Abstract
Many developmental control genes contain stalled RNA Polymerase II (Pol II) in the early Drosophila embryo, including four of the eight Hox genes. Here, we present evidence that the stalled Hox promoters possess an intrinsic insulator activity. The enhancer-blocking activities of these promoters are dependent on general transcription factors that inhibit Pol II elongation, including components of the DSIF and NELF complexes. The activities of conventional insulators are also impaired in embryos containing reduced levels of DSIF and NELF. Thus, promoter-proximal stalling factors might help promote insulator–promoter interactions. We propose that stalled promoters help organize gene complexes within chromosomal loop domains.
Keywords: Hox, Pol II stalling, insulator, NELF, DSIF, long range
Hox genes are responsible for the anterior–posterior patterning of most metazoan embryos (Lewis 1998). They are typically organized in gene complexes containing a series of cis-regulatory DNAs, including enhancers, silencers, and insulator DNAs (Kaufman et al. 1990; Maeda and Karch 2007). In Drosophila, the eight Hox genes are contained within two gene complexes: the Antennapedia complex (ANT-C), which controls the patterning of anterior regions, and the Bithorax complex (BX-C), which controls posterior regions (Sanchez-Herrero et al. 1985; Kaufman et al. 1990; Lewis 1998). The proper spatiotemporal transcription of Hox genes is achieved by the coordinated action of linked cis-regulatory DNAs that are organized in a colinear fashion across the ANT-C and BX-C complexes.
Chromosomal boundary elements, or insulators, are essential for the orderly regulation of Hox gene expression. They are thought to ensure proper cis-regulatory “trafficking,” whereby the correct enhancers interact with the appropriate target promoters (Sipos and Gyurkovics 2005; Maeda and Karch 2007; Bushey et al. 2008). Insulators might also help control the levels of transcription by attenuating enhancer–promoter interactions (Zhou et al. 1996). Insulators are sometimes associated with promoter targeting sequences (PTS) (Zhou and Levine 1999; Akbari et al. 2007; Lin et al. 2007), which can facilitate enhancer–promoter interactions by modulating the activities of neighboring insulators.
Recently, long-range cis-regulatory interactions have been mapped in Drosophila Hox complexes using the DamID technique (Cleard et al. 2006), chromosomal conformation capture (3C) assays (Lanzuolo et al. 2007), and transgenic approaches (Kyrchanova et al. 2008). These studies suggest that the Fab7 and Fab8 insulators are associated with the Abd-B promoter under repressed conditions, even though they map >30–50 kb downstream from the promoter (Cleard et al. 2006; Lanzuolo et al. 2007; Kyrchanova et al. 2008). These long-range interactions depend on the CTCF boundary-binding protein (Kyrchanova et al. 2008), thereby raising the possibility that insulators interact with one another and organize Abd-B cis-regulatory DNAs within chromosomal loop domains. Similarly, the prototypic insulators flanking the heat-shock puff locus, scs and scs′, have also been shown to interact with one another (Blanton et al. 2003). Additional insulator–insulator loops have also been documented (Kravchenko et al. 2005; Kyrchanova et al. 2007). These loops are thought to facilitate the interactions of remote enhancers and silencers with appropriate target promoters. Here, we present evidence that Hox promoters with stalled RNA Polymerase II (Pol II) possess an intrinsic insulator activity, which might help foster the formation of insulator–promoter chromosomal loop domains.
Results and Discussion
Stalled Hox promoters contain insulator activities
Four of the eight Hox genes contained in the ANT-C and BX-C contain stalled Pol II. Interestingly, all four stalled genes map at the boundaries of the two Hox complexes (see Supplemental Fig. 1; Muse et al. 2007; Zeitlinger et al. 2007). In contrast, internal Hox genes (pb, Dfd, and Scr within the ANT-C, and abd-A within the BX-C) lack stalled Pol II. This arrangement of stalled Hox genes raises the possibility that stalling contributes to the chromosomal organization of Hox complexes. All four stalled Hox genes (lab, Antp, Ubx, and Abd-B) were tested for enhancer-blocking activity in transgenic embryos, along with the promoter regions of two nonstalled genes (Scr and abd-A) (Fig. 1). Test promoters were placed 5′ of lacZ and inserted between a divergent white reporter gene and 3′ iab-5 enhancer (IAB5) (see diagrams in Fig. 1).
