SUMMARY
Systemic acquired resistance (SAR) is a broad-spectrum plant immune response involving profound transcriptional changes that are regulated by the co-activator NPR1. Nuclear translocation of NPR1 is a critical regulatory step, but how it is regulated in the nucleus is unknown. Here, we show that turnover of nuclear NPR1 protein plays an important role in modulating its target gene transcription. In the absence of pathogen challenge, NPR1 is continuously cleared from the nucleus by the proteasome, which restricts its co-activator activity to prevent untimely activation of SAR. Surprisingly, inducers of SAR promote turnover of NPR1 by phosphorylation of residues Ser11/Ser15, thereby facilitating its recruitment to a Cullin3-based ubiquitin ligase. Genetic experiments showed that turnover of phosphorylated NPR1 is required for full induction of target genes and establishment of SAR. These in vivo data demonstrate unique dual roles for co-activator turnover in both preventing and stimulating gene transcription to regulate plant immunity.
INTRODUCTION
Immune responses are tightly regulated in all eukaryotes to ensure that they are effective only against invading pathogens but not harmful to selves. In contrast to animals, plants lack a specialized immune system and instead rely on each individual cell for defense. In response to pathogen challenge, plant cells undergo dramatic transcription reprogramming to favor immune responses over normal cellular functions. Failure to do so results in infection. On the other hand, suppressing immune responses in the absence of a pathogen threat is equally important for maintaining plant growth and development. Thus, plants have sophisticated regulatory mechanisms to control defense-related transcription.
An important signal molecule for defense-related transcription in plants is salicylic acid (SA) . Pathogen-induced increases in cellular SA levels or exogenous application of SA leads to profound changes in gene transcription (reviewed in Durrant and Dong, 2004). These changes occur through the activity of the transcription co-activator NPR1 (nonexpressor of pathogenesis-related (PR) genes), a master regulator of plant immunity. Mutations in the NPR1 gene in Arabidopsis block this SA-mediated transcriptional reprogramming and renders the plant completely defective in systemic acquired resistance (SAR), an inducible immune response against a broad-spectrum of pathogens (Cao et al., 1994; Delaney et al., 1995; Wang et al., 2006).
The activity of NPR1 is regulated in part by its subcellular localization (Kinkema et al., 2000). In unchallenged cells NPR1 is predominantly sequestered in the cytoplasm as a high molecular weight oligomeric complex (Mou et al., 2003). The oligomeric complex is formed through redox-sensitive intermolecular disulfide bonds between conserved cysteine residues. Upon pathogen infection, accumulation of SA triggers a change in cellular reduction potential, resulting in partial reduction of NPR1 oligomer to monomer. A bipartite nuclear localization sequence targets the released NPR1 monomer to the nucleus where it functions as a co-activator of gene transcription (Kinkema et al., 2000). Furthermore, NPR1 was found to interact with TGA transcription factors (Després et al., 2000; Zhang et al., 1999; Zhou et al., 2000) whose binding motif has been shown to be essential for SA-responsiveness of the PR-1 gene (Lebel et al., 1998). NPR1 may affect both the DNA binding capacity and the activity of TGA factors (Després et al., 2003; Després et al., 2000; Fan and Dong, 2002; Johnson et al., 2003; Rochon et al., 2006). Besides the PR genes, which encode antimicrobial effectors, NPR1 also directly activates the expression of several WRKY transcription factors with both activator and suppressor activities (Wang et al., 2006). Thus, NPR1 regulates plant immunity through a transcription cascade involving multiple transcription factors.
A major challenge in understanding the function of NPR1 is to uncover the nuclear regulation of this co-activator. Phosphorylation and ubiquitin-mediated proteolysis are prominent post-translational mechanisms that control transcription regulators. In mammalian immunity, the co-factor IκB, which shares structural features with NPR1 (Cao et al., 1997; Ryals et al., 1997), functions to sequester the transcription factor NF-κB in the cytoplasm and prevents it from activating gene expression. In response to pathogen attack, IκB is rapidly phosphorylated and targeted for ubiquitin-mediated proteolysis, allowing NF-κB to localize to the nucleus and activate target genes (Hayden and Ghosh, 2004). Furthermore, transcription factors are often unstable and a significant overlap has been found between transcriptional activation domains and domains that regulate ubiquitin-mediated proteolysis (Salghetti et al., 2000). Recent findings indicate that proteasome-mediated turnover of activators may be essential for their ability to activate transcription (Collins and Tansey, 2006). Whereas activator turnover is thought to stimulate gene transcription, it remains largely unknown if proteolysis plays a role in the regulation of transcription co-activators.
In this study we investigated if the co-activator NPR1 is regulated by post-translational mechanisms. Our findings revealed opposing roles for co-activator proteolysis in the regulation of gene transcription and demonstrate for the first time that multi-cellular organisms employ proteolysis-coupled transcription as a mechanism to control their responses to external stimuli.
RESULTS
NPR1 is subject to proteasome-mediated degradation
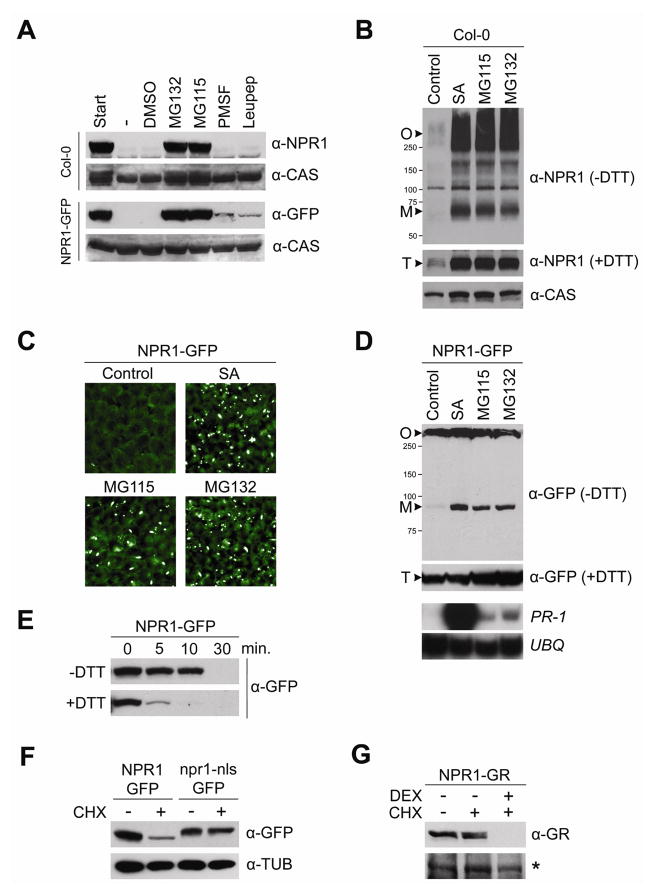
To examine if protein stability plays a role in NPR1 regulation, we performed a cell-free degradation assay (see Supplemental methods) using extracts from wild-type Col-0 plants and previously characterized transgenic 35S::NPR1-GFP plants (Kinkema et al., 2000; Mou et al., 2003). We found that both the endogenous NPR1 and NPR1-GFP were completely degraded within two hours (Figure 1A). To test which cellular mechanism is responsible for this observed degradation, we studied the effect of several proteolysis inhibitors. Whereas addition of the proteasome inhibitors MG115 or MG132 prevented NPR1 and NPR1-GFP degradation, the protease inhibitors leupeptin and PMSF were ineffective in this respect (Figure 1A), indicating that NPR1 degradation specifically requires proteasome activity.
