Abstract
Sequence-specific DNA-binding activators, key regulators of gene expression, stimulate transcription in part by targeting the core promoter recognition TFIID complex and aiding in its recruitment to promoter DNA. Although it has been established that activators can interact with multiple components of TFIID, it is unknown whether common or distinct surfaces within TFIID are targeted by activators and what changes if any in the structure of TFIID may occur upon binding activators. As a first step toward structurally dissecting activator/TFIID interactions, we determined the three-dimensional structures of TFIID bound to three distinct activators (i.e., the tumor suppressor p53 protein, glutamine-rich Sp1 and the oncoprotein c-Jun) and compared their structures as determined by electron microscopy and single-particle reconstruction. By a combination of EM and biochemical mapping analysis, our results uncover distinct contact regions within TFIID bound by each activator. Unlike the coactivator CRSP/Mediator complex that undergoes drastic and global structural changes upon activator binding, instead, a rather confined set of local conserved structural changes were observed when each activator binds holo-TFIID. These results suggest that activator contact may induce unique structural features of TFIID, thus providing nanoscale information on activator-dependent TFIID assembly and transcription initiation.
Keywords: TAF, TFIID, transcription, activator, structure
Expression of protein-coding genes mediated by mammalian RNA polymerase II (Pol II) is a highly coordinated and elaborate process. To accurately transcribe a gene, a preinitation complex (PIC) is required to form at specific regions of the promoter DNA (Thomas and Chiang 2006). One critical step during PIC assembly involves directing the core promoter recognition complex to specific target genes. The core promoter recognition complex TFIID is a principal component within the transcriptional machinery responsible for recognizing and binding specific promoter DNA. Holo-TFIID consists of the TATA-binding protein (TBP) and 10–14 evolutionarily conserved TBP-associated factors (TAFs). Once TFIID alights on core promoter DNA, it directs a sequential recruitment of other general transcription factors including TFIIA, TFIIB, TFIIE, TFIIH, TFIIF, CRSP/Mediator, and Pol II that culminates with activated transcription at specific promoters.
Sequence-specific DNA-binding activators facilitate transcriptional activation at several levels including transcriptional preinitiation, promoter escape, and elongation/reinitiation that are dependent on various parameters including the context and complexity of gene promoters (Stargell and Struhl 1996; Naar et al. 2001). One well-documented mechanism involves activators directly interacting with various components of the PIC to enhance recruitment and to stabilize general transcription factors at specific promoters. Not surprisingly, multiple components within TFIID can serve as targets for mediating activator-stimulated transcription initiation (Wu and Chiang 2001). As such, a number of reports suggest that activators can enhance the kinetics of TFIID recruitment to DNA (Chi and Carey 1996). Conversely, emerging evidence has also revealed that TAF subunits within TFIID can, in some instances, actually help recruit activators to specific promoters to stimulate transcription (Ainbinder et al. 2002; Hilton et al. 2005; Liu et al. 2008). Therefore, it seems reasonable that activators can target TFIID prior to or after binding to DNA, depending on the context and conditions at specific promoters. However, it was not clear whether different activators use the same or distinct surfaces within holo-TFIID during activator/TFIID assembly, even though in vitro biochemical studies revealed that activators can bind to distinct subsets of TAFs in binary binding reactions.
As an alternative and increasingly important mechanism to regulate transcription, activators also have been shown to directly induce structural alterations in certain components of the PIC and/or DNA (Roberts and Green 1994; Chi and Carey 1996; Lieberman et al. 1997; Taatjes et al. 2002). Electron microscopy (EM) and single-particle reconstruction studies have begun to reveal the three-dimensional (3D) structures of mega-Dalton-size macromolecular assemblies including components of the PIC, and these studies have unmasked significant structural features and complex dynamics. For example, EM studies have determined a common structural framework of holo-TFIID complexes that feature a “horseshoe” shape consisting of three large lobes and a well-defined central cavity (Andel et al. 1999; Leurent et al. 2002). Importantly, recent cryo-EM studies have revealed significant structural flexibility within human TFIID with distinct open and closed conformational states that likely define the end points of a potential “breathing” mechanism (Grob et al. 2006). Indeed, we found recently that a cell type-specific TFIID complex preferentially adopts an open conformation that could likely promote synergistic transcription activation by the coordinate actions of two activators, c-Jun and Sp1 (Liu et al. 2008). EM studies have also visualized the highly mobile structure of the 1.2 MDa CRSP/Med complex, which can adopt distinct conformational states when bound to different activators (Naar et al. 2002; Taatjes et al. 2002, 2004). It remained unknown if holo-TFIID would similarly undergo such global structural alterations in the process of PIC assembly aided by specific activator–TFIID interactions. Therefore, it has become of interest to interrogate the surfaces of activator/TFIID association and any potential structural features induced in large multisubunit coactivator TFIID complexes when targeted by activators.
To begin the challenging task of probing the activator/TFIID-mediated transcription initiation assembly at the single-particle level, we chose to examine three distinct activators, the tumor suppressor protein p53, the glutamine-rich activator Sp1, and the proto-oncoprotein c-Jun, when bound to distinct subunits of TFIID. In response to physiological and pathological stimuli, each of these activators direct diverse gene expression programs by activating a number of promoters that participate in various critical cellular processes including cell cycle progression (Hess et al. 2004; Riley et al. 2008; Wierstra 2008). Although a long history of biochemical reports has documented specific activator/TAF interactions in activator-mediated gene initiation, no direct structural evidence exists for stable activator/TFIID assemblies. Given the significant impact of the association between these activators and TFIID in transcriptional regulation, we first set out to assemble the activators individually with holo-TFIID in vitro and subsequently visualize the 3D structures of these activator/TFIID assemblies by EM and single-particle reconstruction. More importantly, we attempted to document potentially distinct activator targeting surfaces within holo-TFIID and any alterations that could be induced within these TFIID complexes with three different activators binding distinct TAF subunits. In addition to locating the position of each activator when bound to holo-TFIID by EM, we also investigated potential TAF–activator contacts using photoactivatable protein cross-linking label transfer assays. By combining both biochemical and structural studies, we gained new insights into the assembly of active intact complexes.