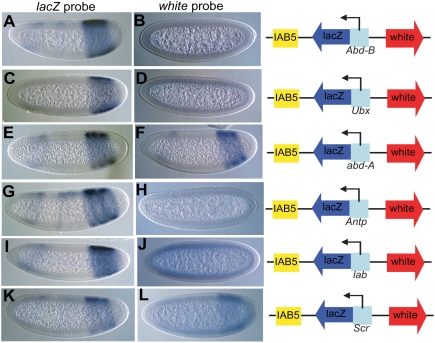
Figure 1.
Paused/stalled Hox gene promoters display insulator activity. The Hox gene promoters were tested for their ability to block enhancer promoter interactions in transgenic assays. The Hox/lacZ fusions were cloned between IAB5 enhancer and white reporter gene. The IAB5-lacZ interaction was seen for all of the transgenic lines for Abd-B/lacZ (A), Ubx/lacZ (C), abd-A/lacZ (E), Antp/lacZ (G), lab/lacZ (I), and Scr/lacZ (K) as the classical PS10-14 staining was observed during lacZ probe in situ. The IAB5–white interaction was blocked in all stalled promoter-lacZ fusions like Abd-B/lacZ (B), Ubx/lacZ (D), Antp/lacZ (H), and lab/lacZ (J) lines as no white in situ signal was observed in the IAB5 pattern. The nonstalled promoter-lacZ fusions like abd-A/lacZ (F) and Scr/lacZ (L), however, did show the IAB5 pattern when stained for white probe, as they did not show any insulator activity. The weak staining in the head regions is due to the P-transformation vector used in these experiments (Small et al. 1992). The embryos are aligned anterior to the left and posterior to the right.
IAB5 regulates Abd-B expression in posterior regions of the early embryo, corresponding to the primordia for parasegments 10–14 (Busturia and Bienz 1993). IAB5 is a robust enhancer, and can activate lacZ and white even when positioned far from the reporter genes. This assay was used to reveal an intrinsic enhancer-blocking activity of the eve promoter region (Ohtsuki and Levine 1998). eve/lacZ fusion genes block the ability of IAB5 to activate a distal CAT reporter gene. However, mutagenized eve promoter sequences lacking a critical proximal GAGA element failed to block IAB5–white interactions (Ohtsuki and Levine 1998). Similarly, the Abd-B proximal promoter (Abd-Bm) and Ubx promoter regions block activation of distal white expression (Fig. 1A–D), whereas the abd-A promoter does not interfere with the activation of white expression in the presumptive abdomen by the IAB5 enhancer (Fig. 1E,F).
These results suggest that the stalled Abd-B proximal promoter and Ubx promoters possess an enhancer-blocking activity, whereas abd-A does not. A similar trend was observed for Hox promoter sequences from the ANT-C. The Antp and lab promoters block IAB5–white interactions (Fig. 1G–J), whereas the Scr promoter (which lacks stalled Pol II) does not interfere with the activation of white expression in the presumptive abdomen (Fig. 1K,L). We also examined stalled genes from the tinman complex (Tin-C), which encode NK homeobox proteins responsible for patterning mesodermal lineages. All of the stalled promoters from the Tin-C contain insulator activities (Supplemental Fig. 2). In contrast, nonstalled promoters from lbl and C15 lack such activities when tested in similar transgenic assays (J Cande, unpubl.). Even the Hsp70 promoter, the classic example of Pol II pausing, displayed insulator activity when tested in similar enhancer-blocking transgenic assays (Supplemental Fig. 3).
Stalled Hox promoters do not display tethering activities
The preceding experiments suggest that stalled Hox gene promoters contain enhancer-blocking activities. However, an alternative possibility is that stalled promoters are “stronger” than the white promoter, and are able to sequester the shared IAB5 enhancer. To distinguish between competition and insulator activities, the IAB5 enhancer was placed between the divergently transcribed white and lacZ reporter genes (Fig. 2). When the white promoter sequence was placed 5′ of the lacZ reporter gene, the shared IAB5 enhancer worked equally well to activate both white and lacZ expression (Fig. 2A,B). Similar results were obtained when the leftward lacZ reporter gene was placed under the control of either the stalled Abd-B (Fig. 2C,D) or Ubx (Fig. 2E,F) promoters. In all of these cases, both white and lacZ are expressed equally well in the presumptive abdomen. These results suggest that stalled promoters do not block enhancer–promoter interactions by a competition mechanism. Rather, they work like insulators and block such interactions only when positioned between the distal enhancer and target promoter.