Figure 1. Proteasome-mediated degradation of NPR1 monomer in the nucleus.
(A) Total protein was extracted from wild-type Col-0 and 35S::NPR1-GFP (in npr1-1) plants in a buffer supporting proteolytic activity. Extracts were untreated (−) or treated with either 2% DMSO (vehicle), 40 μM MG132, 40 μM MG115, 4 mM PMSF, or 40 μM Leupeptin. After 2 hours, proteins were analyzed by reducing SDS-PAGE and Western blotting using anti-NPR1 and anti-GFP antibodies. Detection of a constitutively expressed calcium-sensing receptor (CAS) confirmed equal loading.
(B) Wild-type Col-0 plants were treated for 24 hours with either water (control), 0.5 mM SA, 100 μM MG115, or 100 μM MG132. Total protein was separated by SDS-PAGE in the presence or absence of DTT (50 mM) and analyzed by Western blotting using an anti-NPR1 antibody. Detection of a constitutively expressed calcium-sensing receptor (CAS) confirmed equal loading. Molecular weight standards are indicated. O, oligomer; M, monomer; T, total.
(C) 35S::NPR1-GFP plants were treated as in (B). Leaf tissue was examined using fluorescence microscopy.
(D) 35S::NPR1-GFP plants were treated and analyzed as in (B), except an anti-GFP antibody was used. Simultaneously, mRNA was extracted and analyzed by Northern blotting using gene-specific probes against PR-1 and constitutively expressed Ubiquitin (UBQ).
(E) Total protein was extracted from 35S::NPR1-GFP plants in a buffer supporting proteolytic activity. Extracts were pretreated with (+) or without (−) 5 mM DTT and incubated at room temperature for the time points indicated. Total NPR1-GFP protein was analyzed by reducing SDS-PAGE and Western blotting using an anti-GFP antibody.
(F) 35S::NPR1-GFP (in npr1-1) and 35S::npr1-nls-GFP (in npr1-1) plants were treated with (+) or without (−) 100 μM cycloheximide (CHX) for 24 hours. Total protein was analyzed by reducing SDS-PAGE and Western blotting using an anti-GFP antibody. Detection of constitutively expressed Tubulin (TUB) confirmed equal loading.
(G) Seedlings of 35S::NPR1-GR (in npr1-3) plants were treated for with (+) or without (−) 100 μM CHX and 5 μM dexamethasone (DEX) for 24 hours. Total protein was analyzed by reducing SDS-PAGE and Western blotting using an anti-GR antibody. A non-specific band (*) confirmed equal loading.
NPR1 degradation was then examined in planta by treating wild-type Col-0 plants with MG115 or MG132. Similar to SA-induced plants, inhibition of proteasome activity significantly enhanced the accumulation of both NPR1 oligomer and monomer (Figure 1B). To eliminate the effect of transcriptional regulation on NPR1 protein concentration, we examined the 35S::NPR1-GFP plants, which constitutively express the transgene independent of SA and proteasome inhibitor treatment (Figure S1). As reported previously (Kinkema et al., 2000; Mou et al., 2003), GFP fluorescence was weak in untreated plants due to the oligomeric status of the NPR1-GFP protein (Figures 1C and 1D). SA treatment induced NPR1-GFP monomer formation, resulting in strong GFP fluorescence in the nuclei (Figure 1C). Plants treated with MG115 or MG132 also exhibited readily detectable GFP fluorescence in the nuclei (Figure 1C) and showed a significant increase in total NPR1 protein (Figure 1D; +DTT). As shown in non-reducing Western blot analysis, this increase was predominantly in the form of NPR1 monomer (Figure 1D; -DTT). Consequently, the NPR1 target gene PR-1 was induced in these MG115- or MG132-treated plants, albeit at a level lower than that found in SA-treated plants (Figure 1D). These data indicate that transcriptionally active NPR1 is constantly degraded by the proteasome. Blocking proteasome activity causes ectopic accumulation of NPR1 monomer and spurious expression of its target genes. However, an additional SA-dependent mechanism seems to be required to fully turn on transcriptional activity of NPR1.
NPR1 monomer is degraded in the nucleus
We then investigated whether NPR1 oligomer and monomer are equally sensitive to proteasome-mediated degradation using plant extracts supplemented with or without the reducing agent dithiothreitol (DTT). Addition of 5 mM DTT reduced nearly all NPR1-GFP oligomer to its monomeric form (Figure S2) and consequently accelerated its degradation (Figure 1E), suggesting that NPR1 monomer is preferentially degraded.
Because NPR1 monomer was observed in the nucleus upon proteasome inhibitor treatment, we hypothesized that proteolysis of NPR1 may occur there. To test this hypothesis, we examined the in vivo stability of NPR1-GFP and the nuclear localization sequence (nls) mutant npr1-nls-GFP by treating plants with the protein synthesis inhibitor cycloheximide (CHX). Whereas the amount of NPR1-GFP rapidly decreased in the absence of new protein synthesis, the levels of npr1-nls-GFP, which has been shown before to reside predominantly in the cytoplasm (Kinkema et al., 2000), remained unchanged (Figure 1F). The lack of degradation of npr1-nls-GFP was probably due to its cytosolic mislocalization, not resistance to degradation per se, because the mutant protein was as readily degraded as the wild-type protein in cell-free extracts (data not shown). To confirm this result, we also used plants expressing NPR1 fused to a dexamethasone (DEX)-responsive glucocorticoid receptor (NPR1-GR)(Kinkema et al., 2000). In the absence of DEX, NPR1-GR was retained in the cytosol and its abundance was not affected by CHX treatment (Figure 1G). In the presence of DEX, however, NPR1-GR was nuclear translocated and rapidly degraded. These findings demonstrate that in vivo NPR1 monomer is constitutively degraded in the nucleus by the proteasome. Since blocking entry into the nucleus completely stabilized the protein, NPR1 monomer is probably degraded only in the nucleus.