Results
Activator and TFIID assembly in vitro
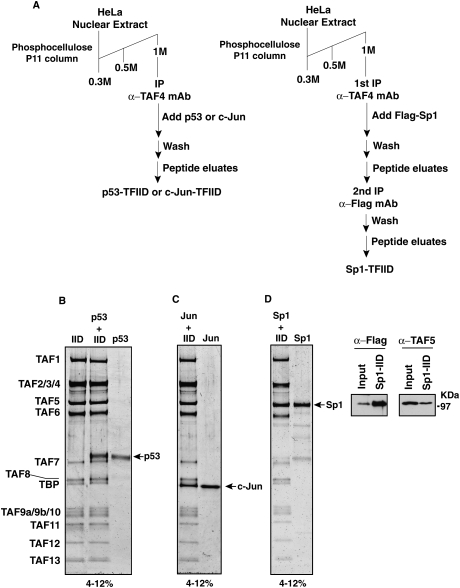
Currently, no 3D structures have been reported for stable activator/TFIID complexes. This paucity of structural information on key activator/TFIID modules is in great part due to several technical hurdles including the difficulty of obtaining homogenous binary activator/TFIID cocomplexes. As a first step toward overcoming these technical challenges to obtain stable activator-bound TFIID, we developed a strategy to assemble nearly 1:1 complexes of TFIID bound to activators in vitro. In order to obtain stable activator-bound TFIID species for structural analysis, we established various activator loading procedures that take advantage of our well-established immunopurification of holo-TFIID (Fig. 1A; Liu et al. 2008). In brief, we carried out highly specific immunoprecipitations from fractionated HeLa nuclear extracts using a peptide-elutable monoclonal antibody against TAF4 (Fig. 1A). The holo-TFIID bound to protein G resin containing covalently conjugated TAF4 mAb was washed extensively to remove loosely associated and nonspecifically bound proteins. Our previous work established that this “affinity purification” of TFIID yielded holo-complexes that were ∼99% pure and contain TBP along with TAFs 1–13 as determined by SDS-PAGE and mass spectrometry (Liu et al. 2008). Next, a 10-fold molar excess of activator (either p53, c-Jun, or Sp1) was incubated with the affinity-purified TFIID complex while still bound to the TAF4 mAb/protein G resin. Unbound activators were removed by extensive washing of the TAF4 affinity resin. A complex containing activator stably bound to TFIID was eluted from the TAF4 antibody affinity column using specific peptides recognized by the TAF4 mAb. The p53- and c-Jun-bound TFIIDs were analyzed by SDS-PAGE/Flamingo staining (Bio-Rad) and designated as p53-IID and c-Jun-IID, respectively (Fig. 1B,C). Since Sp1 comigrates with TAF5 in the SDS-PAGE gels, we were unable to directly determine the extent of Sp1 association with TFIID by gel electrophoresis. To circumvent this problem, we performed a tandem immunoprecipitation using peptide-elutable monoclonal antibodies against both TAF4 and Flag-tagged Sp1, sequentially (Fig. 1A, right panel). Flag-tagged Sp1 protein was incubated with TFIID-bound resins as described above. Following extensive washes, the eluate from the first affinity purification was next incubated with resin conjugated with anti-Flag antibody. The stable Flag-tagged Sp1-bound TFIID was then eluted with a peptide against the Flag epitope. After this sequential affinity purification procedure, we obtained a TFIID complex containing near stoichiometric amounts of Sp1, designated as Sp1-IID, as characterized by SDS-PAGE/Flamingo staining (Bio-Rad) and Western blot analysis using antibodies against either Flag or TAF5 in which we detected the full-length Sp1 stably bound to holo-TFIID (Fig. 1D). Notably, this strategy of activator loading appeared to be highly specific, since proteins such as TFIIB that interact weakly with TFIID failed to be retained on the affinity resin (Supplemental Fig. S1).
Figure 1.
Affinity purification of stable activator/TFIID protein complexes. (A) Schematic representation of the purification procedures of activator/TFIID complexes. P53- or c-Jun-bound TFIID complexes were obtained by immunoprecipitating TFIID from the phosphocellulose 1M KCl fraction using an anti-TAF4 antibody. After extensive washing, a 10-fold molar excess amount of activators p53 or c-Jun was loaded on to the resins containing TFIID. p53- or c-Jun-bound IIDs were obtained after extensive washes followed by an elution step using a specific peptide against TAF4. For Sp1-bound TFIID, Flag-tagged Sp1 was used in the loading step. Stable Sp1-IID complexes were obtained by tandem immunoprecipitations using anti-TAF4 and anti-Flag antibodies. (B,C) Distinct activator/TFIID complexes (i.e., p53-IID and c-Jun-IID) were analyzed by 4%–12% SDS-PAGE (Invitrogen) and visualized by Flamingo Fluorescent Gel Stain (Bio-Rad) and a Typhoon imaging scanning system (GE). (D) Sp1-IID was analyzed by 4%–12% SDS-PAGE (Invitrogen) and visualized by Flamingo Fluorescent Gel Stain (Bio-Rad) and a Typhoon imaging scanning system (GE). Since Flag-tagged Sp1 comigrates with TAF5, the presence of Flag-tagged Sp1 and TAF5 of the Sp1-IID complexes in the anti-Flag eluates was detected by Western blot analysis using anti-Flag (Sigma) and anti-TAF5 antibodies. “Input” represents the anti-TAF4 eluates from the first immunoprecipitation prior to anti-Flag affinity immunoprecipitations and “Sp1-IID” represents the anti-Flag eluates.
Structural architecture of three activator/TFIID assemblies
3D reconstruction of the p53/TFIID assembly
Several important potential mechanisms for activators to enhance TFIID-mediated transcription involve a direct interaction between activators and components within the PIC. In some cases, certain structural changes of basal factors can be induced by activator targeting (Roberts and Green 1994; Chi and Carey 1996; Taatjes et al. 2002). Our earlier cryo-EM study of TFIID had demonstrated an intrinsic conformational plasticity as determined by the open/closed states of TFIID (Grob et al. 2006). Therefore, it has remained an open and intriguing question whether TFIID contacts different activators using a common or distinct surfaces and whether the TFIID/activator assemblies display any structural alterations upon activator binding. To this end, we next determined the 3D structures of the three distinct activator/TFIID assemblies described above (i.e., p53-IID, c-Jun-IID, and Sp1-IID) by EM and single-particle reconstruction.
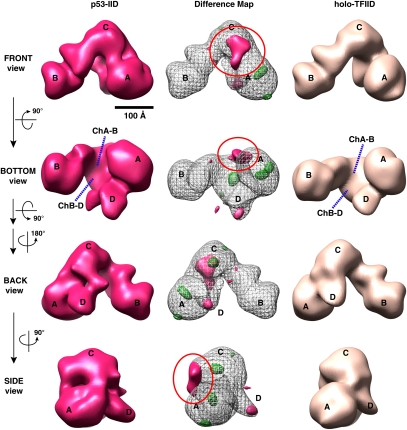
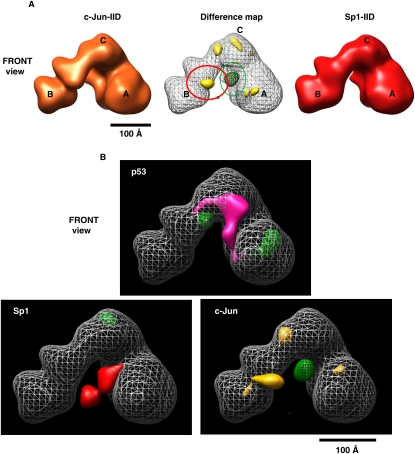
Having isolated three distinct stable activator/TFIID complexes, we first developed optimized negative staining conditions for protein complex preservation and image quality. To minimize potential bias in our single-particle 3D constructions, we chose to use the “average” conformational form of the cyro-TFIIID EM structure filtered to 60 Å as an initial reference for data processing (see the Materials and Methods). We obtained the 3D structure of p53-IID derived from a total of 13,855 particles employing the projection matching method (Grob et al. 2006). The final 3D structure was resolved to 33 Å as determined by Fourier-shell correlation (FSC) with a 0.5 cutoff (Supplemental Fig. S2, FSC curve of p53-IID). The resulting p53-IID 3D reconstruction, filtered at 33 Å, is presented with a similar threshold condition except for a total protein mass of ∼1.2–1.3 MDa as described previously (Fig. 2, left panel; Liu et al. 2008). This structure revealed that the overall architecture of the p53-IID complex is highly reminiscent of free holo-TFIID with its signature trilobed horseshoe shape containing a central cavity (Fig. 2, cf. left and right panels). However, the distances between lobe A and B (ChA-B) as well as between lobe B and D (ChB-D) across the central cavity of p53-IID are considerably shorter than in holo-TFIID (seen in Fig. 2, the bottom view), indicating that the structure of p53-IID appears to be tighter around the central cavity compared with the holo-TFIID structure on average. In fact, we found that the p53-IID structure appears to be more similar to the closed conformational form of cryo-TFIID structure than the open form (Grob et al. 2006), suggesting that binding of p53 to TFIID either selects or promotes a more closed state of the complex. Importantly, a careful comparison of p53-IID to unbound holo-TFIID revealed a prominent extra density region between lobes A and C, suggesting the potential location of the activator p53 (Fig. 2, left panel, front view).
