Abstract
Purpose
We investigated the cytotoxicity and mechanisms of cell death of the broad-spectrum histone deacetylase inhibitor (HDACi), PCI-24781, alone and combined with bortezomib in Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL) cell lines and primary lymphoproliferative (CLL/SLL) cells.
Experimental design
Apoptosis, mitochondrial membrane potential, cell cycle analysis, and reactive oxygen species (ROS) were measured by flow cytometry, while caspase-activation was determined by Western blot. NF-kB-related mRNAs were quantified by RT-PCR, NF-kB-related proteins by Western blotting, and NF-kB DNA binding-activity by electromobility shift assay. Finally, gene expression profiling (GEP) was analyzed.
Results
PCI-24781 induced concentration-dependent apoptosis that was associated with prominent G0/G1 arrest, decreased S-phase, increased p21 protein, and increased ROS in HL and NHL cell lines. Dose-dependent apoptosis with PCI-24781 was also seen among primary CLL/SLL cells. PCI-24781-induced apoptosis was shown to be ROS- and caspase-dependent. Combined PCI-24781/bortezomib treatment resulted in strong synergistic apoptosis in all NHL lines (combination indices: 0.19-0.6) and was additive in HL and primary CLL/SLL cells. Further, PCI-24781/bortezomib resulted in increased caspase cleavage, mitochondrial depolarization, and histone acetylation vs either agent alone. GEP showed that PCI-24781-alone significantly downregulated several antioxidant genes, proteasome components, and NF-kB pathway genes, effects which were enhanced further with bortezomib. RT-PCR confirmed downregulation of NF-kB1 (p105), c-Myc, and IkB-kinase subunits, while NF-kB DNA-binding activity was decreased.
Conclusion
We show that PCI-24781 results in increased ROS and NF-kB inhibition, leading to caspase-dependent apoptosis. We also demonstrate that bortezomib is synergistic with PCI-24781. This combination or PCI-24781 alone has potential therapeutic value in lymphoma.
Keywords: HDAC inhibition, reactive oxygen species, NFκB, apoptosis, lymphoma
Introduction
Lymphoid malignancies are caused in part by genetic and epigenetic deregulation of tumor suppressor genes (1). The process of histone deacetylation is a well-characterized epigenetic modification (2, 3). Histone deacetylases (HDAC) and histone acetylases are enzymes that have been shown to be aberrantly expressed or regulated in malignant tissues, resulting in inhibition of certain tumor suppressor genes, thereby allowing expression of the malignant phenotype. By inhibiting deacetylation of histones and allowing acetyl groups to remain on histones, HDAC inhibitors (HDACi) promote an open chromatin structure that allows gene transcription in relevant tumor suppressor genes that may favor tumor cell apoptosis.
The biological effects of HDACi include reversion of the transformed phenotype, inhibition of proliferation, cell cycle arrest, induction of differentiation, and apoptosis in tumor cell lines (4-6). They have also been shown to generate reactive oxygen species (ROS) in solid tumor and leukemia cells (5, 7-9), which may contribute to the mechanism. The broad spectrum HDACi, PCI-24781 (Pharmacyclics, Inc), is a phenyl hydroxamic acid-based, orally bioavailable compound currently in clinical trials for the treatment of neoplastic diseases (10, 11). It has activity in solid tumors, including colorectal carcinoma in Phase I trials (12) and it is being evaluated in Phase II trials in a variety of malignancies. We evaluated the cytotoxicity and the mechanisms of cell death using the HDACi PCI-24781, in Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL) cell lines and among primary lymphoproliferative cells.
Bortezomib is a proteasome inhibitor that received FDA approval in the United States for relapsed multiple myeloma and more recently for relapsed mantle cell lymphoma, where cell death has been associated with increased ROS (13-15). Inhibition of proteasome activity by bortezomib results in stabilization of IkBa with resultant NF-kB inhibition as well as stabilization of p53 and Bax, leading to apoptosis. In addition, in vitro studies in solid tumors and hematologic malignancies (multiple myeloma and leukemia) have shown synergy when bortezomib and HDACi are combined (16-20). There is however, little information on the activity and mechanism of this combination in lymphoma, with only one prior report in lymphoma (mantle cell) where HDACi has been tested in combination with bortezomib (18). We hypothesized that concomitant exposure of PCI-24781 and bortezomib might enhance apoptosis in other subtypes of lymphoma through ROS-related mechanisms.
We show here that the HDACi, PCI-24781, induced concentration-dependent apoptosis in HL and NHL cells, which was dependent on caspase and ROS production. Further, PCI-24781 exhibited strong synergy when combined with bortezomib, inducing ROS-dependent apoptosis in all NHL cell lines. Cell death induced by PCI-24781, bortezomib, and the combination occurred through interacting mechanisms including downregulation of oxidative stress response and proteasome/NF-kB pathways, which were likely responsible in part for the observed synergy in these NHL cells.
Materials and Methods
Cell lines and reagents
The L428 HL cell line and the NHL cell lines Ramos (Burkitt lymphoma), HF1 (follicular lymphoma), and SUDHL4 (large B-cell lymphoma) were cultured in RPMI1640 (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS), L-glutamine, and penicillin/streptomycin (Invitrogen, Carlsbad, CA). Cells were maintained at 37°C with 5% CO2. Bortezomib was provided by Millennium Pharmaceuticals (Cambridge, MA) and PCI-24781 was provided by Pharmacyclics Inc. (Sunnyvale, CA). Q-VD-OPh was used for pan-caspase inhibition (Calbiochem, San Diego CA), 6-carboxy-2′7′-dichlorodihydrofluorscein (H2DCF-DA) for ROS, JC-1 and valinomycin, (Molecular Probes, Eugene, OR) for mitochondrial membrane potential, and catalase was obtained from Sigma-Aldrich (St. Louis, MO). Antibodies for caspase 8, caspase 9, caspase 3, acetyl histone H3 and H4, PARP (Cell Signaling, Beverly, MA), c-Myc (BD Pharmingen, San Diego, CA), cytochrome C, and p21 (Santa Cruz Biotech, Santa Cruz, CA) were used to study cell death pathways. GAPDH (Millipore Corporation, Temecula, CA) was used as a loading control for Western blotting. Secondary antibodies included horseradish peroxidase conjugated anti-rabbit and anti-mouse immunoglobulin antibodies (Santa Cruz Biotech). AnnexinV-fluorescein isothiocyanate (annexinV-FITC) detection kit (Biosource-Invitrogen, Camarillo, CA) was used to measure apoptosis.