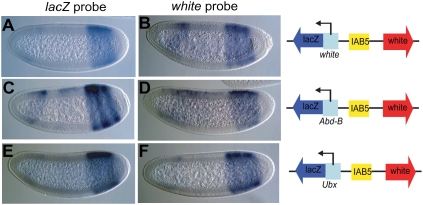
Figure 2.
Abd-B and Ubx promoter insulator activity does not involve tethering. The Abd-B and Ubx stalled promoters were tested for their ability to display promoter competition in a transgenic assay in which the IAB5 enhancer was cloned in between the white reporter and prom/lacZ fusion. As a control, the white/lacZ fusion (A) and white (B) were driven equally well by IAB5 enhancer in a white/lacZ (A,B) transgene. The IAB5-lacZ interaction (C,E) as well as the IAB5–white (D,F) interaction were facilitated in Abd-B/lacZ (C,D) and Ubx/lacZ (E,F) lines, respectively.
Hox promoter-blocking activity requires negative Pol II elongation factors
To determine whether stalled Pol II is important for the enhancer-blocking activities of Ubx and Abd-B, we examined mutant embryos with reduced levels of critical Pol II elongation factors (Fig. 3). Ubx and Abd-B were selected for further studies since we showed recently that optimal expression of both genes depends on the Pol II elongation factors Cdk9 (pTEFb) and Elo-A (Chopra et al. 2009). We reasoned that destabilization of stalled Pol II might reduce the enhancer-blocking activities of the Ubx and Abd-B promoter regions. However, reductions in Cdk9 and Elo-A are expected to stabilize, not destabilize, Pol II stalling since both are positive factors that promote elongation (Saunders et al. 2006). Indeed, reductions in Cdk9 or Elo-A activity do not alter the enhancer-blocking activities of the Ubx and Abd-B promoters (Supplemental Fig. 4).
Figure 3.
Negative elongation factors are required for Hox gene promoter insulator activity. The stalled promoter insulator lines were tested in elongation factor mutant backgrounds. (A–H) The Abd-B/lacZ line was tested in Nelf-E/+ (A,B), Nelf-A/+ (C,D), Spt4/+ (E,F), and Spt5/+ (G,H) backgrounds, and there was loss of insulator activity of the Abd-B promoter in these mutant backgrounds as seen by the appearance of the IAB5 pattern in situ signal when white probe was used (B,D,F,H). (I–P) Similar disruption of the insulator activity was observed for the Ubx/lacZ transgene in different mutant backgrounds of Nelf-E/+ (I,J), Nelf-A/+ (K,L), Spt4/+ (M,N), and Spt5/+ (O,P). (J,L,N,P) The white probe displayed the IAB5 pattern staining in these elongation mutant backgrounds.