Figure 2.
The 3D EM structure of p53-IID. 3D reconstructions of p53-IID (left), holo-TFIID (right), and a 3D density difference map (center) between p53-IID and TFIID are shown in four different views (front, bottom, back, and side). The major lobes are labeled A, B, and C, and a smaller domain labeled D lobe (see the back view). Channels within the central cavity between the A and B (ChA–B) or the B and D (ChB–D) lobes are shown with the dotted blue lines. The center column shows the difference map between p53-IID and holo-TFIID reconstructions, with solid magenta representing positive differences and mesh green representing negative differences relative to the holo-TFIID reconstructions. The gray mesh corresponds to the 3D structure of p53-IID with the A, B, C, and D lobes indicated. The most prominent difference region denoted by the red circle is located at lobe C, weakly connected to lobe A, and was interpreted as the location of the p53 protein. Two small extra density pockets are located in lobe D (back view). The missing densities in p53-IID are predominantly located at lobes A and C. Bar, 100 Å.
To further analyze this structure and locate the putative p53-binding site, a 3D density difference map was calculated between the newly defined p53-IID complex and free holo-TFIID structures (Liu et al. 2008) by using volumes that were both filtered to 33 Å. The structural features shown in magenta represent the positive density difference when the 3D volume of free holo-TFIID is subtracted from the reference volume of p53-IID (Fig. 2, middle panel). The negative density difference is represented in green at the same density threshold. The most likely location of the tumor suppressor p53 protein when bound to holo-TFIID was assigned to a prominent positive density located between lobes A and C (best seen in the front view in Figure 2; Supplemental Movie S1). This important feature revealed by the positive density difference map suggests that p53 likely attaches to both the A and C lobes, and in doing so may stabilize a structure resembling the “closed” conformation of the complex. A few additional small positive and negative densities are also located in lobes A, C, and D, possibly due to different partitioning resulting from the open/closed conformational states of these two complexes. There were no detectable differences in lobe B, which appears to remain in a fixed position with respect to lobe C. The significance of the observed differences between p53-bound and unbound holo-TFIID structures was further verified by additional analysis using FSC calculations of mixed particle data sets as previously described (Supplemental Fig. S2A; Liu et al. 2008).
The p53-IID data set may contain a minor population of TFIID particles lacking activators that might average out some potential differences between p53-IID and free TFIID reconstructions. To reduce this potential “noise” and improve our 3D reconstructions, we also sorted out particles using our newly refined p53-IID 3D structure filtered to 45 Å as an additional reference in addition to the TFIID reference volume for projection matching refinement (Grob et al. 2006). The resulting p53-IID 3D reconstruction, filtered at 31 Å, is very similar to the original p53-IID reconstruction from the unsorted particle data set (Supplemental Fig. S2B; details in the Supplemental Material). To confirm our results and avoid possible variations in staining, we calculated the difference density maps between the newly defined p53-IID and holo-TFIID generated from the same particle data set. These studies again revealed a significant positive density located at the lobe A/C interface where we propose p53 binds TFIID (Supplemental Fig. S2B). Because there is no commensurate “negative differences” associated with the “positive differences” attributed to p53 binding, it is unlikely that the latter corresponds to a conformational change. Thus, our data strongly suggest that the main positive density difference we observed is most likely attributed to an additional density bound to holo-TFIID instead of movement between lobes A/C that might have resulted from the breathing activity of TFIID (Grob et al. 2006).
3D reconstruction of the Sp1/TFIID assembly
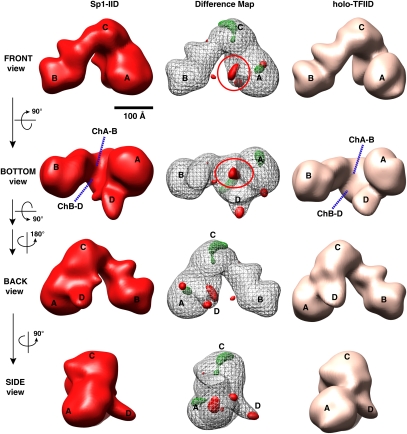
Next, we used an identical experimental approach to determine the structure of Sp1-IID and to examine potential structural differences with respect to the unbound holo-TFIID. The final 3D structure of Sp1-IID using a total of 17,824 particles was resolved to 33 Å as determined by FSC with a 0.5 cutoff (Supplemental Fig. S3, the FSC curve of Sp1-IID). The overall structural architecture of Sp1-IID is similar to the structures of both p53-bound IID and free TFIID (cf. Figs. 2 and 3, both left and right panels), demonstrating that Sp1-IID also retains the global structure of free holo-TFIID. However, similar to p53-IID, a few prominent altered features can also be seen with the Sp1-IID structure. A slightly tighter central cavity with a shorter distance of ChB–D is observed in the Sp1-IID structure on average compared with the free holo-TFIID structure on average (Fig. 3, bottom view), once again revealing features that more closely resemble the closed conformational state of free holo-TFIID. In addition, there appears to be a significant extra density located in lobe A, facing the central cavity that potentially represents bound Sp1 (Fig. 3, left panel, front and bottom views).
Figure 3.
The 3D EM structure of Sp1-IID. 3D reconstructions of Sp1-IID (left), holo-TFIID (right), and 3D density difference map (center) between Sp1-IID and TFIID are shown as in Figure 2 with the major lobes and channels as previously indicated. The channel within the central cavity between lobes A and B (ChA–B) of Sp1-IID is significantly narrower than that of holo-TFIID (see bottom view). The center column shows the difference map between Sp1-IID and holo-TFIID, with solid red representing positive differences, mesh green representing negative differences with respect to the holo-TFIID reconstruction, and the gray mesh corresponding to the 3D structure of Sp1-IID. The solid red circle denotes the most significant positive difference, and is assigned as the position of Sp1 contact within TFIID. There are missing densities in Sp1-IID located at the top of lobe C and within lobe A. Bar, 100 Å.