Primary CLL/SLL cells
Following informed consent, peripheral blood was drawn from four patients with CLL/SLL. Patient #1 was a 78 year-old man with newly diagnosed CLL/SLL (absolute lymphocyte count (ALC) 95.2 K/uL, hemoglobin 10.4 g/dL, no thrombocytopenia, presence of bulky (>7.5cm) lymphadenopathy, while FISH studies showed trisomy 12 in 48% of nuclei and a 13q deletion of both chromosomes 13 in 92% of nuclei). Patients #2 and #3 were 46- and 68-year-old men with newly diagnosed CLL/SLL both with 11q deletion and both with ALC of >75.0 K/uL without anemia or thrombocytopenia. Patient #4 was a 61-year-old female with relapsed CLL/SLL with 17p deletion with ALC of 122.0 K/uL and platelets of 20.0 K/uL. All peripheral blood was diluted 1:1 with PBS (Ca2+ and Mg2+ free) and was layered on top of Ficoll-Paque Plus (Sigma). Samples were then centrifuged at 150 × g for 20 min at room temperature; the buffy coat layer was removed and centrifuged again. Isolated peripheral blood mononuclear cells (PBMC) were then re-suspended in RPMI + 1% fetal bovine serum to 1.5 × 106 cells/mL.
Quantification of apoptosis
All cell lines and CLL/SLL cells were incubated with PCI-24781 and /or bortezomib for 24-48 hours. Cell viability was examined morphologically after staining with trypan blue and by analysis of apoptosis using fluorescence activated cell sorting (FACS) after staining with annexinV-FITC and propidium iodide (PI). In brief, after treatment, 1×106 cells were washed with phosphate buffered saline (PBS) and then labeled with annexinV-FITC /PI in the binding buffer according to manufacturer’s protocol. Fluorescent signals of FITC and PI were detected at 518nm and 620nm, respectively, on a Beckman Coulter FACS instrument (Fullerton, CA). The data were analyzed with Flow Jo software (Tree Star, Ashland, OR). For each analysis 20,000 events were recorded.
Measurement of ROS
Intracellular ROS concentration was determined by using cell permeable dyes as described previously (21, 22). In brief, cells were washed with PBS and re-suspended in 1ml of RPMI containing 5μM H2DCF-DA and incubated at 37°C for 30 minutes in the dark. ROS were measured by oxidation of H2DCFDA to DCF. Fluorescence intensity was read by flow cytometry on the FL1 channel.
Western blot analysis
Cells were centrifuged, washed with cold PBS, and lysed on ice for 30 minutes in lysis buffer containing protease and phosphatase inhibitors. Protein concentrations were determined with the Bio-Rad protein assay kit (Bio-Rad, Hercules,CA). Total protein (50μg) was electrophoresed on 12% SDS polyacrylamide gels and bands were visualized by chemiluminescence (Amersham Biosciences, Buckinghamshire, United Kingdom).
Measurement of mitochondrial membrane potential (MMP)
MMP was measured by flow cytometry using JC-1 staining. Cells were washed with Hank’s buffered salt solution (HBBS) and incubated with 4 μg/ml JC-1 dye in HBSS for 15 minutes at 37°C in an incubator. Cells were washed with HBSS and immediately subjected to flow cytometric analysis.
Cell cycle analysis
Distinct phases of the cell cycle were distinguished by PI flow cytometry. Cells were washed in ice cold PBS, fixed in 70% ethanol, and stained for 30 minutes at 37°C with PI (50μg/ml PI in hypotonic sodium citrate solution containing 50μg/ml RNase) followed by flow cytometric analysis. The percentage of cells in G1, S, and G2/M phases were determined using the cell cycle analysis program Modfit LT (Verity Software, Topsham, ME).
Protein extracts and electrophoretic mobility shift assay (EMSA)
EMSA was performed using a gel-shift kit from Panomics (Fremont, CA). In brief, cellular extracts were prepared as described earlier and protein concentrations were determined using Bio-Rad’s protein assay reagent. The cellular extracts were then incubated with a biotin labeled NF-kB probe for 30 minutes at 15°C. The extracts were electrophoresed on a 6% polyacrylamide gel and transferred to a Hybond nylon membrane (Pall Corporation, Ann Arbor, MI). The membrane was blocked for 15 minutes in blocking buffer followed by 15 min incubation with streptavidin-HRP antibody in the blocking buffer. Membranes were washed three times, developed using the detection kit and visualized using hyperfilm ECL (Amersham Biosciences, Buckinghamshire, United Kingdom).
RT-PCR analysis
Taqman Gene Expression Assays for selected genes were obtained from Applied Biosystems Inc. (Foster City, CA). One-step RT-PCR was carried out in triplicate on 25ng of total RNA from each sample on an ABI PRISM 7300 instrument according to the manufacturer’s standard protocols. The mRNA levels for each gene were normalized to the amount of RNA in the well as measured in parallel using Ribogreen (Molecular Probes, Eugene, OR). The treated samples were then normalized to the vehicle control at that time point.
Gene expression profiling
The RNA expression profile was analyzed on custom Codelink oligonucleotide arrays (GE Biosciences, Piscataway, NJ), each containing 1857 gene probes, representing cellular cancer-related pathways, selected from the Codelink Human Genome arrays used in previous work (11, 23). cRNA probes were prepared from the total RNA isolated from treated and control cells, and hybridized to the arrays using standard protocols (Codelink Protocol v2.1). Arrays were hybridized for 18 hours at 37°C, washed and detected with Strepatavidin-Alexa 647. They were scanned with a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA) and the images were processed with Codelink 4.0 Batch Processing software. The data were then analyzed in Genespring (Agilent Inc., Santa Clara, CA); only genes passing quality filters (“G”) and p-value cutoff of 0.05 were used in the analyses.