Cullin3/CSN-mediated degradation of transcriptionally active NPR1 prevents inappropriate activation of SAR
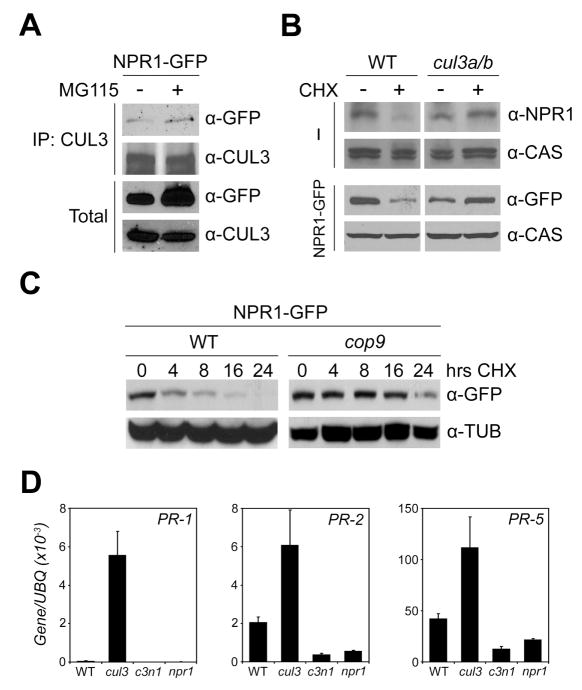
NPR1 contains a BTB/POZ (broad-complex, tramtrack, and bric-à-brac/poxvirus, zinc finger) domain, which is found in proteins that function as substrate adapters of CUL3-based ubiquitin ligases for the degradation of specific substrates (Petroski and Deshaies, 2005). Interestingly, BTB-containing proteins themselves may also be substrates for these CUL3 complexes (Luke-Glaser et al., 2007; Pintard et al., 2003). Since NPR1 is degraded by the proteasome, we tested the possibility that NPR1 is a substrate of CUL3-based ubiquitin ligases using co-immunoprecipitation experiments between NPR1-GFP and CUL3A. Even though previously reported yeast two-hybrid analysis found no direct interaction between CUL3A and NPR1 (Dieterle et al., 2005), NPR1-GFP could be pulled down with an antibody against CUL3A (Figure 2A). This suggests that CUL3 and NPR1 may interact indirectly through an adaptor protein.
Figure 2. NPR1 is constitutively targeted for degradation by a CUL3-based ubiquitin ligase.
(A) 35S::NPR1-GFP (in npr1-1) plants were treated with (+) or without (−) MG115 (100 μM) for 24 hours. Protein extracts were immunoprecipitated (IP) with an antibody against CUL3A. Total and immunoprecipitated proteins were analyzed by Western blotting using anti-GFP and anti-CUL3A antibodies.
(B) Wild-type (WT) and cul3a cul3b plants in the absence (−) or presence of the 35S::NPR1-GFP transgene (NPR1-GFP) were treated with (+) or without (−) 100 μM CHX for 24 hours. Total protein was analyzed by reducing SDS-PAGE and Western blotting using an antibody against the endogenous NPR1 or GFP. Detection of a constitutively expressed calcium-sensing receptor (CAS) confirmed equal loading.
(C) 35S::NPR1-GFP was expressed in wild-type (WT) and cop9 plants. Plants were treated with 100 μM CHX for the indicated hours. Total protein was analyzed by reducing SDS-PAGE and Western blotting using an anti-GFP antibody. Detection of constitutively expressed Tubulin (TUB) confirmed equal loading.
(D) mRNA was extracted from the wild-type (WT), npr1, cul3a cul3b (cul3) double, and cul3a cul3b npr1 (c3n1) triple mutant plants. The expression of PR-1, PR-2, and PR-5 was analyzed using qPCR and normalized against constitutively expressed UBQ. Error bars represent SD (n = 3).
To validate the NPR1-CUL3 interaction genetically, we generated a double mutant between Arabidopsis cul3a and cul3b, both of which are T-DNA insertion mutants. It has been shown previously that a cul3a cul3b double knock-out is embryonic lethal (Figueroa et al., 2005; Thomann et al., 2005). To overcome this obstacle we generated a cul3a cul3b double mutant using a knockout cul3a allele and a ~50% knockdown cul3b allele (Figure S3), which resulted in viable adult plants. Compared to the wild type, NPR1 mRNA levels were reduced in the cul3a cul3b mutant (Figure S4). Nevertheless, steady-state NPR1 protein levels were comparable in wild-type and mutant plants (Figure 2B), suggesting that NPR1 protein is more stable in absence of CUL3. To further test this, wild-type Col-0 and cul3a cul3b plants were treated with CHX to block new protein synthesis. CHX treatment led to a decrease in the amount of NPR1 in the wild type, yet NPR1 levels did not decrease in cul3a cul3b plants (Figure 2B). Similar results were obtained for NPR1-GFP protein expressed in wild-type and cul3a cul3b plants (Figure 2B).
The COP9 signalosome (CSN) has been shown to regulate the stability and activity of CUL proteins by cycles of (de)neddylation, a process in which lysine residues are modified by the ubiquitin-like molecule Nedd8 (Petroski and Deshaies, 2005). To investigate if the CSN is also involved in NPR1 degradation, the 35S::NPR1-GFP transgene was introduced into the cop9 mutant background through genetic crosses. As shown in Figure 2C, NPR1-GFP degradation was dramatically blocked by the cop9 mutation.
If degradation of transcriptionally active NPR1 monomer is to keep SAR inactive in unchallenged plants, we expected this regulation to be compromised in the cul3a cul3b mutant. Indeed, compared to wild-type plants, unchallenged cul3a cul3b mutants showed high constitutive expression of the NPR1 target genes PR-1, PR-2, and PR-5 (Figure 2D). Importantly, this constitutive PR gene expression was NPR1-dependent, because it was completely lost in the cul3a cul3b npr1-1 triple mutant (Figure 2D). In accordance with the observed constitutive PR gene activation, cul3a cul3b plants exhibited elevated levels of resistance against the virulent bacterial leaf-pathogen Pseudomonas syringae pv. maculicola (Psm) ES4326 (Figure S5). Collectively, these findings demonstrate that in unchallenged plants, CUL3/CSN-mediated degradation of transcriptionally active NPR1 monomer prevents costly activation of PR genes and SAR.
SA-induced transcription of NPR1 target genes and SAR require proteasome activity
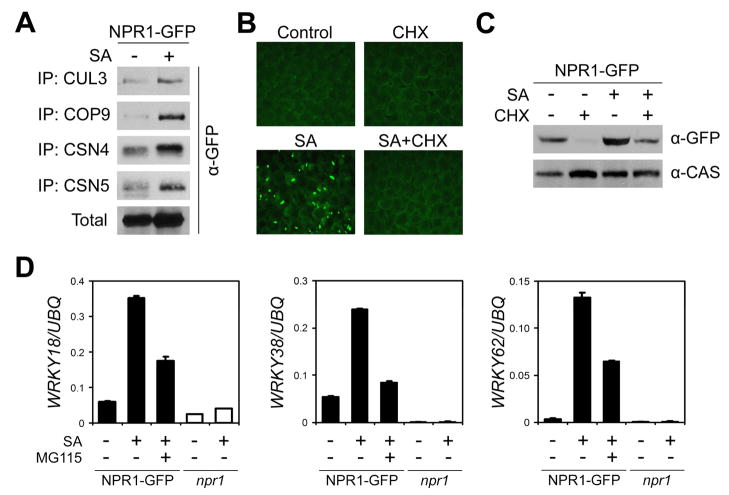
SA induces the release of transcriptionally active NPR1 monomer, which regulates the expression of many defense genes. This prompted us to investigate the effect of SA on NPR1 degradation. Unexpectedly, co-immunoprecipitation experiments indicated that in SA treated plants, more NPR1-GFP was pulled down with CUL3A and three components of the CSN complex (COP9, CSN4, CSN5; Figure 3A). To reconcile this with the fact that SA treatment leads to accumulation of NPR1-GFP monomer in the nucleus (Figure 3B), we hypothesized that this accumulation resulted from a significant increase in NPR1-GFP import into the nucleus rather than a reduction in protein degradation. Indeed, SA-induced monomer was completely absent when de novo protein synthesis was inhibited by CHX (Figure 3B). Accordingly, Western blot analysis indicated that SA treatment did not rescue NPR1-GFP protein from degradation (Figure 3C). These data indicate that proteolysis of NPR1 still occurs after SAR induction despite the fact that NPR1 is a positive regulator of this response.