In order to identify the putative site of Sp1 binding within TFIID and map significant density differences between the Sp1-IID and free holo-TFIID structures, we calculated a 3D density difference map using both volumes represented as described in Figure 2 (Fig. 3, middle panel), except that the positive density differences are shown in red. The density difference map shows a significant extra density attached to Lobe A that extends across the central cavity that most likely corresponds to Sp1 (refer to Supplemental Movie S3). We again observed additional minor positive differences in lobes D and A, features also seen in the p53-IID structure, while some missing density differences can be seen located at lobes C and A, with lobe B again being mostly unaffected (Fig. 3, middle panel). To gauge the significance of these observed differences between Sp1-IID and free holo-TFIID, we again performed FSC calculations using mixed particle data sets as described previously (Liu et al. 2008). The FSC curve from the mixed data sets resolved to 43 Å, which is much poorer than the resolution of Sp1-IID and TFIID, thus confirming the significance of the structural changes seen between Sp1-bound TFIID and free TFIID (Supplemental Fig. S3A). In addition, we applied the same particle sorting algorithm for Sp1-IID as we did for p53-IID and performed the density difference map calculation as described above for p53-IID. This analysis adds further support to our conclusion that Sp1 is likely positioned at lobe A within the central cavity (Supplemental Fig. S3B; details in the Supplemental Material).
3D reconstruction of the c-Jun/TFIID assembly
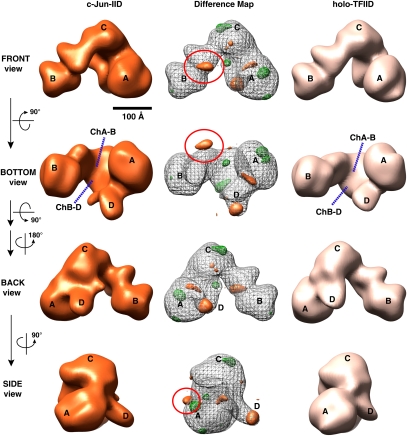
Finally, we determined the 3D structure of c-Jun-bound TFIID, thus allowing us to compare three distinct activator–TFIID structures as a means of potentially mapping both unique and common features induced by activator–TFIID interactions. We analyzed 14,434 particles using the identical strategy described above to obtain a 33 Å c-Jun-IID structure as determined by FSC with a 0.5 cutoff (Fig. 4, left panel; the FSC curve in Supplemental Fig. S4A). The structure of c-Jun-IID was the most similar to free holo-TFIID.
Figure 4.
The 3D EM structure of c-Jun-IID. 3D reconstructions of c-Jun-IID (left), holo-TFIID (right), and 3D density difference map (center) between c-Jun-IID and TFIID are presented as in Figures 2 and 3. The distance within the central cavity between lobes A and B (ChA–B) of c-Jun-IID is similar to the one of holo-TFIID (bottom view). The center column shows the difference map between c-Jun-IID and holo-TFIID, with orange representing positive differences, green representing negative differences, and the gray mesh corresponding to the 3D structure of c-Jun-IID. The orange circle denotes the significant positive difference and is predicted as the position of c-Jun contact within TFIID. Two positive differences are located at the tip of Lobe D and the junction between lobes A and D. Similar to p53- and Sp1-IID, missing densities in c-Jun-IID are located at the top of lobe C and within Lobe A. Bar, 100 Å.
However, a careful comparison of these two large structures by density difference map analysis again revealed several prominently altered features (Fig. 4, middle panel). First, one significant extra density is now located near Lobe B in the c-Jun-IID structure and likely represents the activator c-Jun bound to TFIID (Fig. 4, front view; Supplemental Movie S4). Similar to both p53-IID and Sp1-IID assemblies, the c-Jun-IID structure exhibits an extended D lobe when compared with free holo-TFIID complexes (Fig. 4, back view). There are also substantial pockets of density missing in lobes A and C in c-Jun-IID as was observed in the p53-IID and Sp1-IID structures (cf. Figs. 2, 3, and 4).
To further verify the distinct new density difference observed in lobe B, we again performed the mixed FSC calculations (Supplemental Fig. S4A) and the sorting algorithm as described above (Supplemental Fig. S4B; details in the Supplemental Material). The results validate the density difference between c-Jun-IID and free holo-TFIID. Furthermore, we calculated an additional density difference map between the 3D reconstructions of both c-Jun-IID and Sp1-IID complexes as described above (Fig. 5A). The positive difference shown in yellow represents the extra density located in c-Jun-IID but not in Sp1-IID. Indeed, a significant positive density “knob” can be seen situated at the same location in lobe B, as shown in Figure 4 (best seen in the front view, orange circle). These results lend further support to the notion that the significant extra density in lobe B of c-Jun-IID likely corresponds to c-Jun bound to TFIID.
Figure 5.
The significance of extra density by additional density difference map analysis. (A) 3D reconstructions of c-Jun-IID (left), Sp1-IID (right), and 3D density difference map (center) of c-Jun-IID minus Sp1-IID are shown with the major lobes and channels as previously indicated in Figures 2–4. The yellow and green mesh represents positive and negative differences, respectively, with the gray mesh corresponding to the 3D structure of c-Jun-IID (shown at center). The orange circle denotes the significant positive difference on the surface of TFIID and is predicted as the position of c-Jun contact within TFIID. (B) Summed 3D density difference maps of each activator for p53-IID (top center), Sp1-IID (bottom left), and c-Jun-IID (bottom right). The summed positive density differences are shown in magenta (p53), red (Sp1), and yellow (c-Jun) and confirm the points of contact for each respective activator on TFIID. The green mesh represents the sum of the negative differences. Bar, 100 Å.
Thus far, we determined three activator–TFIID structures and calculated their corresponding density difference maps when compared with holo-TFIID. To reduce the nonsignificant density variations between different reconstructions and strengthen the specific differences that we observed, we computed the sum of all density differences. In the case of p53, we summed up the density differences calculated from p53-IID minus holo-TFIID together with both the p53-IID minus c-Jun-IID and the p53-IID minus Sp1-IID (Fig. 5B). This same formula was applied to the other two activator–TFIID complexes (Fig. 5B). The resulting structures clearly confirm our previous observations (Supplemental Figs. S2–S4). Taken together, our 3D EM reconstructions indicate that each activator very likely targets a different surface within TFIID. Remarkably, while none of these activators caused dramatic global structural changes, all three activators induced a common set of small but consistent and significant structural changes in the conformational state of the mega-Dalton TFIID complex.