Statistics
For all apoptosis experiments (annexinV/PI and MMP), values represent the mean from three independent studies done in triplicate. Differences in groups were assessed by student t test and were considered statistically significant at p<0.05, <0.01, and <0.001. For the experiments combining bortezomib and PCI-24781, synergy was determined using isobologram analysis based on the method of Chou and Talay using the Calcusyn (Biosoft, Ferguson,MO) software program (24). This method is based on the equation: CI=(D)1/(Dx)1+(D)2/(Dx)2, where D1 and D2 are concentrations of drug 1 and drug 2 that have x effect when used in combination, and (Dx)1 and (Dx)2 are the concentrations of drug 1 and drug 2 that have the same x effect when used alone.
Results
PCI-24781 time- and concentration-dependent apoptosis in HL and NHL cell lines
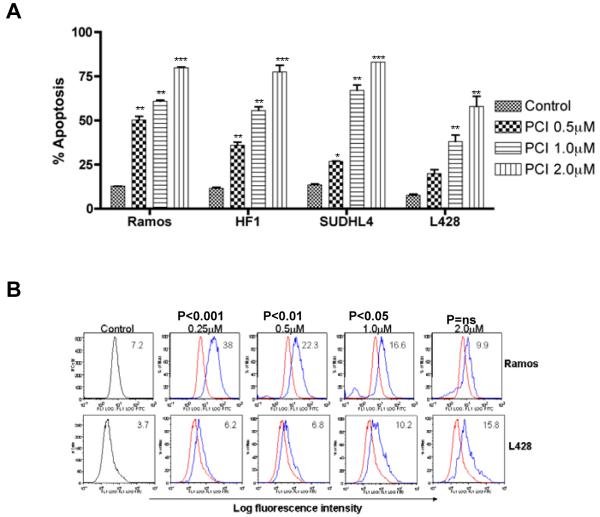
The four lymphoma cell lines were exposed to increasing concentrations (0.5μM to 2μM) of PCI-24781 for 48 hours. PCI-24781 induced apoptosis in all cell lines in a concentration-dependent manner (Figure 1A). The IC50 of PCI-24781 was 0.5μM for Ramos, 0.8μM for SUDHL4, 0.9μM for HF1, and 1.4μM for L428. Apoptosis was also time-dependent, with increasing cell death from 24 through 72 hours (data not shown). Several reports have indicated that the activity of HDACi occurs through production of ROS (5, 7-9). A four-fold increase in ROS was seen here in Ramos and L428 cells following 24 hour exposure of PCI-24781 (Figure 1B). Similar ROS production was also demonstrated in SUDHL4 and HF1 cells (data not shown).
Figure 1. PCI-24781 concentration-dependent apoptosis in lymphoma cell lines.
A) Ramos, HF1, SUDHL4, and L428 lymphoma cell lines were cultured with increasing concentrations (0.5μM to 2.0μM) of PCI-24781 for 48 hours. Percentage apoptosis was measured by annexinV/propidium iodide staining and analyzed by flow cytometry. B) Ramos and L428 cell lines were cultured with indicated concentrations of PCI-24781 for 24 hours and ROS was measured. Peak shift to right denotes an increase in ROS production. Red line: no PCI-24781, blue line: treated with PCI-24781. A shift towards the right indicates more ROS. *P<0.05, **P<0.01, and ***P<0.001 compared with control (RPMI). Abbreviation: PCI, PCI-24781.
Bortezomib concentration-dependent apoptosis
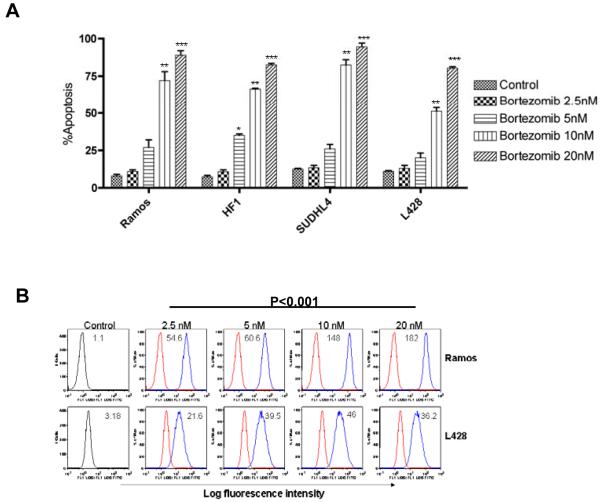
Concentration-dependent apoptosis was seen in all lymphoma cell lines following 48-hour exposure with increasing concentrations of bortezomib (Figure 2A). The IC50 for bortezomib was 20nM for L428 and 10nM for all three NHL cell lines. We next investigated whether apoptosis induced by bortezomib was associated with ROS production. As shown in Figure 2B, treatment of cells with bortezomib resulted in over 10-fold increase in ROS in a concentration-dependent manner in Ramos and L428 cells.
Figure 2. Bortezomib in lymphoma cell lines.
A) Bortezomib induced concentration-dependent apoptosis in Ramos, HF1 SUDHL-4, and L-428. Cells were treated with indicated increasing concentrations of bortezomib, and at 48 hours, the percentage of apoptotic cells was determined by annexinV/propidium iodide staining and measured by flow cytometry. B) ROS levels were measured by flow cytometry in Ramos and L428 cells following incubation of cells with bortezomib for 6 hours followed by staining with H2DCF-DA. Red line: without treatment; blue line: treated with bortezomib. *P<0.05 and **P<0.01 compared with control (RPMI). A shift towards the right indicates more ROS.