Figure 3. SA-induced transcription of NPR1 target genes requires the proteasome.
(A) 35S::NPR1-GFP (in npr1-1) plants were treated with (+) or without (−) 0.5 mM SA for 24 hours. Protein extracts were immunoprecipitated (IP) using antibodies against CUL3, COP9, CSN4, and CSN5. Total and immunoprecipitated proteins were analyzed by Western blotting using an anti-GFP antibody.
(B) 35S::NPR1-GFP (in npr1-2) plants were treated with (+) or without (−) 0.5 mM SA and 100 μM CHX for 24 hours. Leaf tissue was examined by fluorescence microscopy.
(C) 35S::NPR1-GFP (in npr1-2) plants were treated as in (B). Total protein was analyzed by reducing SDS-PAGE and Western blotting using an anti-GFP antibody. Detection of a constitutively expressed calcium-sensing receptor (CAS) confirmed equal loading.
(D) 35S::NPR1-GFP (in npr1-2) and npr1-2 plants were treated with (+) or without (−) 0.5 mM SA and 100 μM MG115 for 28 hours. The expression of three WRKY genes was analyzed using qPCR and normalized with constitutively expressed UBQ. Error bars represent SD (n = 3).
We then examined whether proteasome activity affects induction of the NPR1 target genes WRKY18, WRKY38, and WRKY62 (Wang et al., 2006). The NPR1-dependency of these target genes was clearly demonstrated by their complete lack of responsiveness to SA treatment in the npr1 mutant (Figure 3D). Transformation of 35S::NPR1-GFP into the npr1 mutant restored the SA-mediated transcription of the WRKY genes. Whereas treatment with MG115 alone resulted in weak NPR1-dependent activation of the WRKY genes (Figure S6), the SA-mediated induction of these genes was strongly inhibited in the presence of MG115 (Figure 3D and S6), indicating that SA-induced transcription of these WRKY genes requires both NPR1 and the proteasome activity. SA-induced expression of PR-1, another NPR1 target (Wang et al., 2005), was only modestly affected by MG115 treatment (Figure S7), suggesting that its activation is less dependent on the proteasome. Since the NPR1-GFP transgene is constitutively expressed, independent of SA and MG115 (Figure S1), the observed reduction in NPR1 target gene expression was specifically due to the change in NPR1 protein stability, not expression.
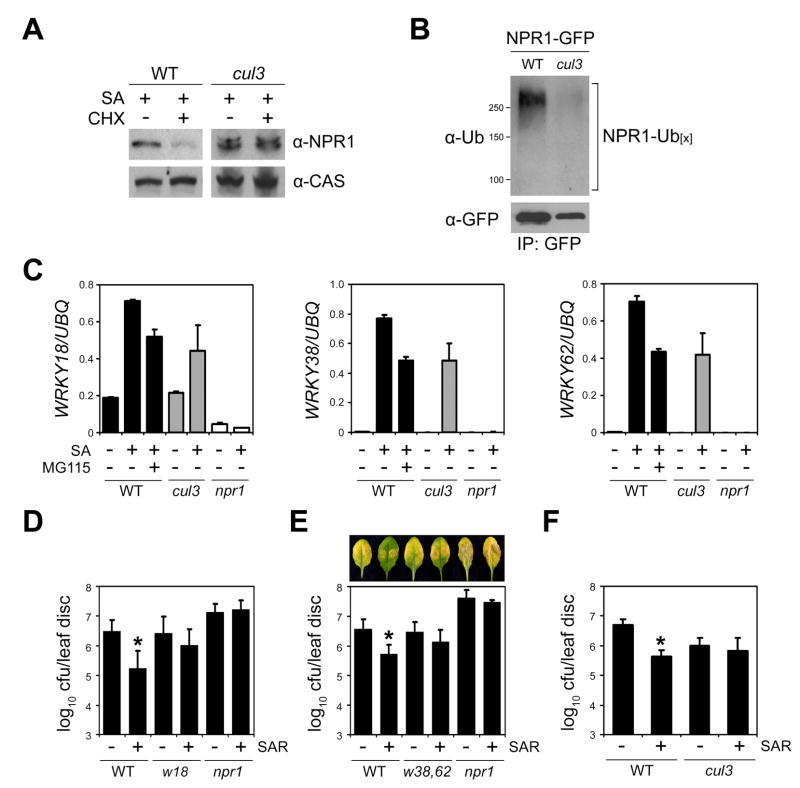
Because degradation of NPR1 requires CUL3 in uninduced plants, we examined its requirement for NPR1 degradation in SA-induced plants. Blocking protein synthesis with CHX in SA-treated wild-type plants strongly reduced endogenous NPR1 protein levels, whereas NPR1 levels remained constant in the cul3a cul3b mutant (Figure 4A). Moreover, the NPR1-GFP protein expressed in SA-treated wild-type plants was highly poly-ubiquitinylated (Figure 4B). This modification was significantly reduced in the cul3a cul3b mutant background (Figure 4B).
Figure 4. SA-induced transcription of NPR1 target genes requires CUL3.
(A) Wild-type (WT) and cul3a cul3b (cul3) plants were treated with 0.5 mM SA for 16 hours. Subsequently, plants were SA-treated for an additional 4 hours in the absence (−) or presence (+) of 100 μM CHX. Total protein was analyzed by reducing SDS-PAGE and Western blotting using an anti-NPR1 antibody. Detection of a constitutively expressed calcium-sensing receptor (CAS) confirmed equal loading.
(B) Wild-type (WT) and cul3a cul3b (cul3) plants carrying the 35S::NPR1-GFP transgene were treated with 0.5 mM SA and 100 μM MG115 for 8 hours. Protein extracts were immunoprecipitated (IP) using an anti-GFP antibody. Immunoprecipitated proteins were analyzed by Western blotting using anti-Ubiquitin (Ub) and anti-GFP antibodies. Molecular weight standards are indicated.
(C) Wild-type, cul3a cul3b (cul3), and npr1-2 plants were treated with (+) or without (−) 0.5 mM SA and 100 μM MG115 for 28 hours. The expression of three WRKY genes was analyzed using qPCR and normalized with constitutively expressed UBQ. Error bars represent SD (n = 3).
(D) Induction of SAR against Psm ES4326 in wrky18 (w18) plants. Cfu, colony-forming units. Error bars represent 95% confidence limits (n = 8). Asterisks indicate statistically significant differences between the control and SAR treatment in each genotype (Tukey–Kramer ANOVA test; α = 0.05, n = 8). See Supplemental Data for details.
(E) Induction of SAR against Psm ES4326 disease symptoms and growth in wrky38 wrky62 (w38, 62) plants was carried out as in (D).