Each activator contacts distinct TAFs within TFIID
Most previous biochemical studies have examined direct contacts between activators and isolated TAF subunits of TFIID rather than activator contacts in the context of holo-IID complex. For example, p53 is capable of binding to TAF1, TAF3, TAF6, TAF9, and TBP when tested as binary binding partners in isolation (Martin et al. 1993; Lu and Levine 1995; Thut et al. 1995; Li et al. 2004; Bereczki et al. 2008). Likewise, the oncoprotein c-Jun has been reported to associate with TAF7, TAF1, or TBP in isolation (Franklin et al. 1995; Munz et al. 2003; Lively et al. 2004), while the glutamine-rich activator Sp1 was found to bind TAF4 and TAF7 (Lively et al. 2004). It seems reasonable to propose that, depending on the context of the binding region, different activators could be targeting multiple TAFs. Our unexpected and intriguing finding with each activator binding to distinct regions within holo-TFIID prompted us to further use a complementary biochemical strategy to determine the TAF subunits targeted by each activator (i.e., p53, c-Jun, and Sp1) when in association with highly purified native TFIID complexes. To map potential activator-TAF contacts in the context of intact holo-TFIID, we performed photo-cross-linking label transfer reactions using the trifunctional reagent Sulfo-SBED (S-SBED) (Supplemental Fig. S5A), which has been used previously to determine the targets of activators within various transcription complexes (Brown et al. 2001; Neely et al. 2002)
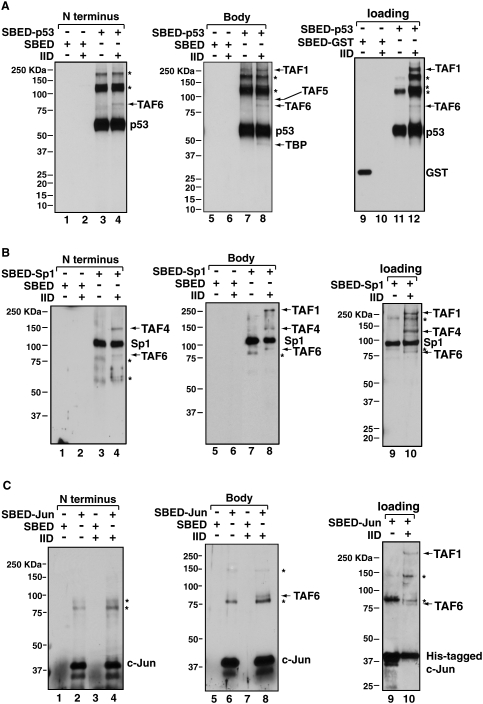
First, we labeled p53, c-Jun, or Sp1 each with S-SBED, respectively. Since the activation domains of these three activators reside within the N-terminal half of each protein, we chose to label activators with a low pH (∼7.0) buffer that allows the sulfonated ester group to react primarily with the N-terminal primary amine group, thus achieving an N-terminal labeling. To examine potential TAF contacts with additional domains of our activators, we also labeled each activator with S-SBED using a higher pH buffer to target internal tertiary amines (lysines), thus biasing the reaction toward “body labeling.” After removal of unreacted S-SBED, we incubated our highly purified TFIID with S-SBED-labeled p53, c-Jun, or Sp1 to initiate formation of activator/TFIID assemblies (Supplemental Fig. S5B). The samples were then exposed to UV to activate the aryl azide group that will cross-link to nearby TAFs. After cleavage of the disulfide bond within S-SBED with DTT, the biotin moiety will be “transferred” from the activator to any adjacent TAF within 21 Å (Brown et al. 2001; Neely et al. 2002). The resulting biotin-tagged TAFs in the holo-TFIID were disrupted by SDS, subjected to gel electrophoresesis and examined by immunoblotting using an antibody against biotin.
Our analysis of the p53-TFIID complex showed that the N-terminal activation domain of p53 most likely contacted TAF6 (Fig. 6A, lane 4). Since p53 contains an oligomerization domain, not surprisingly, p53 cross-linked with itself easily and several forms of p53 in addition to the monomer were observed in our assays. When we body-labeled the lysines throughout p53, two additional subunits, TAF1 and TAF5, plus TAF6, were tagged with biotin while TBP was weakly cross-linked (see Fig. 6A, lane 8; also refer to a longer exposure in Supplemental Fig. S6). These specifically cross-linked TAFs were confirmed by Western blot analysis using the same blots that were stripped and reprobed with antibodies against TAF1, TAF5, TAF6, and TBP (Supplemental Fig. S7). However, since there was a background band present in the p53-alone sample around 230 kDa where TAF1 migrates on the gel (Fig. 6B, lanes 7,8), we had to perform two additional experiments to confirm the cross-linking of TAF1. One approach was to pass SBED-labeled activators or GST over our TFIID affinity resins in a strategy analogous to that used for our EM work, followed by cross-linking of the activator/TFIID complexes while still bound to the column (Fig. 6C). This data confirmed that TAF1 was cross-linked when p53 was bound to TFIID on the affinity resins compared with the control GST samples (Fig. 6C, cf. lanes 10 and 12). Another approach was to specifically label the C terminus of p53 with the MTS label transfer reagent (Pierce) followed by an identical label transfer experiment as described in Figure 6A. We found that TAF1was again selectively cross-linked to the MTS-labeled C terminus of p53 (data not shown). Furthermore, these specifically cross-linked TAFs were confirmed by Western blot analysis using the same blots that were stripped and reprobed with antibodies against TAF1, TAF5, and TAF6 (data not shown).
Figure 6.
Activator contacts within TFIID by label transfer assays. Activators p53 (A), Sp1 (B), and c-Jun (C), were labeled at the N terminus (left panel, marked as N Terminus) and internal lysines (center panel, marked as body) with the heterotrifunctional SBED cross-linker. Activators and a control GST protein labeled at internal lysines were also loaded onto our TFIID affinity columns (right panel, marked as loading, lanes 9–12). Reactions containing cross-linker alone (lanes 1,2,5,6) and labeled activators/GST (lanes 3,4,7–9,11) were mixed with TFIID (lanes 2,4,6,8,10,12) and exposed to UV light. TFIID subunits that are within 21 Å of the S-SBED were covalently cross-linked to the activator. After cleaving the cross-linker with DTT, biotin was transferred from the activator to the cross-linked TFIID subunit. SDS-PAGE and immunoblotting using anti-biotin antibodies reveals TFIID subunits that contact distinct regions of the three activators. The stars represent nonspecific cross-linked bands in the blots.
Next, we found that the N-terminally labeled Sp1 cross-linked predominantly to a band corresponding to either TAF2, TAF3, or TAF4, which all comigrate in SDS-PAGE gels, in addition to very weak cross-linking to TAF6 (Fig. 6B, lane 4). TAF4 is most likely a main target of Sp1 in this cross-linking assay given the previously measured strong binary association between the Sp1 activation domain and TAF4 (Rojo-Niersbach et al. 1999; Wang et al. 2007). Using body-labeled Sp1 revealed that TAF1, TAF4, and TAF6 are located within 21 Å of Sp1 bound to TFIID. Label transfer experiments of SBED-labeled Sp1 bound to the TFIID affinity resin confirmed that TAF1, TAF4, and TAF6 cross-link to Sp1 (Fig. 6B, lane 10). These specifically cross-linked TAFs were further identified by Western blot analysis using the same blots that were stripped and reprobed with antibodies against TAF1, TAF4, and TAF6 (data not shown).
In the case of c-Jun, surprisingly, there were no TAFs that strongly cross-linked to the N-terminally labeled protein (Fig. 6C, lane 4). We confirmed the above result with a number of strategies and conditions (data not shown). In contrast, the body-labeled c-Jun clearly targeted TAF6 when c-Jun was bound to TFIID (Fig. 6C, lane 8). Because these two conditions of labeling c-Jun picked up different background bands that may complicate our findings, we performed an additional label transfer experiment with SBED-labeled c-Jun bound to the TFIID affinity resin. The results suggest that TAF1 could also cross-link to c-Jun in addition to TAF6 (Fig. 6C, lane 10). The identity of the cross-linked TAFs was ascertained by Western blotting using anti-TAF1 and TAF6 antibodies (data not shown). Taken together, these label transfer results suggest that these three different activators indeed bind to distinct regions of TFIID, likely making contact with different subsets of TAFs that are targeted by each of the activators (p53, Sp1, and c-Jun). This biochemical label transfer analysis thus provided an independent means of confirming our EM reconstructions of activator/TFIID assemblies that also revealed these three activators contacting different surfaces within holo-TFIID.
