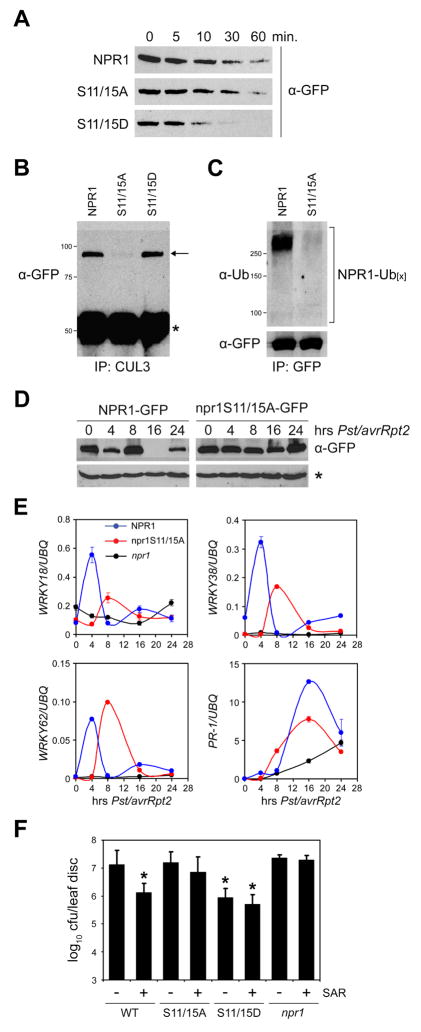
Figure 6. Phosphorylation stimulates NPR1 turnover and is required for SAR.
(A) Total protein was extracted from 35S::NPR1-GFP (in npr1-2), 35S::npr1S11/15A-GFP (in npr1-2), and 35S::npr1S11/15D-GFP (in npr1-2) plants in a buffer supporting proteasome activity. Extracts were incubated at room temperature for the time points indicated. Proteins were analyzed by reducing SDS-PAGE and Western blotting using an anti-GFP antibody.
(B) 35S::NPR1-GFP, 35S::npr1S11/15A-GFP, and 35S::npr1S11/15D-GFP plants were treated with a combination of 0.5 mM SA and 100 μM MG115. Protein extracts were immunoprecipitated (IP) with an anti-CUL3 antibody. Immunoprecipitated proteins were analyzed by Western blotting using an anti-GFP antibody. Molecular weight standards are indicated. Arrow indicates NPR1; asterisk indicates a cross-reacting IgG chain.
(C) 35S::NPR1-GFP and 35S::npr1S11/15A-GFP plants were treated as in (B). Protein extracts were immunoprecipitated (IP) with an anti-GFP antibody. Immunoprecipitated proteins were analyzed by Western blotting using anti-Ubiquitin and anti-GFP antibodies. Molecular weight standards are indicated.
(D) The leaf-halves of 35S::NPR1-GFP and 35S::npr1S11/15A-GFP plants were inoculated with avirulent Pst DC3000/avrRpt2 (OD600=0.02). At the indicated time points protein was extracted from the uninoculated leaf halves and subjected to reducing SDS-PAGE and Western blot analysis using an anti-GFP antibody. A non-specific band (*) indicated equal loading.
(E) 35S::NPR1-GFP, 35S::npr1S11/15A-GFP, and npr1-2 plants were treated as described in (D). mRNA was extracted from the uninoculated halves of the inoculated leaves and analyzed for the expression of WRKY18, WRKY38, WRKY62, and PR-1 using qPCR. Expression was normalized against constitutively expressed UBQ. Error bars represent SD (n = 3).
(F) Induction of SAR against Psm ES4326 in 35S::NPR1-GFP, 35S::npr1S11/15A-GFP, 35S::npr1S11/15D-GFP, and npr1-2 plants. Cfu, colony-forming units. Error bars represent 95% confidence limits (n = 8). Asterisks indicate statistically significant differences compared with uninduced 35S::NPR1-GFP plants (Tukey–Kramer ANOVA test; α = 0.05, n = 8).