(F) Induction of SAR against Psm ES4326 growth in cul3a cul3b plants was carried out as in (D).
To investigate the role of CUL3 in activation of NPR1-dependent gene transcription, we examined expression of the NPR1 targets in the cul3a cul3b mutant. SA-induced transcription of all three WRKY genes was partially compromised in this mutant (Figure 4C). Importantly, after SA treatment, the levels of gene expression in cul3a cul3b plants were comparable to those observed in wild-type plants treated with both SA and MG115 (Figure 4C), confirming the role of CUL3 in this proteasome-dependent gene expression. To examine the impact of impaired WRKY gene expression on SAR, we used the previously described wrky18 mutant (Wang et al., 2006) and generated a double mutant of the two closely related and likely redundant WRKY38 and WRKY62 genes. In wrky18 and wrky38 wrky62 mutants, NPR1-dependent SAR against Psm ES4326, triggered after local inoculation of avirulent P.s. pv. tomato (Pst) DC3000/ avrRpt2, was partially compromised (Figures 4D and 4E). Thus, NPR1-mediated activation of these WRKY genes is essential for induction of SAR.
Because knocking out NPR1-target WRKY genes impairs SAR, we predicted that the cul3a cul3b mutant would also be defective in SAR due to the failure to fully induce these genes. Indeed, cul3a cul3b plants failed to activate SAR against Psm ES4326 (Figure 4F), even though it had higher levels of basal resistance against this pathogen due to elevated PR gene expression (Figure 2D). Taken together, these data suggest that SA-induced, CUL3-mediated turnover of the co-activator NPR1 may stimulate target gene transcription and is required for activation of SAR.
NPR1 is phosphorylated at Ser11 and Ser15 in the nucleus
Targeting substrates to the proteasome is often regulated by post-translational modifications, such as phosphorylation. To examine if NPR1 is phosphorylated, we treated 35S::NPR1-GFP plants with or without SA, extracted total protein, and applied the extracts onto a column that specifically binds phosphoproteins (see Experimental Procedures). We found that NPR1-GFP could bind to the column as significant amounts of the protein were eluted, especially from extracts of SA-treated plants (Figure 5A). Indeed, using an antibody against phosphorylated serine/threonine residues, we found that SA treatment strongly increased the level of NPR1 phosphorylation (Figure 5B). NPR1 contains an N-terminal phosphodegron motif that is highly conserved among NPR1 orthologues in different plant species (Figure 5C). Phosphodegrons are degradation motifs found in many proteasome-regulated substrates, including IκB (Hayden and Ghosh, 2004). To study if Ser11 and Ser15 of the IκB-like phosphodegron motif in NPR1 are phosphorylated in vivo, we generated an antibody that specifically recognizes phosphorylated Ser11/15 (Figure S8; see Experimental Procedures). Little Ser11/15 phosphorylation of the endogenous NPR1 and NPR1-GFP was observed in untreated plants, whereas SA treatment greatly enhanced phosphorylation (Figures 5D and 5E). Moreover, SA-induced Ser11/15 phosphorylation of NPR1 occurs in the nucleus, as this modification was completely abolished in the cytoplasmic npr1-nls-GFP mutant protein (Figure 5E).
Figure 5. NPR1 is phosphorylated at Ser11 and Ser15 in the nucleus.
(A) 35S::NPR1-GFP (in npr1-1) plants were treated with (+) or without (−) 0.5 mM SA for 24 hours. Total protein was loaded onto a column that specifically binds phosphoproteins. Bound proteins were eluted and the amount of NPR1-GFP determined by Western blotting with an anti-GFP antibody.
(B) 35S::NPR1-GFP plants were treated as in (A). Total protein was extracted and immunoprecipitated (IP) with an anti-GFP antibody. Immunoprecipitated proteins were analyzed by Western blotting using an antibody that recognizes phosphorylated serine and threonine residues (α-pS/pT) as well as an anti-GFP antibody.
(C) Sequence alignment of NPR1 proteins from different plant species. The IκB-like phosphodegron motif (DSxxxS; x, any amino acid) is indicated. Le, Lycopersicum esculentum; Nt, Nicotiana tabacum; Bv, Beta vulgaris; Os, Oryza sativa; At, Arabidopsis thaliana.
(D) Wild-type Col-0 plants were treated with (+) or without (−) 0.5 mM SA for 24 hours. Total protein was analyzed by reducing SDS-PAGE and Western blotting. NPR1 phosphorylation was specifically detected with an antibody against phosphorylated Ser11 and Ser15 residues (α-pSer11/15). Equal loading was verified using an anti-NPR1 and anti-Tubulin (TUB) antibody.
(E) 35S::NPR1-GFP (in npr1-1) and 35S::npr1-nls-GFP (in npr1-1) and npr1-1 plants were treated with (+) or without (−) 0.5 mM SA for 24 hours. Total protein was analyzed by reducing SDS-PAGE and Western blotting using anti-pSer11/15 and anti-GFP antibodies. A non-specific band (*) indicated equal loading.
Phosphorylation facilitates NPR1 turnover and promotes its transcriptional activity in SAR
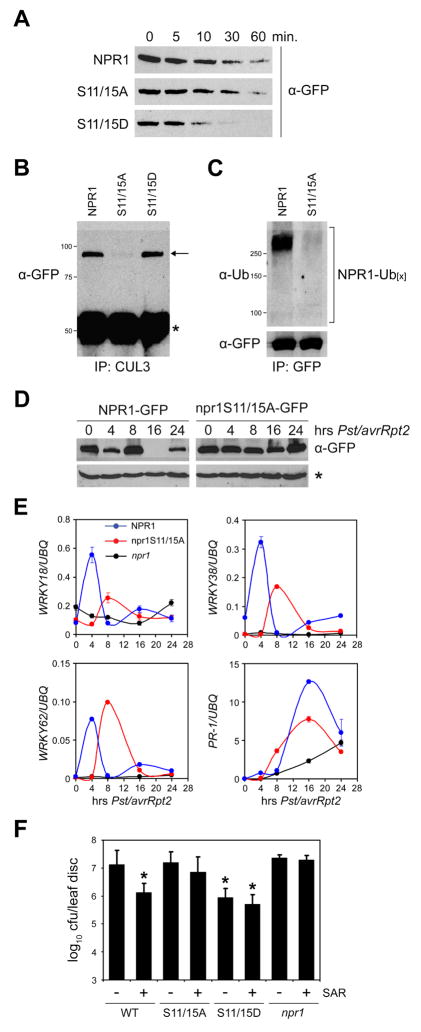
Since NPR1 phosphorylation and degradation both occur in the nucleus, we investigated if SA-induced phosphorylation of the IκB-like phosphodegron motif was coupled to NPR1 turnover. Ser11 and Ser15 of NPR1-GFP were replaced with non-phosphorylatable alanines (S11/15A) or phosphomimic aspartic acids (S11/15D). The resulting 35S::npr1S11/15A-GFP and 35S::npr1S11/15D-GFP constructs were then transformed into npr1-2 plants. In a cell-free degradation assay, the npr1S11/15D-GFP protein showed an increased degradation rate as compared to both NPR1-GFP (not phosphorylated in the absence of an inducer) and npr1S11/15A-GFP (Figure 6A). To further investigate this, plants were treated with a combination of SA and MG115 and co-immunoprecipitation experiments were performed. As shown in Figure 6B, both the NPR1-GFP (phosphorylated in the presence of an inducer) and npr1S11/15D-GFP proteins were readily pulled down with CUL3A, whereas only a small amount of npr1S11/15A-GFP protein was recovered. Moreover, poly-ubiquitinylation of npr1S11/15A-GFP was markedly reduced compared to NPR1-GFP (Figure 6C). Thus, SA-induced phosphorylation of NPR1 facilitates its interaction with the CUL3-based ubiquitin ligase and stimulates turnover.