PCI-24781 combined with bortezomib
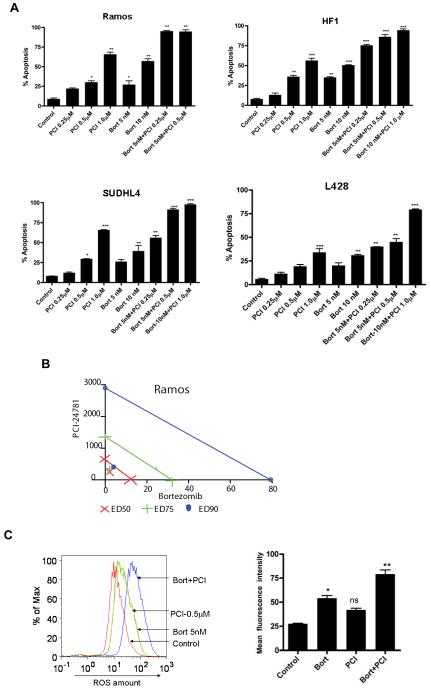
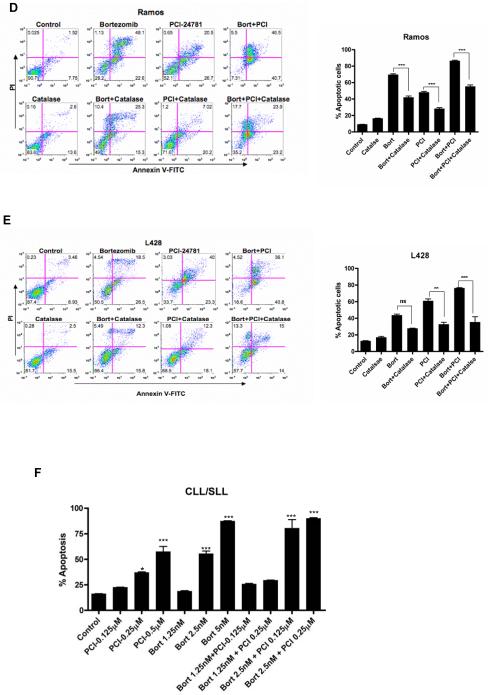
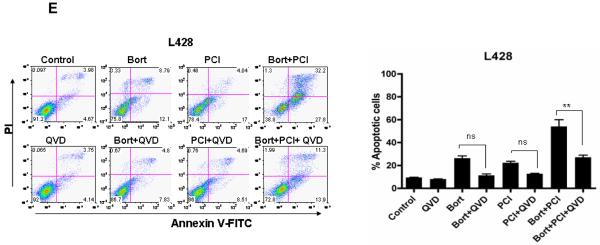
At 48 hours, all cell lines exhibited a significant increase in apoptosis with the combined PCI-24781/bortezomib as shown in Figure 3A. Combined treatment with 0.5μM PCI-24781 and 5nM bortezomib resulted in synergistic apoptosis in all three NHL cell lines, while the effects were additive or synergistic depending on concentrations of the drugs used in the L428 HL cell line (Figure 3A). As shown by isobologram analyses (Figure 3C), Ramos cells displayed stronger synergy (CI= 0.19 for 0.5μM PCI-24781 and 5nM bortezomib) compared with other cell lines (CI= 0.3 to 0.6). In L428 cells, combination index values indicated synergy with 10nM bortezomib and 1μM PCI-24781 (CI=0.6), while 5nM bortezomib and 0.5μM PCI-24781 was additive (CI of 1.1; data not shown). An increase in ROS was also observed with the combination of PCI-24781/bortezomib in Ramos as shown in Figure 3C. Cells were co-incubated with catalase, a free radical scavenger that degrades hydrogen peroxide. In Ramos (Figure 3D) and L428 (Figure 3E), apoptosis induced by PCI-24781, bortezomib, and PCI-24781/bortezomib combination were all blocked in the presence of catalase, suggesting that the effects on apoptosis are in part ROS-mediated (Figure 3D and 3E). A similar effect of abrogated apoptosis by catalase was observed in HF1 and SUDHL4 cells (Figure S1). Primary CLL/SLL cells were exposed to increasing concentrations (0.125μM to 2.0μM) of PCI-24781 for 48 hours. PCI-24781 induced concentration-dependent apoptosis with an associated IC50 of 0.5μM in CLL/SLL primary cells. Bortezomib alone also induced apoptosis at 5nM, while the combination of PCI-24781 and bortezomib resulted in additive cell death (Figure 3F).
Figure 3. Combined PCI-24781 with bortezomib induced synergistic apoptosis that was ROS-dependent.
A) Co-treatment with bortezomib enhanced PCI-24781-related apoptosis. All cell lines were treated with varying concentrations of PCI-24781 (0.25μM to 1.0μM) and bortezomib (5nM to 10nM) either alone or in combination as indicated for 48 hours. Percentage of apoptotic cells was measured by flow cytometry after Annexin-V/propidium iodide (PI) staining. B) Isobolograms of the Ramos cell line treated with both PCI-24781 and bortezomib. Data points (blue circle, green plus, red x) indicate specific PCI-24781 and bortezomib concentrations at which apoptosis was 50%, 75% or 90% as indicated, after 48 hours. Lines connecting cytotoxic IC50, IC75, or IC90 for each drug signify a linear relationship between the drugs. Data points below each respective straight colored line indicate synergistic cytotoxicity. C) ROS production in Ramos. Ramos cells were treated with the indicated concentrations of bortezomib and PCI-24781 for 16 hours followed by staining with H2DCF-DA analyzed by flow cytometry. Red line: no drug treatment, blue line: with bortezomib, green line: with PCI-24781, and yellow line: bortezomib/PCI-24781 combined. Apoptosis in Ramos (D) and L-428 (E) was ROS-dependent. Catalase inhibited bortezomib and PCI-24781-induced apoptosis in Ramos and L428. Cell lines were treated with 4000 units of catalase for 2 hours following incubation with the 10nM bortezomib or 0.5μM (Ramos)/1.0μM (L428) PCI-24781 or combination bortezomib/PCI (Ramos 5nM bortezomib + 0.5μM PCI and L428 with 10nM bortezomib + 1μM PCI) for 48 hours. F) Apoptosis in primary CLL/SLL cells. For all experiments, the percentage of apoptotic cells was determined by annexinV/PI staining followed by flow cytometric analysis. *P<0.05, **P<0.01, and ***P<0.001. Unless designated, P values for single-agents reflect bortezomib or PCI-24781 vs control; P values for combinations reflect bortezomib/PCI-24781 vs matching single-agent concentrations.
