10497–507. doi: 10.1523/JNEUROSCI.2566-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, et al. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron. 2004;41:625–38. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- 17.Yuferov V, Kroslak T, Laforge KS, Zhou Y, Ho A, Kreek MJ. Differential gene expression in the rat caudate putamen after “binge” cocaine administration: advantage of triplicate microarray analysis. Synapse. 2003;48:157–69. doi: 10.1002/syn.10198. [DOI] [PubMed] [Google Scholar]
- 18.Yuferov V, Nielsen D, Butelman E, Kreek MJ. Microarray studies of psychostimulant-induced changes in gene expression. Addict Biol. 2005;10:101–18. doi: 10.1080/13556210412331308976. [DOI] [PubMed] [Google Scholar]
- 19.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 20.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 21.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–6. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 22.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–53. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–5. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 27.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–99. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 28.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–33. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 30.Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–26. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 31.Xu W, Edmondson DG, Roth SY. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol. 1998;18:5659–69. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–59. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 33.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–72. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine AA, Guan Z, Barco A, Xu S, Kandel ER, Schwartz JH. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci U S A. 2005;102:19186–91. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Mem. 2005;12:111–9. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001;58:693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 38.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–82. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 39.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–40. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–18. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 43.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–73. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A. 2004;101:15064–9. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chawla S, Vanhoutte P, Arnold FJ, Huang CL, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–9. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 46.McKinsey T, Zhang C, Olson E. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–5. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–8. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–38. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 49.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 50.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalivas PW. Glutamate systems in cocaine addiction. Curr Opin Pharmacol. 2004;4:23–9. doi: 10.1016/j.coph.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Freeman WM, Patel KM, Brucklacher RM, Lull ME, Erwin M, Morgan D, et al. Persistent alterations in mesolimbic gene expression with abstinence from cocaine self-administration. Neuropsychopharmacology. 2007 doi: 10.1038/sj.npp.1301577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–14. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–29. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 55.Brami-Cherrier K, Valjent E, Herve D, Darragh J, Corvol JC, Pages C, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–54. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stipanovich A, Valjent E, Matamales M, Nishi A, Ahn JH, Maroteaux M, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008 doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–82. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 58.Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–59. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 59.Renthal W, Carle TL, Maze I, Covington HE, 3rd, Truong HT, Alibhai I, et al. DeltaFosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28:7344–9. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder GL, et al. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376–80. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- 61.Grimm JW, Lu L, Hayashi T, Hope BT, Su TP, Shaham Y. Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: implications for incubation of cocaine craving. J Neurosci. 2003;23:742–7. doi: 10.1523/JNEUROSCI.23-03-00742.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lobo IA, Harris RA. GABA(A) receptors and alcohol. Pharmacol Biochem Behav. 2008 doi: 10.1016/j.pbb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahadev K, Vemuri MC. Effect of ethanol on chromatin and nonhistone nuclear proteins in rat brain. Neurochem Res. 1998;23:1179–84. doi: 10.1023/a:1020778018149. [DOI] [PubMed] [Google Scholar]
- 64.Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–37. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Krishnan HR, Ghezzi A, Yin JC, Atkinson NS. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol. 2007;5:2342–53. doi: 10.1371/journal.pbio.0050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghezzi A, Al-Hasan YM, Larios LE, Bohm RA, Atkinson NS. slo K(+) channel gene regulation mediates rapid drug tolerance. Proc Natl Acad Sci U S A. 2004;101:17276–81. doi: 10.1073/pnas.0405584101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar A, Sikder D, Renthal W, Covington HE, 3rd, Chakravarty S, Choi KH, et al. Genome-wide epigenetic changes underlying chronic cocaine-induced neuroadaptations in the mouse nucleus accumbens. Soc Neurosci Abs. 2007:767.5. [Google Scholar]
- 68.Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci. 2004;24:5603–10. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–9. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 70.Carlezon WA, Jr, Thome J, Olson VG, Lane-Ladd SB, Brodkin ES, Hiroi N, et al. Regulation of cocaine reward by CREB. Science. 1998;282:2272–5. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- 71.Walters CL, Blendy JA. Different requirements for cAMP response element binding protein in positive and negative reinforcing properties of drugs of abuse. J Neurosci. 2001;21:9438–44. doi: 10.1523/JNEUROSCI.21-23-09438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006;13:609–17. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–13. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 74.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–9. [PubMed] [Google Scholar]
- 75.Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, et al. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci U S A. 1998;95:3003–7. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kalda A, Heidmets LT, Shen HY, Zharkovsky A, Chen JF. Histone deacetylase inhibitors modulates the induction and expression of amphetamine-induced behavioral sensitization partially through an associated learning of the environment in mice. Behav Brain Res. 2007;181:76–84. doi: 10.1016/j.bbr.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schroeder FA, Penta KL, Matevossian A, Jones SR, Konradi C, Tapper AR, et al. Drug-induced activation of dopamine D(1) receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology. 2008 doi: 10.1038/npp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 79.Renthal W, Terzi D, Olson EN, Nestler EJ, Zachariou V. Histone deacetylase 5 controls analgesic responsiveness to tricyclic antidepressants in a mouse model of neurophatic pain. Soc Neurosci Abs. 2008 [Google Scholar]
- 80.Zhang C, McKinsey T, Chang S, Antos C, Hill J, Olson E. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–88. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–8. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 82.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 83.Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 84.Tsankova N, Berton O, Renthal W, Kumar A, Neve R, Nestler E. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 85.Rashid AJ, So CH, Kong MM, Furtak T, El-Ghundi M, Cheng R, et al. D1-D2 dopamine receptor heterooligomers with unique pharmacology are coupled to rapid activation of Gq/11 in the striatum. Proc Natl Acad Sci U S A. 2007;104:654–9. doi: 10.1073/pnas.0604049104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–52. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 87.Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, et al. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100–12. doi: 10.1093/nar/gkl1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith AE, Hurd PJ, Bannister AJ, Kouzarides T, Ford KG. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J Biol Chem. 2008;283:9878–85. doi: 10.1074/jbc.M710393200. [DOI] [PubMed] [Google Scholar]