Figure 6. Phosphorylation stimulates NPR1 turnover and is required for SAR.
(A) Total protein was extracted from 35S::NPR1-GFP (in npr1-2), 35S::npr1S11/15A-GFP (in npr1-2), and 35S::npr1S11/15D-GFP (in npr1-2) plants in a buffer supporting proteasome activity. Extracts were incubated at room temperature for the time points indicated. Proteins were analyzed by reducing SDS-PAGE and Western blotting using an anti-GFP antibody.
(B) 35S::NPR1-GFP, 35S::npr1S11/15A-GFP, and 35S::npr1S11/15D-GFP plants were treated with a combination of 0.5 mM SA and 100 μM MG115. Protein extracts were immunoprecipitated (IP) with an anti-CUL3 antibody. Immunoprecipitated proteins were analyzed by Western blotting using an anti-GFP antibody. Molecular weight standards are indicated. Arrow indicates NPR1; asterisk indicates a cross-reacting IgG chain.
(C) 35S::NPR1-GFP and 35S::npr1S11/15A-GFP plants were treated as in (B). Protein extracts were immunoprecipitated (IP) with an anti-GFP antibody. Immunoprecipitated proteins were analyzed by Western blotting using anti-Ubiquitin and anti-GFP antibodies. Molecular weight standards are indicated.
(D) The leaf-halves of 35S::NPR1-GFP and 35S::npr1S11/15A-GFP plants were inoculated with avirulent Pst DC3000/avrRpt2 (OD600=0.02). At the indicated time points protein was extracted from the uninoculated leaf halves and subjected to reducing SDS-PAGE and Western blot analysis using an anti-GFP antibody. A non-specific band (*) indicated equal loading.
(E) 35S::NPR1-GFP, 35S::npr1S11/15A-GFP, and npr1-2 plants were treated as described in (D). mRNA was extracted from the uninoculated halves of the inoculated leaves and analyzed for the expression of WRKY18, WRKY38, WRKY62, and PR-1 using qPCR. Expression was normalized against constitutively expressed UBQ. Error bars represent SD (n = 3).
(F) Induction of SAR against Psm ES4326 in 35S::NPR1-GFP, 35S::npr1S11/15A-GFP, 35S::npr1S11/15D-GFP, and npr1-2 plants. Cfu, colony-forming units. Error bars represent 95% confidence limits (n = 8). Asterisks indicate statistically significant differences compared with uninduced 35S::NPR1-GFP plants (Tukey–Kramer ANOVA test; α = 0.05, n = 8).
To test our hypothesis that phosphorylation-mediated turnover of NPR1 is required for activation of gene expression and SAR, we first examined the stability of NPR1-GFP and npr1S11/15A-GFP proteins in systemic tissues during the time course of SAR induction. As shown in the Western blot in Figure 6D, the constitutively expressed NPR1-GFP protein showed a remarkable biphasic degradation pattern at 4 and 16 hours post inoculation with avirulent Pst DC3000/avrRpt2. In contrast, this degradation pattern was significantly diminished for the npr1S11/15A-GFP protein. Corresponding to NPR1 protein measurements, the transcription profiles of the NPR1 target genes WRKY18, WRKY38, WRKY62 and PR-1 were also analyzed in systemic tissues (Figure 6E). Consistent with the notion that NPR1 turnover stimulates the expression of these target genes, induction of WRKY gene transcription coincided with a decrease in NPR1-GFP protein levels 4 hours post inoculation (Figures 6D and 6E). In 35S::npr1S11/15A-GFP plants, however, induction of WRKY gene transcription was weakened and/or delayed, corresponding to the slow turnover rate of the npr1S11/15A-GFP protein (Figures 6D and 6E). In 35S::NPR1-GFP plants, PR-1 gene expression was also strongly induced during the second NPR1 turnover phase (Figure 6E). Notably, induction of this gene was significantly reduced in 35S::npr1S11/15A-GFP plants. Similar patterns were also observed when these plants were treated with SA (data not shown), but protein fluctuations were the most profound in systemic tissue during biological induction of SAR. Together with the finding that mutations in Ser11 and Ser15 do not affect NPR1’s ability to interact with transcription factors (Figure S9), these data demonstrate that turnover of phosphorylated NPR1 is required for full-scale expression of its target genes.
We next tested the ability of 35S::npr1S11/15A-GFP plants to mount SAR. As controls, pre-inoculation of 35S::NPR1-GFP plants with avirulent Pst DC3000/avrRpt2 protected the plants against virulent Psm ES4326 infection three days after. This protection was compromised in npr1 plants (Figure 6F). Interestingly, 35S::npr1S11/15A-GFP plants also failed to efficiently induce SAR, indicating that turnover of phosphorylated NPR1 is required for the onset of SAR.
The result from the npr1S11/15A-GFP mutant was further corroborated using 35S::npr1S11/15D-GFP transgenic plants. Consistent with the increased degradation rate of the npr1S11/15D-GFP protein (Figure 6A), mock-treated 35S::npr1S11/15D-GFP plants exhibited elevated levels of resistance that were comparable to SAR-induced 35S::NPR1-GFP plants (Figure 6F). Interestingly, this might not be due to elevated basal expression of NPR1 target genes, but rather due to higher levels of target gene expression after induction in the 35S::npr1S11/15D-GFP plants (Figure S10). This is consistent with the fact that NPR1 phosphorylation occurs only after pathogen challenge and suggests that the npr1S11/15D-GFP mutant is probably potentiated for this regulatory step. Indeed, SAR induction in 35S::npr1S11/15D-GFP did not further enhance resistance against Psm ES4326 (Figure 6F). These data demonstrate that the turnover of phosphorylated NPR1 is an important regulatory switch in inducing SAR.
DISCUSSION
Proteasome-mediated protein degradation plays a pivotal role in many plant growth and developmental processes, including photomorphogenesis and plant hormone signaling (Smalle and Vierstra, 2004). In these instances the proteasome either degrades activators to suppress transcription or degrades repressor proteins to activate gene expression. Our study discovered that proteasome-mediated degradation of NPR1 not only prevents untimely gene activation, but also plays an essential role in stimulating gene expression during plant immune responses.