To investigate the link between Pol II stalling and enhancer blocking, we examined two negative elongation factors: NELF and DSIF (Wada et al. 1998; Yamaguchi et al. 1998; Kaplan et al. 2000; Lee et al. 2008). The NELF-E protein binds to the short nascent transcripts protruding from the active site of Pol II after transcription initiation and promoter clearance, and thereby inhibits Pol II elongation (Wu et al. 2005; Lee et al. 2008). Both NELF and DSIF are thought to help stabilize Pol II at the pause site, typically 20–50 base pairs (bp) downstream from the transcription start site (Saunders et al. 2006; Gilchrist et al. 2008; Lee et al. 2008). Since Pol II elongation factors are encoded by essential genes, it is not possible to examine the lacZ/white reporter genes in homozygous mutant embryos. Instead, the transgenes were expressed in embryos derived from heterozygous females, and thereby contain half the normal levels of NELF and DSIF (Spt) subunits (Fig. 3). Reductions in Nelf-E (Fig. 3A,B,I,J), Nelf-A (Fig. 3C,D,K,L), Spt4 (Fig. 3E,F,M,N), and Spt5 (Fig. 3G,H,O,P) cause clear disruptions in the enhancer-blocking activities of both the Ubx and Abd-B promoters, as seen by the strong activation of the distal white reporter gene (Fig. 3B,D,F,H,J,L,N,P). In contrast, white expression is blocked when the same transgenes are expressed in a wild-type background (see Fig. 1B,D). The simplest interpretation of these results is that reduced levels of the NELF and DSIF inhibitory complexes destabilize stalled Pol II at the pause site. Reduced pausing results in diminished enhancer-blocking activities. There is a similar loss in the enhancer-blocking activities of the eve promoter (Ohtsuki and Levine 1998) and Fab7 insulator (Schweinsberg et al. 2004) when the transgenes are expressed in embryos containing reduced levels of the GAGA factor, Trl (Farkas et al. 1994). It is conceivable that the GAGA factor also contributes to the enhancer-blocking activity of the Ubx promoter since Trl/+ embryos display augmented expression of white (data not shown).
In principle, the augmented expression of the white reporter gene might not result from the impaired function of the stalled insulators, but might arise from enhanced activity of the white promoter. To investigate this issue, we performed Pol II chromatin immunoprecipitation (ChIP) assays, coupled with quantitative PCR (qPCR) assays (Supplemental Fig. 5). In DSIF and NELF mutant embryos, there is no increase in Pol II levels at either the white promoter or intronic regions as compared with wild-type embryos (Supplemental Fig. 5). These results suggest that augmented expression of white is due to diminished insulator activities of stalled promoters in embryos containing reduced levels of negative Pol II elongation factors.
Negative elongation factors are essential for Fab7 and Fab8 function
It has been suggested that insulators might work, at least in part, via promoter mimicry (Geyer 1997). To explore this issue, we examined the impact of reductions in NELF and DSIF on the activities of two known insulators—Fab7 (Gyurkovics et al. 1990) and Fab8 (Barges et al. 2000)—from the BX-C. Previously published transgenic lines were used that contain Fab7 or Fab8 inserted between the IAB5 and 2XPE (twist) enhancers (Zhou et al. 1996) attached to a leftward lacZ reporter gene and rightward white reporter (see the diagrams in Fig. 4). In wild-type embryos, the reporter genes are activated only by the proximal enhancer. Thus, white is activated solely in the mesoderm by the 2XPE enhancer (Fig. 4B,L), while lacZ is activated in the presumptive abdomen by IAB5 (Fig. 4A,K). The distal enhancers are blocked by the Fab7 or Fab8 insulators. Consequently, IAB5 fails to activate white and the 2XPE enhancer fails to activate lacZ.
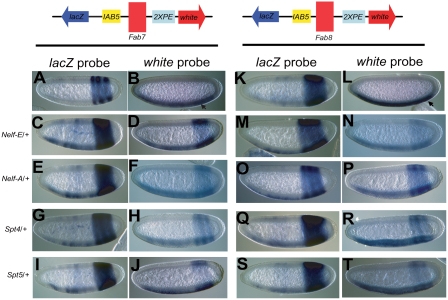
Figure 4.
Negative elongation factors are required for enhancer-blocking activity of putative insulators like Fab7 and Fab8. Enhancer-blocking lines containing Fab7 (A–J) and Fab8 (K–T) insulators, cloned between divergently placed IAB5/lacZ and 2XPE/white reporters, were tested in elongation factor mutant backgrounds. The Fab7 (A,B) and Fab8 (K,L) line shows lacZ staining in IAB5 pattern (A,K) and white in twist (2XPE) pattern (arrow) (B,L), respectively. In Nelf-E/+ (C,M), Nelf-A/+ (E,O), Spt4/+ (G,Q), and Spt5/+ (I,S) backgrounds the lacZ in situ now displays both the IAB5 as well as the 2XPE patterns for both Fab7 and Fab8 lines, respectively. A similar dual IAB5 and 2XPE pattern was observed for the white in situ in Nelf-E/+ (D,N), Nelf-A/+ (F,P), Spt4/+ (H,R), and Spt5/+ (J,T) backgrounds for Fab7 and Fab8 lines, respectively. The embryos are aligned anterior to the left, posterior to the right, dorsal up, and ventral down.