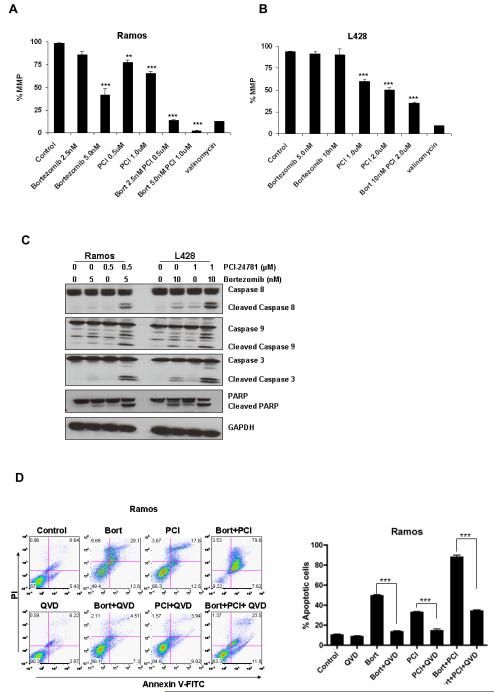
PCI-24781/bortezomib-induced apoptosis is associated with early mitochondrial events (ΔΨm), caspase activation, and PARP cleavage
Mitochondria play a crucial role in the regulation of programmed cell death (25). The release of proteins from the inter-membrane space of mitochondria is a pivotal event in the initiation of the intrinsic cascade of apoptosis (26). Ramos cells showed 60% loss of MMP (ΔΨm) with 5nM bortezomib and <20% with 2.5nM (Figure 4A), while PCI-24781 alone showed 25%-30% ΔΨm (0.5μM-1μM). However, the combination of bortezomib (2.5nM) and PCI-24781 (0.5μM) resulted in over 80% ΔΨm (P<0.01 combination vs single agents). L428 cells showed minimal ΔΨm following bortezomib treatment, while 50%-60% ΔΨm was observed with PCI-24781 alone (Figure 4B). Higher concentrations of PCI-24781 alone were needed in L428 compared with Ramos, while the combination resulted in over 75% ΔΨm.. Similar loss of MMP after treatment of cells with bortezomib and PCI-24781 alone or with the combination was also observed in HF1 and SUDHL4 cells (Figure S2).
Figure 4. ΔΨm and caspase-dependent apoptosis with PCI-24781 and/or bortezomib.
A) Ramos and B) L428 cells were treated with indicated concentrations of bortezomib and PCI-24781 for 24 hours. The percentage of cells exhibiting loss of mitochondrial membrane potential (ΔΨm) was determined by JC-1 staining followed by flow cytometric analysis. C) Western blot analysis of cleaved caspases 3, 9, 8, and PARP activation in Ramos and L428 cells. Cells were treated with the indicated concentrations of bortezomib or PCI-24781 for 24 hours. D) The pan-caspase inhibitor, Q-VD-OPh, inhibited bortezomib/PCI-24781-induced apoptosis in Ramos and E) L428 cells. Ramos and L428 cells were treated with either 5 nM (Ramos) and 10 nM (L428) bortezomib or 0.5μM (Ramos)/1.0μM PCI or combined bortezomib/PCI-24781 (5nM bortezomib and 0.5μM PCI-24781 in Ramos and 10nM bortezomib and 1μM PCI-24781 in L428). For 48 hours alone (control) or with 4-hour pretreatment with 50μM Q-VD-OPh. Apoptotic cells were detected by annexinV/propidium iodide staining and measured by flow cytometry. *P<0.05, **P<0.01, and ***P<0.001. Unless designated, P values for single-agents reflect bortezomib or PCI-24781 vs control; P values for combinations reflect bortezomib/PCI-24781 vs matching single-agent concentrations. PCI, PCI-24781; Bort, bortezomib. MMP, mitochondrial membrane potential; ns, not significant; QVD, Q-VD-OPh.
The involvement of caspases in PCI-24781 and bortezomib-induced apoptosis was assessed by assessment of cleaved caspases and PARP by western blotting. As shown in Figure 4C, both agents induced caspase 8 and 9 cleavage when used alone, while the combination of bortezomib and PCI-24781 resulted in markedly increased cleaved caspase 8 and caspase 9 compared with either agent alone (Figure 4C). Cleavage of caspase 3 and PARP was also observed following treatment of cells with bortezomib or PCI-24781. Activation of caspases and PARP was also observed in HF1 and SUDHL4 cells (Figure S3) following treatment with bortezomib and PCI combination. To assess the importance of caspase activation in bortezomib and/or PCI-24781-induced cell death, cells were co-incubated with the broad spectrum caspase inhibitor, Q-VD-OPh. Figure 4D shows that PCI-24781/bortezomib-indcued cell death in L428 and Ramos cells was in part caspase-dependent. Inhibition of apoptosis with pan caspase inhibitor was also observed in HF1 and SUDHL4 cells (Figure S4).
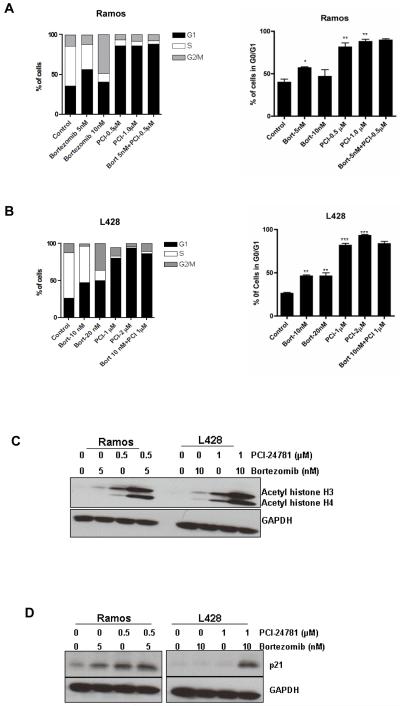
Apoptosis is associated with cell cycle arrest and p21 upregulation
Concentration-dependent G2/M arrest occurred following treatment of Ramos and L428 cells with bortezomib (Figures 5A and 5B) that was accompanied by a decreased number of cells within the S and G1 phases. Treatment with PCI-24781 resulted in G0/G1 arrest with a decrease in G2/M and S phase cell population in both Ramos and L428 (Figures 5A and 5B). HF1 and SUDHL4 cells have also shown G0/G1 arrest following treatment of cells with PCI-24781 (Figure S5). The combination of bortezomib and PCI-24781 mimicked was similar to the effects of bortezomib or PCI-24781 alone, although PCI-24781 alone resulted in >75% G0/G1 arrest.. The biologic effects of HDACi are thought to be related in part by modifications of the acetylation state of histones. Hyperacetylation of histone H3 and H4 was observed following PCI-24781 treatment (Figure 5C). Interestingly, bortezomib also provoked a small increase in the acetylation of histone H4, although to a lesser extent. However, the combination of PCI-24781 and bortezomib resulted in a significant increase in histone acetylation (Figure 5C). The promoter of the transcription of the CDK inhibitor for p21 (CDKN1A) is regulated by histone acetylation status,(27) and up-regulation of p21 has been reported with HDAC inhibitors (28, 29). We also observed increased protein levels of p21 with PCI-24781, and more so with the combination (Figure 5D). A significant increase in histone H3 acetylation was observed in HF1 and SUDHL4 cells. p21 was also upregulated in these cells following treatment with the combination of bortezomib and PCI-24781 (Figure S6)
Figure 5. Bortezomib and PCI-24781 cell cycle arrest and histone acetylation.