Discussion
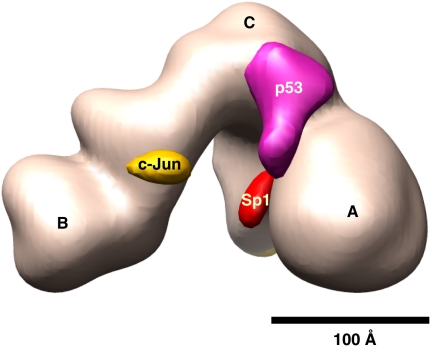
Our 3D density difference maps generated from reconstructions of the three independent activator/TFIID assemblies (i.e., p53-IID, Sp1-IID, and c-Jun-IID) and free holo-TFIID have served as a method to map the most likely contact sites of these activators within the native TBP–TAF complex. Remarkably, each activator contacts TFIID via select TAF interfaces within TFIID (presented as a model in Fig. 7). The unique and localized arrangements of these three activators contacting different surfaces of TFIID could be indicative of the wide diversity of potential activator contact points within TFIID that would be dependent on both the specificity of activation domains as well as core promoter DNA sequences appended to target gene promoters. It is also possible, however, that these distinct activator–TFIID contacts can form a common scaffold when TFIID binds to the core promoter DNA.
Figure 7.
A representative model of distinct surfaces within TFIID that are targeted by activators. Overlay of 3D density difference maps of p53-IID/holo-TFIID, Sp1-IID/holo-TFIID, and c-Jun-IID/holo-TFIID. The holo-TFIID structure (light pink) along with positive density differences for p53 (magenta), Sp1 (red), and c-Jun (orange) are shown, suggesting the location of potential distinct activator contacts. Bar, 100 Å.
It is well established that activators including p53, Sp1, and c-Jun frequently work synergistically with each other or other activators to potentiate selective gene expression programs in response to a variety of stimuli in vivo (Chinenov and Kerppola 2001; Wierstra 2008). Therefore, combinatorial mechanisms of promoter activation might favor distinct nonoverlapping activator-binding sites within TFIID, which can be achieved by specific interactions between selective TAF subunits and activators. Indeed, it was established that TAF1 and TAF4 serve as coactivators for Sp1, while TAF1, TAF6, and TAF 9 mediate p53-dependent transactivation (Rojo-Niersbach et al. 1999; Hilton et al. 2005; Wang et al. 2007) and TAF1 and TAF7 subunits are thought to be coactivators for c-Jun (Munz et al. 2003; Lively et al. 2004). Since activators make sequence-specific contacts with the DNA template at various positions upstream of the core promoter, it is also plausible that activators bound to unique surfaces of TFIID can influence specific structures of a promoter as the DNA traverses along TFIID resulting in distinct activator/promoter DNA structures.
Our activator mapping results also complement and structurally extend the functional relevance of previous biochemical and immunomapping studies of TFIID. For example, our label transfer studies show that the N-terminal activation domain of p53 contacts TAF6 (Fig. 6A), confirming previous biochemical evidence showing that amino acids 1–42 of p53 contact TAF6/9 (Thut et al. 1995). In support of this observation, the p53-IID 3D structure indicates that p53 contacts TFIID at lobes A and C where TAF6/9 are located as determined by EM immunomapping (Leurent et al. 2002). In addition, previous studies have shown that both TBP and TAF1 can directly contact p53 in the absence of additional TFIID subunits (Li et al. 2004). Interestingly, body-labeled p53 cross-linked to TAF1, TAF5, and weakly to TBP, thus extending the immunomapping studies that determined the locations of TBP and the N terminus of TAF1 at lobe C (Andel et al. 1999; Leurent et al. 2002, 2004). Thus, our EM activator mapping studies show a significant interface between p53 and specific TAFs located at lobes A and C of TFIID. Likewise, our Sp1 label transfer results confirmed previous biochemical data showing a direct interaction between TAF4 and the N-terminal glutamine-rich domains of Sp1 (Rojo-Niersbach et al. 1999; Wang et al. 2007). In addition to TAF4, we identified TAF6 as weakly cross-linked to Sp1, suggesting that TAF6 may also be in the vicinity but perhaps more distal to the N terminus of Sp1. The largest TFIID subunit, TAF1, was cross-linked when body-labeled Sp1 was used. This result is not entirely unexpected, since previous studies found that TAF1 is required for Sp1-dependent transactivation (Hilton et al. 2005), possibly through a direct interaction between TAF1 and Sp1 as shown in Figure 6B.
In comparison with p53 and Sp1, body-labeled c-Jun was shown to contact TAF1 and TAF6 in our label transfer studies with no subunits contacting the N-terminal activation domain of c-Jun (Fig. 6C). This N-terminal activation domain of c-Jun may be structurally flexible or predominantly unstructured and is apparently positioned away from TFIID contacts. Indeed, successful structural studies of c-Jun thus far have been limited to the C-terminal leucine zipper DNA-binding region when bound to DNA (Glover and Harrison 1995; Junius et al. 1996; Chen et al. 1998). Previous biochemical assays have shown that the C-terminal basic leucine zipper DNA-binding region also contacts the N terminus of TAF1(Lively et al. 2004).
It is worth noting that the extra density representing c-Jun and the other activator polypeptides in our EM studies may not reflect the full-expected size of the activators. This is due to the presence of large unstructured regions in these proteins that are averaged out during structural analysis. As activators contain multiple molten globular domains that likely interact with different partners, one would expect a high degree of structural disorder in the domains that are not in direct contact with TFIID. Thus, the extra density associated with each activator determined from the single-particle reconstructions likely only represents minimally the most stably associated portion of activators bound to TFIID. This common situation would invariably lead to underrepresenting the actual size of the activator in a manner not unlike crystal structures of domains with flexible loops that become “invisible” in the crystal structure.
Based on EM immunomapping, there are two copies of TAF6 within TFIID, wherein one copy resides in lobe A and another in lobe B (Leurent et al. 2002). Collectively, our studies suggest that two distinct activators (p53 and c-Jun) strongly contact the two different TAF6 subunits that are each located in different lobes of TFIID. It is unknown how p53 or c-Jun discriminates between TAF6 on lobe A versus B when binding to TFIID. In the future, it will be interesting to investigate if these two activators can bind to a single TFIID molecule simultaneously and decipher 3D structures of TFIID assemblies bound to select endogenous promoter DNA sequences in the presence and absence of distinct activators that are engaged in synergistic transcriptional activation.