Very different results are observed when the transgenes are crossed into mutant embryos containing reduced levels of NELF or DSIF (Spt) subunits. There is a loss in the enhancer-blocking activities of the Fab7 and Fab8 insulators and, as a result, white and lacZ display composite patterns of expression in the mesoderm and abdomen since they are now activated by both enhancers (Fig. 4C–J,M–T). These results suggest that negative Pol II elongation factors are required for the enhancer-blocking activities of the Fab7 and Fab8 insulators.
We propose that insulators interact with stalled promoters to form higher-order chromatin loop domains (Fig. 5), similar to those created by insulator–insulator interactions (Blanton et al. 2003; Vazquez et al. 2006; Kyrchanova et al. 2007; Lanzuolo et al. 2007; Bartkuhn et al. 2009). Perhaps proteins that bind insulators interact with components of the Pol II complex at stalled genes. Indeed, the recent documentation that the BEAF insulator protein binds to many of the same sites as NELF is consistent with a physical link between stalled Pol II and insulators (Jiang et al. 2009). The resulting chromatin loops can prevent the inappropriate activation of stalled genes by enhancers associated with neighboring loci (Fig. 5). As discussed earlier, stalled Hox genes are located at the boundaries of the ANT-C and BX-C (Supplemental Fig. 1). This arrangement might help ensure that cis-regulatory sequences located outside the complexes do not fortuitously interact with genes contained inside the complex and vice versa. The demonstration that stalled Hox promoters possess an intrinsic insulator activity adds to the intricacy of the chromosomal landscapes (Spitz et al. 2003) that control Hox gene expression in both arthropods and vertebrates.
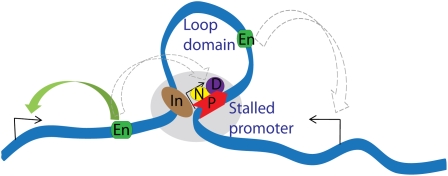
Figure 5.
Stalled Hox promoters may help promote higher-order chromatin organization within the Hox loci. Our results suggest that the stalled promoters contain intrinsic insulator activity that requires NELF (N) and DSIF (D) proteins, and this may help define higher-order loops within gene complexes such as the Hox complex. The stalled Pol II (P) along with the NELF and DSIF complex may interact with putative insulator (In) sequences, as seen for the Abd-B promoter (stalled) and the Fab7 (insulator). Our experiments also suggest that that putative insulator sequences also require NELF and DSIF proteins, and this could be due to sharing of these proteins via the formation of higher-order loops. Such loop domains may help in proper regulation of genes and prevent any aberrant activation from neighboring enhancers (En) (shown by dashed arrow), thus favoring proper gene regulations at the higher-order level.
Materials and methods
Plasmid construction and molecular analysis
The iab-5 enhancer was amplified as a 1-kb fragment from the genome and cloned into the enhancer-blocker vector. The enhancer-blocker vector was engineered to retain white and lacZ as markers and the backbone was similar to as described earlier (Ohtsuki and Levine 1998). The paused promoters were selected based on Pol II-binding signals from the ChIP–chip study and usually flanked 100 bp either side of transcription start site of the gene. The paused promoter sequences (ranging 180–220 bp) were PCR-amplified (primers sequences available on request) and subcloned into a pBSKS vector containing a lacZ fragment to make a promoter-lacZ fusion. This promoter-lacZ fragment was then subcloned into the pCaSper-based enhancer-blocker vectors to get final constructs that were w-prom/lacZ-IAB5-Casper (Zhou et al. 1996; Ohtsuki and Levine 1998). To make the w-IAB5-prom/lacZ vector, the IAB5 enhancer was excised from the w-prom-lacZ-IAB5-Casper vector and cloned between w and prom-lacZ reporters. All of the cloned fragments were verified by sequencing.