A) Ramos and B) L428 cells were treated with the indicated concentrations of bortezomib or PCI-24781 and the combination for 24 hours and then stained with annexinV/propidium iodide and their cell cycle profiles were examined. On the right side is the comparison of G1 fraction in all the population. C) Western blot showing histone hyperacetylation and P21 upregualtion (D). Cells were treated with 5nM (Ramos) or 10nM (L428) of bortezomib and 0.5μM (Ramos) or 1.0μM (L428) of PCI-24781 for 16 hours. (D)The level of acetyl histone H3/H4 and p21 protein was measured using antibody as described in the Methods. GAPDH was used as internal control for all Western blots. *P<0.05, **P<0.01, and ***P<0.001. P values for single-agents reflect bortezomib or PCI-24781 vs control. Combination bortezomib/PCI-24781 vs single-agents was not significant. PCI, PCI-24781; Bort, bortezomib.
Gene expression profiling following PCI-24781 and/or bortezomib
Gene expression profiling using the pathway analysis chip revealed a subset of genes whose expression was altered in response to 0.25μM PCI-24781 and/or 3nM bortezomib in Ramos cells. These concentrations were chosen since higher concentrations led to increased cell death at the 24-hour time point. The Codelink oligonucleotide microarray used also included genes in other pathways of interest, including those previously shown to be affected by PCI-24781 treatment (11). Statistical analyses of the data revealed a CV of 11.52% between four replicates, leading to a minimum detectable fold change of 1.3 according to the Codelink Analysis software (data not shown). Selected genes meeting the p-value cutoff of 0.05 are shown in Table 1.
Table 1.
Selected genes from expression analysis following 24-hour treatment with PCI-24781, bortezomib, or the combination (in Ramos cells)*
| Downregulated genes | 0.25uM PCI/3nM Bort | |||
|---|---|---|---|---|
| Accn # | Name | PCI | Bort | Comb |
| Cell cycle related | ||||
| NM_000075 | cyclin-dependent kinase 4 (CDK4) | 0.49 | 0.83 | 0.37 |
| NM_001237 | cyclin A2 (CCNA2) | 0.43 | 0.87 | 0.37 |
| NM_001950 | E2F transcription factor 4, p107/p130-binding (E2F4) | 0.48 | 0.79 | 0.40 |
| NM_001951 | E2F transcription factor 5, p130-binding (E2F5) | 0.46 | 0.98 | 0.43 |
| NM_003903 | CDC16 cell division cycle 16 homolog (S cerevisiae) (CDC16) | 0.61 | 0.78 | 0.43 |
| NM_031966 | cyclin B1 (CCNB1) | 0.55 | 0.90 | 0.43 |
| NM_001760 | cyclin D3 (CCND3) | 0.48 | 1.02 | 0.46 |
| NM_001255 | CDC20 cell division cycle 20 homolog (S cerevisiae) (CDC20) | 0.61 | 0.82 | 0.46 |
| NM_001262 | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) (CDKN2C) | 0.61 | 1.15 | 0.56 |
| NM_001238 | cyclin E1 (CCNE1) | 0.56 | 1.05 | 0.60 |
| NM_001239 | cyclin H (CCNH) | 0.74 | 0.90 | 0.64 |
| NM_004701 | cyclin B2 (CCNB2) | 0.90 | 0.95 | 0.67 |
| NM_001240 | cyclin T1 (CCNT1) | 0.67 | 1.16 | 0.71 |
| NM_001761 | cyclin F (CCNF) | 0.77 | 0.93 | 0.74 |
| NM_001258 | cyclin-dependent kinase 3 (CDK3) | 0.60 | 1.14 | 0.80 |
| NM_004702 | cyclin E2 (CCNE2) | 0.76 | 1.40 | 0.82 |
| NM_001759 | cyclin D2 (CCND2) | 0.85 | 0.87 | 0.86 |
| Proteasome | ||||
| NM_017518 | 26S proteasome-associated UCH interacting protein 1 (UIP1) | 0.36 | 0.79 | 0.29 |
| NM_002811 | proteasome 26S subunit, non-ATPase, 7 (PSMD7) | 0.68 | 1.44 | 0.57 |
| NM_005047 | proteasome 26S subunit, non-ATPase, 5 (PSMD5) | 0.61 | 1.04 | 0.70 |
| NM_002812 | proteasome 26S subunit, non-ATPase, 8 (PSMD8) | 0.60 | 1.13 | 0.70 |
| NM_002794 | proteasome subunit, beta type, 2 (PSMB2) | 0.75 | 1.26 | 0.83 |
| NM_002790 | proteasome subunit, alpha type, 5 (PSMA5) | 0.74 | 1.43 | 0.92 |
| Oxidative Stress | ||||
| NM_006440 | thioredoxin reductase 2 (TXNRD2) | 0.36 | 0.83 | 0.24 |
| NM_145177 | dehydrogenase/reductase (SDR family) X-linked (DHRSX) | 0.37 | 0.87 | 0.40 |
| NM_012473 | thioredoxin 2 (TXN2) | 0.60 | 0.94 | 0.53 |
| NM_145792 | microsomal glutathione S-transferase 1 (MGST1) | 0.66 | 0.83 | 0.54 |
| NM_002134 | heme oxygenase (decycling) 2 (HMOX2) | 0.66 | 0.89 | 0.57 |
| NM_012331 | methionine sulfoxide reductase A (MSRA) | 0.74 | 0.86 | 0.72 |
| NM_000637 | glutathione reductase (GSR) | 0.61 | 1.46 | 0.80 |
| NM_001752 | catalase (CAT) | 0.93 | 0.72 | 0.83 |
| NM_147149 | glutathione S-transferase M4 (GSTM4) | 0.89 | 0.92 | 0.87 |
| NM_182743 | thioredoxin reductase 1 (TXNRD1) | 0.81 | 1.69 | 0.98 |