NPR1 monomer is constitutively cleared from the nucleus by a CUL3-based ubiquitin ligase to restrict its transcription co-activator activity (Figures 1 and 2). A similar regulatory mechanism was observed recently for the yeast transcription factor GAL4, a regulator of galactose metabolism (Muratani et al., 2005). In the absence of galactose, GAL4 activity is limited by proteasome-mediated destruction, preventing wasteful activation of metabolic genes. Proteasome-mediated degradation of transcription factors was also found to prevent inappropriate transcription at tissue specific gene loci in embryonic stem cells (Szutorisz et al., 2006). Importantly, our data suggests that the proteasome may restrict gene transcription not only by destruction of transcription factors, but also by proteolysis of co-activators to prevent assembly of active transcriptional complexes (Figure 7). In the case of NPR1, this mechanism renders SAR inactive to avoid detrimental fitness costs associated with constitutive defense (Heidel et al., 2004; van Hulten et al., 2006).
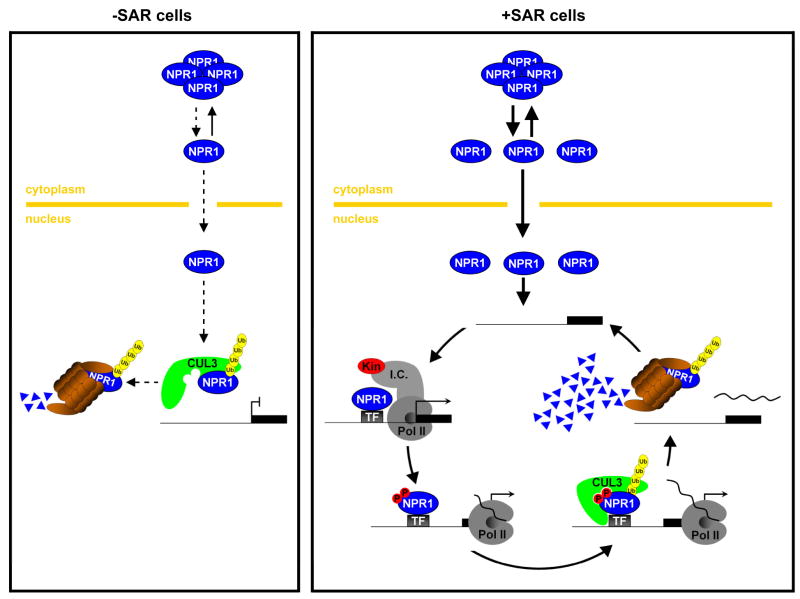
Figure 7. Working model for the dual role of the proteasome in preventing and stimulating NPR1 target gene transcription.
In uninduced cells (left panel), NPR1 monomer constitutively translocates, at a low rate (dashed lines), to the nucleus where it is targeted to the proteasome by CUL3-based E3 ligase-mediated ubiquitinylation (Ub). This prevents the activation of NPR1 target genes. In SAR-induced cells (right panel), a large amount of NPR1 monomer translocates to the nucleus where it interacts with transcription factors (TF) to initiate target gene transcription by recruiting the transcription initiation complex (I.C.) and RNA polymerase II (PolII). As a consequence NPR1 may be phosphorylated (P) by a kinase (Kin) that is associated with the I.C. and PolII. A CUL3-based ligase with high affinity for phosphorylated NPR1, possibly distinct from the one involved in turnover of non-phosphorylated NPR1 in uninduced cells, rapidly ubiquitinylates and targets NPR1 for degradation by the proteasome. Clearance of “exhausted” phosphorylated NPR1 from the target gene promoter allows “fresh” non-phosphorylated NPR1 to reinitiate the transcription cycle, thereby directly linking the rate of NPR1 degradation to the amplitude of target gene transcription.
To our surprise, activation of SAR did not prevent CUL3-mediated degradation of NPR1. Instead, blocking NPR1 turnover by inhibition of proteasome activity, genetically knocking down CUL3 activity, and mutating the IκB-like phosphodegron, all compromised transcription of the NPR1 target genes WRKY18, WRKY38, and WRKY62 (Figures 3, 4, and 6), indicating that NPR1 turnover also stimulates transcription. Compared to the WRKY genes, expression of PR-1 was less dependent on the proteasome (Figure S7), suggesting that NPR1 target genes may require different rates of NPR1 turnover. Recent reports have demonstrated that in yeast and human cells, transcription factors are often unstable and their instability correlates with their ability to induce target gene transcription (Kim et al., 2003; Lipford et al., 2005; Muratani et al., 2005; Reid et al., 2003; von der Lehr et al., 2003). It is thought that turnover of transcription factors promotes gene expression by continuously delivering ‘fresh’ activator to gene promoters. This may be necessary to sustain a high ratio of transcriptional active over inactive activator (Collins and Tansey, 2006). Although mono-ubiquitinylation of co-activators has been reported to stimulate their activity (Wu et al., 2007), a role for co-activator proteolysis in gene transcription has not been established. In fact, it was previously reported that the turnover of co-activators associated with the human estrogen receptor-α did not stimulate estrogen receptor-mediated transcription (Lonard et al., 2000). Our findings suggest, however, that gene activation by proteolysis is not limited to transcription factors, but also includes transcription co-activators like NPR1 (Figure 7).
Turnover of NPR1 is limited by its nuclear translocation (Figure 1). We recently reported that S-nitrosylation of Cys156 in NPR1 facilitates oligomer formation in the cytoplasm. Mutating Cys156 to alanine (C156A) inhibited this regulatory step and caused depletion of the npr1C156A protein in response to SA treatment (Tada et al., 2008). Here, we found that the C156A mutation does not affect the protein turnover rate in our cell-free degradation assay (Figure S11). Instead, the SA-induced instability of this mutant protein in planta was reversed by inhibition of the proteasome activity (Figure S12). Thus, nuclear turnover of NPR1 presented in this report underlined the importance of S-nitrosylation-mediated oligomerization in the cytoplasm to maintain NPR1 homeostasis. Upon SAR induction, a large amount of NPR1 monomer is released from the cytoplasmic oligomer, translocated into the nucleus, and subsequently turned over. To maintain protein homeostasis, oligomerization of NPR1 in the cytoplasm is facilitated by a pathogen-induced increase in cellular GSNO levels (Tada et al., 2008). NPR1 oligomerization and monomer release occur sequentially according to the SA-induced biphasic redox changes (Tada et al., 2008), which may be responsible for the observed fluctuations in NPR1-GFP levels after avirulent pathogen inoculation (Figure 6). Because a constitutive promoter was used to drive the expression of NPR1-GFP, we were able to detect these dynamic changes in protein stability. Such fluctuations are much harder to detect for the endogenous NPR1 protein as NPR1 gene transcription also fluctuates with the redox changes (data not shown).