Fly strain culturing and transgenics
All fly stocks were cultured at 25°C in standard cornmeal medium. The transgenic flies were created at BestGene, Inc., or were injected as per standard protocol (Small et al. 1992). At least three independent lines were analyzed for each of the constructs for all the experiments (see Supplemental Table 1). For the crosses, virgin females of elongation factor mutants were crossed to males of the transgenic fly, followed by in situ hybridization. The elongation factor mutations were obtained from the Bloomington Stock Center, Exilexis, and Drosdel deletion project. The fly stocks used were y1 w1118; Spt5MGE-3/SM1; Psn143/TM6B (Spt5), y1 w[67c23]; Pn,Spt4[k05316]/CyO (Spt4), y1 w[67c23]; P{w[+mC] y[+mDint2] = EPgy2} rdx[EY06724]/TM3,Sb (Nelf-E), c00768/TM6b,Tb (Nelf-E), y1; and ry506 P{SUPor-P}NELF-AKG09483/TM3, Sb1 Ser1 (Nelf-A).
In situ hybridization
All probe templates were obtained from PCR-amplified genomic fragments cloned into pBluescript vector (Fermentas) and confirmed by sequencing. PCR primers were designed by using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), and a list of primers used is available on request. For each template, antisense RNA probes were in vitro transcribed by using T7 or SP6 RNA polymerase and digoxigenin-UTP (Roche Molecular Biochemicals). Embryos were collected for 4 h and aged for an additional 2 h. Fixed embryos were hybridized with the riboprobes as described elsewhere (Jiang et al. 1991).
ChIP and qPCR
The ChIPs using RNA Pol II antibodies followed by qPCR were performed as described elsewhere (Chopra et al. 2009). In brief, a cocktail of Pol II antibodies 8WG16 and H14 was used for the ChIP assays on chromatin prepared from wild-type or mutant embryos.
Acknowledgments
We thank J. Zhou for providing the Fab7 and Fab8 transgenic fly stocks, and the Bloomington and Exelixis Stock Centers for mutant strains. We also thank J.T. Lis for critical feedback on the manuscript, and P. Paliwal, B. Haley, D. Hendrix, and Y. Goltsev for helpful discussions and technical advice. This work was supported by a grant from the NIH to M.L. (GM34431).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1807309.
Supplemental material is available at http://www.genesdev.org.
References
- Akbari OS, Schiller BJ, Goetz SE, Ho MC, Bae E, Drewell RA. The abdominal-B promoter tethering element mediates promoter-enhancer specificity at the Drosophila bithorax complex. Fly (Austin) 2007;1:337–339. doi: 10.4161/fly.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes & Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG. Chromatin insulators: Regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia A, Bienz M. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila . Curr Biol. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Kosman D, Ip YT, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes & Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of BEAF binding sites in Drosophila links BEAF to transcription. Mol Cell Biol. 2009 doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster . Genes & Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman TC, Seeger MA, Olsen G. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster . Adv Genet. 1990;27:309–362. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- Kravchenko E, Savitskaya E, Kravchuk O, Parshikov A, Georgiev P, Savitsky M. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol Cell Biol. 2005;25:9283–9291. doi: 10.1128/MCB.25.21.9283-9291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: Effects of insulator pairing on enhancer-promoter communication. Mol Cell Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol Cell Biol. 2008;28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila . Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. The bithorax complex: The first fifty years. Int J Dev Biol. 1998;42:403–415. [PubMed] [Google Scholar]
- Lin Q, Chen Q, Lin L, Smith S, Zhou J. Promoter targeting sequence mediates enhancer interference in the Drosophila embryo. Proc Natl Acad Sci. 2007;104:3237–3242. doi: 10.1073/pnas.0605730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Making connections: Boundaries and insulators in Drosophila . Curr Opin Genet Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes & Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos L, Gyurkovics H. Long-distance interactions between enhancers and promoters. FEBS J. 2005;272:3253–3259. doi: 10.1111/j.1742-4658.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Vazquez J, Muller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome–chromosome interactions in Drosophila . Mol Biol Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Lee C, Fan R, Smith MJ, Yamaguchi Y, Handa H, Gilmour DS. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 2005;33:1269–1279. doi: 10.1093/nar/gki274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Handa H. Interplay between positive and negative elongation factors: Drawing a new view of DRB. Genes Cells. 1998;3:9–15. doi: 10.1046/j.1365-2443.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567–575. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]
- Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer–promoter interactions in the Drosophila embryo. Genes & Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]