| HDACs & Histones | ||||
| NM_016596 | histone deacetylase 7A (HDAC7A) | 0.35 | 1.06 | 0.31 |
| NM_004964 | histone deacetylase 1 (HDAC1) | 0.74 | 0.89 | 0.55 |
| NM_018486 | histone deacetylase 8 (HDAC8) | 0.76 | 1.02 | 0.64 |
| NM_001527 | histone deacetylase 2 (HDAC2) | 0.94 | 0.97 | 0.87 |
| NF-kB target genes | ||||
| BC022556 | myc target 1 (MYCT1) | 0.12 | 0.92 | 0.08 |
| NM_001556 | IkB kinase beta (IKBKB) | 0.60 | 1.13 | 0.32 |
| NM_032778 | MYC induced nuclear antigen (MINA) | 0.40 | 0.87 | 0.33 |
| NM_002983 | chemokine (C-C motif) ligand 3 (CCL3) | 0.46 | 0.85 | 0.42 |
| NM_003998 | NFkB p105 (NFKB1) | 0.47 | 1.19 | 0.43 |
| NM_002467 | v-myc viral oncogene homolog (MYC) | 0.80 | 0.95 | 0.67 |
| NM_006273 | chemokine (C-C motif) ligand 7 (CCL7) | 0.51 | 0.61 | 0.69 |
| NM_006509 | Rel B, NFkB subunit (RELB) | 0.32 | 1.27 | 0.72 |
| NM_014002 | IkB kinase epsilon (IKBKE) | 0.59 | 0.88 | 0.76 |
| NM_000600 | interleukin 6 (interferon, beta 2) (IL6) | 0.84 | 1.28 | 0.81 |
| NM_002503 | IkB beta (NFKBIB) | 0.80 | 1.22 | 0.82 |
| BC033522 | Rel A, NFkB p65 subunit | 0.92 | 1.04 | 0.85 |
| Apoptosis | ||||
| NM_001168 | baculoviral IAP repeat-containing 5 (survivin) (BIRC5) | 0.56 | 0.83 | 0.54 |
| NM_003879 | CASP8 and FADD-like apoptosis regulator (CFLAR)(c-FLIP) | 0.60 | 1.03 | 0.56 |
| NM_016252 | baculoviral IAP repeat-containing 6 (apollon) (BIRC6) | 0.75 | 1.10 | 0.72 |
| Upregulated genes | 0.25uM PCI/3nM Bort | |||
|---|---|---|---|---|
| Accn # | Name | PCI | Bort | Comb |
| Apoptosis & TNFR related | ||||
| NM_000177 | gelsolin (GSN) | 6.99 | 1.04 | 7.56 |
| NM_032974 | caspase 10, apoptosis-related cysteine protease (CASP10) | 7.99 | 0.81 | 6.16 |
| NM_006573 | tumor necrosis factor (ligand) superfamily, member 13b (TNFSF13B) | 3.59 | 1.91 | 3.38 |
| NM_016639 | tumor necrosis factor receptor superfamily, member 12A (TNFRSF12A) | 2.02 | 0.79 | 2.44 |
| NM_018647 | tumor necrosis factor receptor superfamily, member 19 (TNFRSF19) | -0.11 | 1.85 | 2.23 |
| NM_003842 | tumor necrosis factor receptor superfamily, member 10b (TNFRSF10B) | 2.61 | 1.15 | 2.22 |
| NM_014417 | BCL2 binding component 3 (BBC3)(PUMA) | 0.95 | 1.69 | 1.85 |
| NM_138621 | BCL2-like 11 (apoptosis facilitator) (BCL2L11) | 1.43 | 0.94 | 1.79 |
| NM_000043 | Fas (TNFRSF6) (FAS) | 1.72 | 1.46 | 1.73 |
| NM_006290 | tumor necrosis factor, alpha-induced protein 3 (TNFAIP3) | 1.35 | 0.95 | 1.69 |
| NM_003300 | TNF receptor-associated factor 3 (TRAF3) | 2.80 | 0.78 | 1.62 |
| NM_000594 | tumor necrosis factor (TNF superfamily, member 2) (TNF) | 0.99 | 0.94 | 1.31 |
| Cell Cycle Inhibitors | ||||
| NM_000389 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) (CDKN1A) | 25.87 | 0.92 | 28.45 |
| NM_000076 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) (CDKN1C) | 6.21 | 0.46 | 16.18 |
| L36844 | p15INK4B CDK inhibitory protein (CDKN2B) | 5.26 | 2.54 | 4.49 |
| NM_004064 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) (CDKN1B) | 4.34 | 2.15 | 3.51 |
| Cytoskeletal | ||||
| NM_178012 | tubulin, beta 2B (TUBB2B) | 14.66 | 0.87 | 16.45 |
| BC056264 | histone 1, H2bg (HIST1H2BF) | 6.42 | 1.17 | 5.65 |
| NM_021052 | histone 1, H2ae (HIST1H2AE) | 2.23 | 0.91 | 3.76 |
| NM_021065 | histone 1, H2ad (HIST1H2AD) | 1.47 | 0.69 | 2.69 |
| NM_003543 | histone 1, H4h (HIST1H4H) | 1.01 | 1.20 | 1.88 |
| NM_001015053 | histone deacetylase 5 (HDAC5) | 1.22 | 1.09 | 1.71 |
| NM_003509 | histone 1, H2ai (HIST1H2AI) | 1.12 | 0.80 | 1.45 |
| NM_003516 | histone 2, H2aa (HIST2H2AA) | 1.13 | 1.00 | 1.41 |
| NM_003528 | histone 2, H2be (HIST2H2BE) | 2.11 | 0.84 | 1.23 |
| Oxidative Stress | ||||
| NM_002738 | protein kinase C, beta 1 (PRKCB1) | 14.00 | 0.86 | 11.85 |
| NM_002133 | heme oxygenase (decycling) 1 (HMOX1) | 6.52 | 1.82 | 7.53 |
| NM_004417 | dual specificity phosphatase 1 (DUSP1) | 3.49 | 0.67 | 5.54 |
| NM_005346 | heat shock 70kDa protein 1B (HSPA1B) | 2.23 | 3.74 | 5.27 |
| Other | ||||
| NM_002206 | integrin, alpha 7 (ITGA7) | 15.21 | 1.47 | 15.01 |
| NM_002228 | v-jun sarcoma virus 17 oncogene homolog (JUN) | 4.06 | 0.94 | 12.14 |
| NM_000584 | interleukin 8 (IL8) | 3.18 | 1.88 | 6.41 |
| NM_000582 | osteopontin, early T-lymphocyte activation 1 (SPP1) | 3.67 | 0.82 | 5.07 |
| NM_004862 | lipopolysaccharide-induced TNF factor (LITAF) | 2.26 | 1.20 | 2.08 |
Changes are expressed as ratios of treated to control cell levels.