NPR1 contains a conserved N-terminal phosphodegron motif (Figure 5C) that is found in many unstable transcriptional regulators, including IκB, β-Catenin, c-Myc, c-Jun, and SRC-3 (Hayden and Ghosh, 2004; Karin and Ben-Neriah, 2000; Wu et al., 2007). Phosphorylation of the phosphodegron motif signals the destruction of these regulators by recruiting CUL1-based ubiquitin ligases. We showed that SA-induced phosphorylation of Ser11 and Ser15 in the phosphodegron motif of NPR1 also promotes its poly-ubiquitinylation and degradation (Figures 5 and 6). Although site-specific (de)phosphorylation has been found to regulate substrate recruitment to CUL1-based ubiquitin ligases (Petroski and Deshaies, 2005), the mechanisms that control substrate delivery to CUL3 complexes are not yet well understood. We demonstrated that the NPR1 phosphodegron regulates interaction with a CUL3-based ubiquitin ligase and promotes NPR1 turnover (Figures 6A-6D), suggesting that phosphorylation may also be a common mechanism to target substrates for CUL3-mediated proteolysis. Similar to CUL1-substrate interaction, phosphorylation of Ser11/15 may create or stabilize a binding site for the CUL3-based ubiquitin ligase (Figure 7). The role of Ser11/15 phosphorylation-mediated degradation as a key regulatory switch for SAR was clearly revealed by the diminished transcription of NPR1 target genes and the failure to induce resistance in 35S::npr1S11/15A-GFP mutant plants (Figures 6E and 6F). This conclusion was further supported by the phenotype of the 35S::npr1S11/15D-GFP mutant, which has a higher basal level of resistance but is also insensitive to SAR induction (Figure 6F).
Turnover of NPR1 plays dual roles in regulating the transcription of target genes. Whereas CUL3-mediated degradation prevented NPR1 from initiating transcription in uninduced cells, it was necessary for full-scale activation of transcription in SAR-induced cells. Thus, NPR1 may be targeted for degradation by the CUL3-based ligase in distinct ways. Indeed, we identified phosphorylation of NPR1 as an important functional switch for its CUL3-mediated transcription activity: phosphorylation was required for SAR-induced turnover of NPR1, but was dispensable for basal turnover (Figure 6). The mechanism by which phosphorylation may promote the activity of proteasome-regulated activators was investigated for the yeast activator GCN4, an essential regulator of amino acid biosynthesis genes. Inhibition of the proteasome reduced the ability of GCN4 to recruit RNA polymerase II to its target promoters (Lipford et al., 2005). Interestingly, degradation of GCN4 is signaled by SRB10, a cyclin-dependent protein kinase that is intimately associated with RNA polymerase II (Chi et al., 2001; Liao et al., 1995), suggesting that GCN4 is degraded after it has initiated transcription. This supports a model in which phosphorylation labels an activator as “exhausted” once it has initiated transcription. Subsequent removal of the activator by proteasome-mediated degradation may allow “fresh” activator to bind the target promoter and re-initiate transcription (Collins and Tansey, 2006; Kodadek et al., 2006; Lipford et al., 2005). As shown in the working model in Figure 7, we hypothesize that a similar mechanism may regulate the SA-induced activity of NPR1 as its phosphorylation is inducible (Figure 5), specifically occurs in the nucleus (Figure 5E), and stimulates turnover-mediated gene transcription (Figure 6). Whereas SAR-induced phosphorylated NPR1 is probably turned over after interaction with the target promoter, the uninduced non-phosphorylated NPR1 may be degraded before binding to the target gene promoter (Figure 7). Where exactly in the nucleus these events occur will be the subject of future investigation. Moreover, the identity of the substrate adaptor for the NPR1-CUL3 interaction has yet to be revealed and whether the same adaptor binds to non-phosphorylated NPR1 and phosphorylated NPR1 needs to be determined. Regardless of these specifics, this study clearly demonstrates that phosphorylation-mediated turnover of distinct components of transcriptional complexes (e.g. transcription factors, co-activators) may be a common mechanism by which single-cellular as well as multi-cellular organisms regulate gene transcription.
EXPERIMENTAL PROCEDURES
See Supplemental Data for details.
Chemical induction and pathogen infection
The roots of soil-grown plants were submerged in 0.5 mM SA, 100 μM cycloheximide and 100 μM MG115 or MG132. Alternatively, 12-day-old MS-grown seedlings were submerged in 100 μM cycloheximide and 5 μM dexamethasone. Pathogen infections were essentially performed as described (Wang et al., 2006).
Protein analysis
Protein analysis was performed essentially as described (Fan and Dong, 2002; Mou et al., 2003). For cell-free degradation assays protein was extracted in 25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM NaCl, 10 mM ATP, and with or without 5 mM DTT. After centrifugation (14,000 g, 10 min, 4°C) supernatants were incubated at room temperature and the reactions terminated with SDS sample buffer and incubation at 70°C (10 min). Inhibitors (40 μM MG115, 40 μM MG132, 4 mM PMSF, 40 μM Leupeptin) were applied using 0.2% DMSO as vehicle.
RNA analysis
RNA analysis was performed as described (Cao et al., 1994; Wang et al., 2006). Briefly, cDNA was produced by first strand synthesis using oligo(dT) primer and Reverse Transcriptase. Real-time PCR was carried out using the Quantitect SYBR Green PCR kit (Qiagen) and gene-specific primers in a LightCycler (Roche).
Supplementary Material
Acknowledgments
We thank Dr. Zhen-Ming Pei for providing the anti-CAS antibodies, Xudong Zhang and Dr. Karolina Pajerowska-Mukhtar for technical assistance, Dr. Yue Xiong for helpful discussions, and Drs. Meng Chen, James Siedow, Tai-ping Sun, and Tso-Pang Yao for critically reading of the manuscript. This work was supported by a grant from NIH (1R01-GM69594) to X. D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003;15:2181–2191. doi: 10.1105/tpc.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Thomann A, Renou JP, Parmentier Y, Cognat V, Lemonnier G, Muller R, Shen WH, Kretsch T, Genschik P. Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 2005;41:386–399. doi: 10.1111/j.1365-313X.2004.02302.x. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Fan W, Dong X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell. 2002;14:1377–1389. doi: 10.1105/tpc.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, et al. Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell. 2005;17:1180–1195. doi: 10.1105/tpc.105.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Clarke JD, Antonovics J, Dong X. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics. 2004;168:2197–2206. doi: 10.1534/genetics.104.032193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell. 2003;15:1846–1858. doi: 10.1105/tpc.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12:2339–2350. doi: 10.1105/tpc.12.12.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodadek T, Sikder D, Nalley K. Keeping transcriptional activators under control. Cell. 2006;127:261–264. doi: 10.1016/j.cell.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJJ, Young RA. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Nawaz Z, Smith CL, O'Malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–948. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- Luke-Glaser S, Roy M, Larsen B, Le Bihan T, Metalnikov P, Tyers M, Peter M, Pintard L. CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol Cell Biol. 2007;27:4526–4540. doi: 10.1128/MCB.01724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Després C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell. 2006;18:3670–3685. doi: 10.1105/tpc.106.046953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner HY, Johnson J, Delaney TP, Jesse T, Vos P, et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti SE, Muratani M, Wijnen H, Futcher B, Tansey WP. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc Natl Acad Sci USA. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Dong X. S-nitrosylation and thioredoxins regulate conformational changes of NPR1 in plant innate immunity. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A, Brukhin V, Dieterle M, Gheyeselinck J, Vantard M, Grossniklaus U, Genschik P. Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 2005;43:437–448. doi: 10.1111/j.1365-313X.2005.02467.x. [DOI] [PubMed] [Google Scholar]
- van Hulten M, Pelser M, van Loon LC, Pieterse CMJ, Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:5602–5607. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- Wu RC, Feng Q, Lonard DM, O'Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant-Microbe Interact. 2000;13:191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.