It is of note that unlike the radical, diverse, and global structural changes observed with CRSP/Mediator complexes upon activator binding (Naar et al. 2002; Taatjes et al. 2002, 2004), TFIID largely retains its overall architecture when bound by three different activators (Figs. 2–4). Interestingly, we found that two of the activator/IID structures, p53-IID and Sp1-IID assemblies appear to be more constricted around the central cavity with narrower ChB–D and ChA–B channels, while the third structure, c-Jun-IID, remains most similar to free holo-TFIID. In particular, the p53-IID structure more closely resembles the closed conformational state of our previous cryo-TFIID structure (Grob et al. 2006). To test if p53-bound TFIID mimics the most closed conformational form of holo-TFIID, we performed 3D reconstructions using either the most closed or “open” cryo-TFIID structures as an initial reference volume for refinement. Interestingly, we found that both newly refined 3D structures generated from either the closed or open reference volume are fairly similar, with possibly a partial occupancy of p53 on lobe A (data not shown). These findings suggest that the overall p53-TFIID structure tends to move toward the closed conformation with moderate movement at the outer tips of lobes A and B (Supplemental Fig. S2B), even though p53-IID is predominantly observed in an intermediate average conformational form between the most closed and open forms (Grob et al. 2006). Perhaps factors contacting lobe A or C can induce certain coordinated movements within lobes that lead to a closed conformation of TFIID.
Although TFIID largely retains its prototypic global architecture upon activator binding, we observed several common localized structural changes induced upon activator binding in our 3D reconstruction. For example, we observed a prominent and consistent induced extra density protrusion located in lobe D when each of the three different activators binds TFIID (Figs. 2–4, BACK, BOTTOM, and SIDE views). Given that all these activators are represented by distinct densities with unique sizes and shapes within the bound TFIID structure, and the fact that we and others have demonstrated that they each can target different subunits within TFIID by a number of independent biochemical assays, it seems reasonable to assign “unique and significant” extra densities located at distinct sites as representing the different bound activators. In contrast, the common similarly sized extra density seen at lobe D of each activator–IID structure most likely represents a conserved conformational change induced by these three different activators. Interestingly, this protrusion in lobe D resides distal to each of the activator-binding sites, suggesting that these three activators may potentially induce a long-range internal conformational change within TFIID. It would be intriguing to identify which TAF subunits are located at the tip of lobe D and eventually determine the function, if any, of this extended lobe in activator-induced transcription initiation. However, despite the potential significance of these structural changes induced by activators, it is premature to speculate regarding their functional importance.
The low yields of human TFIID purified from HeLa cells have constrained our initial studies to negative stain EM. An important and challenging next step will be to pursue cryo-EM-derived activator/TFIID structures. It will also be of great interest in the future to ascertain if these activator-induced changes can accommodate or even facilitate DNA contacts that may stabilize the binding of TFIID to DNA during PIC assembly. Eventually, it will be important to unravel the enigmatic relationship between promoter recognition, transcriptional activity, and the closed/open conformations of TFIID during TFIID-mediated transcription initiation.
Materials and methods
Reagents and protein purification
Details of reagents and protein purification of activator/TFIID complexes used in this study are described in the Supplemental Material.
EM and single-particle 3D reconstructions
Four microliters (10–20 ng in total amount) of the fresh activator/TFIID assemblies was applied directly onto a thin carbon film supported by holey carbon on a 400-mesh copper grid (Electron Microscopy Sciences), which was freshly glow-discharged. After exchange with 3% Trehalose in 0.1 M KCl/TEM buffer (50 mM Tris-HCl, 0.1 mM EDTA, 2 mM MgCl2 at final pH 7.9), the sample grid was stained in five successive 75-μL drops of 1% Uranyl formate.
The image data were collected with a Tecnai 12 transmission EM (FEI) operated at 120 KeV and at a magnification of 30,000× with a defocus range of −0.85 μm to −1.5 μm. All the images were collected on SO-163 films (Kodak) and developed with D-19 developer (Kodak). The micrographs were digitized with a Super Coolscan 8000 scanner (Nikon) at a pixel size of 12.7 μm, resulting in a 4.23 Å per pixel size at the specimen scale.
A total of 13,855 particles (for p53-IID), 14,000 particles (for c-Jun-IID), or 17,824 particles (for Sp1-IID) were manually selected using Boxer (EMAN) (Ludtke et al. 1999) and then extracted to a particle window size of 119 × 119 pixels and converted to the SPIDER (system for processing of image data from electron microscopy and related fields) format for further data processing by SPIDER. The particles were then normalized prior to the 3D reconstruction using SPIDER's projection matching approach (Frank et al. 1996).
The average conformation of TFIID from cryo-EM data (Macromolecular Structure Database accession no. 1195) was filtered to 60 Å as a reference for the first cycle of projection matching for the p53-IID, c-Jun-IID, and Sp1-IID particle data in SPIDER. New 3D volumes reconstructed from each data set were used as references for all subsequent cycles of alignment (angular step sizes ranging from an initial 20° to a final 2°). The resolution throughout the refinement was determined by the 0.5 cutoff in the FSC curve. The 3D structures shown for p53-IID, c-Jun-IID, and Sp1-IID assemblies were filtered at a final resolution as indicated in the Result section. As for the difference map, the activator/TFIID and free TFIID density maps are defined as Volume 1 and Volume 2 or as indicated in the figures (i.e., Figure 5), respectively. Both 3D densities were normalized and subtracted from one another. The positive densities resulting from the difference [Volume 1–Volume 2] are shown either in magenta (Fig. 2), solid red (Fig. 3), or solid orange (Fig. 4). The negative densities were all shown in mesh green. The density difference maps are presented with a strong threshold (>3σ). In Figure 5A, the c-Jun-IID and Sp1-IID density maps are defined as Volume 1 and Volume 2. The positive density differences resulting from the difference maps [Volume 1–Volume 2] are shown in yellow (Fig. 5A). The negative densities were shown in mesh green. The summed 3D difference maps were calculated using the following formula: (p53-IID minus holo-IID) + (p53-IID minus c-Jun-IID) + (p53-IID minus Sp1-IID) (Fig. 5A, top center), (Sp1-IID minus holo-IID) + (Sp1-IID minus p53-IID) + (Sp1-IID minus c-Jun-IID) (Fig. 5A, bottom left), and (c-Jun-IID minus holo-IID) + (c-Jun-IID minus p53-IID) + (c-Jun-IID minus Sp1-IID) (Fig. 5A, bottom right). The positive densities in the summed 3D difference maps are colored magenta (p53), red (Sp1), and yellow (c-Jun) in Figure 5A, and the negative densities were shown in mesh green. All the 3D reconstructions were represented as isodensity surfaces using the University of California at San Francisco Chimera package (Pettersen et al. 2004) from the Resource for Biocomputing, Visualization, and Informatics at the University of California at San Francisco (supported by NIH P41 RR-01081).
To assess the significance of the differences observed, the FSC was calculated between halves of the activator/TFIID and free holo-TFIID datasets as previously described (Liu et al. 2008). The point of the 0.5 cutoff indicated a significant correlation between the two data sets only up to 43 Å, much poorer than the resolution of the reconstructions, indicating that the differences between the structures are overall significantly above the level of the noise.
Label transfer assays
Zero micrograms (as a mock control) or 1 μg of activators (i.e., c-Jun or Sp1) were incubated with the 0.5 mM final concentration of cross-linking reagent S-SBED (Thermo Scientific) following the manufacturer's instructions, which reacts with lysines using 100 mM KCl/HMG buffer (pH 7.9, [20 mM HEPES, 2 mM MgCl2, 10% glycerol]) or the N terminus of the proteins using 1 × PBS (pH 7.2). The reactions were incubated at room temperature in the dark for 15 min. Excess unreacted cross-linker was removed using Bio-Spin 6 chromatography columns (Bio-Rad). As for p53, the S-SBED-labeling conditions were identical as described, except that the His-tagged p53 proteins or buffer alone were bound to 15 μL of the nickel resins (Qiagen) prior to addition of the cross-linker and eluted off the resins with 40 μL of elution buffer (0.3 M Imidazole, 138 mM KCl, 20 mM HEPES, 2 mM MgCl2, 10% glycerol, 0.1% NP-40) after extensive washes to remove unreacted S-SBED cross-linker. Eighty nanograms of S-SBED-labeled activator proteins were incubated with 60 ng of TFIID at 30°C for 45 min. The mixture was then exposed to UV light (365 nm) at room temperature for 10 min to activate the aryl azide moiety on the SBED-labeled activators and covalently cross-link any protein within 21 Å of the activators. The reactions were treated with DTT to cleave the disulphide bond, and thus transfer the biotin moiety from activators to the cross-linked subunits of TFIID. The biotin-labeled activator-interacting TAFs were detected by SDS-PAGE and subjected to immunoblotting using anti-biotin antibody (Rockland). The identity of biotin-labeled TAFs was determined based on their known migration in SDS-PAGE gels and confirmed by Western blotting analysis using antibodies against the specific TAFs as indicated in the figures. Each reaction was repeated at least three times with the representative data shown. Details for the SBED labeling, loading of derivatized activators onto a TFIID affinity column and cross-linking can be found in the Supplemental Material.
Acknowledgments
We thank S. Zheng for providing TAF4 mAb supernatant, D. King for peptides, the Tjian laboratory tissue culture facility technicians, M. Haggart for assistance, U. Schulze-Gahmen for insect cells, and the Tjian laboratory members. We also thank S. Lipscomb, V. Ramey, and H. Wang for helpful advice and technical support. We are grateful to J. Yao, Z. Zhang, and U. Schulze-Gahmen for critical comments of the manuscript. This work was supported by NIH National Cancer Institute PO1 CA112181 (R.T. and E.N.), and NIH General Medical Sciences RO1 GM63072 (E.N.). R.T. and E.N. are Howard Hughes Medical Institute Investigators. R.T. is the President of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1790709.
Supplemental material is available at http://www.genesdev.org.
References
- Ainbinder E, Revach M, Wolstein O, Moshonov S, Diamant N, Dikstein R. Mechanism of rapid transcriptional induction of tumor necrosis factor α-responsive genes by NF-κB. Mol Cell Biol. 2002;22:6354–6362. doi: 10.1128/MCB.22.18.6354-6362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andel F, III, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID–IIA–IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- Bereczki O, Ujfaludi Z, Pardi N, Nagy Z, Tora L, Boros IM, Balint E. TATA binding protein associated factor 3 (TAF3) interacts with p53 and inhibits its function. BMC Mol Biol. 2008;9:57. doi: 10.1186/1471-2199-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Chi T, Carey M. Assembly of the isomerized TFIIA–TFIID–TATA ternary complex is necessary and sufficient for gene activation. Genes & Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos–Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Franklin CC, McCulloch AV, Kraft AS. In vitro association between the Jun protein family and the general transcription factors, TBP and TFIIB. Biochem J. 1995;305:967–974. doi: 10.1042/bj3050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Grob P, Cruse MJ, Inouye C, Peris M, Penczek PA, Tjian R, Nogales E. Cryo-electron microscopy studies of human TFIID: Conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: Quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Hilton TL, Li Y, Dunphy EL, Wang EH. TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol Cell Biol. 2005;25:4321–4332. doi: 10.1128/MCB.25.10.4321-4332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junius FK, O'Donoghue SI, Nilges M, Weiss AS, King GF. High resolution NMR solution structure of the leucine zipper domain of the c-Jun homodimer. J Biol Chem. 1996;271:13663–13667. doi: 10.1074/jbc.271.23.13663. [DOI] [PubMed] [Google Scholar]
- Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. Mapping histone fold TAFs within yeast TFIID. EMBO J. 2002;21:3424–3433. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leurent C, Sanders SL, Demeny MA, Garbett KA, Ruhlmann C, Weil PA, Tora L, Schultz P. Mapping key functional sites within yeast TFIID. EMBO J. 2004;23:719–727. doi: 10.1038/sj.emboj.7600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Li AG, Sheppard HM, Liu X. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: A role for TAF1 in cell G1 progression. Mol Cell. 2004;13:867–878. doi: 10.1016/s1097-2765(04)00123-6. [DOI] [PubMed] [Google Scholar]
- Lieberman PM, Ozer J, Gursel DB. Requirement for transcription factor IIA (TFIIA)–TFIID recruitment by an activator depends on promoter structure and template competition. Mol Cell Biol. 1997;17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WL, Coleman RA, Grob P, King DS, Florens L, Washburn MP, Geles KG, Yang JL, Ramey V, Nogales E, et al. Structural changes in TAF4b–TFIID correlate with promoter selectivity. Mol Cell. 2008;29:81–91. doi: 10.1016/j.molcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively TN, Nguyen TN, Galasinski SK, Goodrich JA. The basic leucine zipper domain of c-Jun functions in transcriptional activation through interaction with the N terminus of human TATA-binding protein-associated factor-1 (human TAF(II)250) J Biol Chem. 2004;279:26257–26265. doi: 10.1074/jbc.M400892200. [DOI] [PubMed] [Google Scholar]
- Lu H, Levine AJ. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Martin DW, Munoz RM, Subler MA, Deb S. p53 binds to the TATA-binding protein–TATA complex. J Biol Chem. 1993;268:13062–13067. [PubMed] [Google Scholar]
- Munz C, Psichari E, Mandilis D, Lavigne AC, Spiliotaki M, Oehler T, Davidson I, Tora L, Angel P, Pintzas A. TAF7 (TAFII55) plays a role in the transcription activation by c-Jun. J Biol Chem. 2003;278:21510–21516. doi: 10.1074/jbc.M212764200. [DOI] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes & Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Brown CE, Howe L, Workman JL. Transcription activator interactions with multiple SWI/SNF subunits. Mol Cell Biol. 2002;22:1615–1625. doi: 10.1128/MCB.22.6.1615-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Roberts SG, Green MR. Activator-induced conformational change in general transcription factor TFIIB. Nature. 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- Rojo-Niersbach E, Furukawa T, Tanese N. Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators. J Biol Chem. 1999;274:33778–33784. doi: 10.1074/jbc.274.47.33778. [DOI] [PubMed] [Google Scholar]
- Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: Two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Naar AM, Andel F, III, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Schneider-Poetsch T, Tjian R. Distinct conformational states of nuclear receptor-bound CRSP–Med complexes. Nat Struct Mol Biol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Thut CJ, Chen JL, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- Wang X, Truckses DM, Takada S, Matsumura T, Tanese N, Jacobson RH. Conserved region I of human coactivator TAF4 binds to a short hydrophobic motif present in transcriptional regulators. Proc Natl Acad Sci. 2007;104:7839–7844. doi: 10.1073/pnas.0608570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I. Sp1: Emerging roles—Beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J Biol Chem. 2001;276:34235–34243. doi: 10.1074/jbc.M102463200. [DOI] [PubMed] [Google Scholar]