Abbreviations: PCI, PCI-24781; Bort, bortezomib; comb, combination of PCI-24781 and bortezomib (0.25μM and 3nM, respectively).
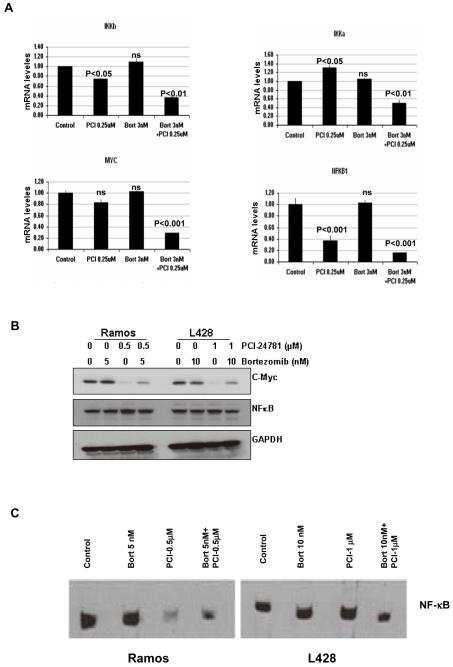
The classical targets of both drugs were affected, including proteasome components and several HDACs (HDACs 1, 2, 7, 8) with bortezomib and PCI-24781 treatment, respectively. In addition, significant downregulation of genes in several pathways including cell cycle, proteasome, oxidative stress, and apoptosis were observed in response to PCI-24781 alone; these effects were enhanced in combination with bortezomib. In particular, it was observed that several anti-oxidant genes were downregulated by PCI-24781 alone and in combination with bortezomib, including thioredoxin-2 and thioredoxin reductase-2, heme oxygenase 2, catalase, glutathione reductase, and several glutathione reductases (Table 1). Some of these pathways have been previously linked to induction of apoptosis by these compounds. A marker for ROS induction, heme oxygenase 1 (HMOX-1), was also increased, but unlike HMOX-2, this gene may serve to facilitate apoptosis. It is likely that transcriptional control of these anti-oxidant genes by PCI-24781 enhances the ROS accumulation and ROS-dependent apoptosis observed in combination with bortezomib.
Interestingly, PCI-24781 appeared to induce downregulation of the proteasome complex and many NF-kB target genes of both the canonical and alternative pathways (i.e., c-Myc, IkB-kinase (IKK)-β, NFkB1, and Rel B) as well as related chemokines and cytokines (including CCL3, CCL7, and IL-6). Several of these genes were further downregulated by PCI-24781/bortezomib combination (c-Myc, myc-regulated genes, and the IKK catalytic subunits) supporting the mechanism of proteasome and NF-kB inhibition for synergy of this combination. Expression of the non-canonical (alternative) pathway components, NF-kB-inducing kinase (NIK) and the NF-kB subunit p52, were not affected by PCI-24781 and/or bortezomib (data not shown). Large increases were also observed in the levels of CDK inhibitors, including p21, consistent with the results before.
Inhibition of NF-kB
We measured the changes in mRNA and protein levels of several NF-kB targets. Accordingly, quantitative RT-PCR analysis of known NF-kB targets including NFkB1 (p105, the precursor of the NF-kB subunit p50), c-Myc, and the two IKK catalytic subunits IKKα (IKK1) and IKKβ (IKK2) were measured. PCI-24781 alone markedly decreased NF-kB1, and to a lesser extent c-Myc and IKKβ in Ramos cells (Figure 6A). A significant decrease in the mRNA levels o NF-kB1, c-Myc and IKK, following exposure to bortezomib or PCI-24781 or the combination was observed in L428 cells (Supplementary Figure S7). Moreover, a notable decrease in all four of these transcripts was seen with PCI-24781/bortezomib in combination. Finally, we analyzed the NF-kB subunit p65 (Rel A) and c-Myc protein levels in response to bortezomib and PCI-24781 alone and in combination, by Western blotting (Figure 6B). NF-kB p65 protein levels did not change significantly, consistent with the gene expression results, whereas c-Myc protein was decreased by PCI-24781 alone and PCI-24781/bortezomib. Similar effect of bortezomib and PCI-24781 was also observed in HF1 and SUDHL4 cells (Supplementary Figure S7). To further determine the effect that combined exposure of bortezomib and PCI-24781 has on NF-kB DNA binding activity, EMSA was performed (Figure 6C). A decrease in NF-kB activity was observed with 10nM-20nM bortezomib and 1μM-2μM PCI-24781 alone and in combination in Ramos and L428 cells. These findings support the concept that NF-kB signaling is a key component in the cell death pathways induced by PCI-24781 alone and in combination with bortezomib.
Figure 6. Effect of PCI-24781 and/or bortezomib on NFκB mRNA and protein and downstream targets.
A) NF-kB1 (p105), c-Myc, IKKα, and IKKβ mRNAs were quantified by RT-PCR. Ramos cells were treated with the indicated concentrations of bortezomib and PCI-24781 for 24 hours. B) Western blot of c-Myc and NF-kB p65 (RelA) protein expression. Ramos and L428 cells were treated with indicated concentrations of bortezomib and of PCI-24781 as single agents and in combination for 24 hours. C) DNA binding activity of NF-kB by electromobility shift assay (EMSA). Ramos and L428 cells were treated with the indicated concentrations of bortezomib and PCI-24781 for 24 hours. Whole cell lysates were analyzed for NF-kB DNA binding activity by EMSA. IKK, IkB-kinase; *P<0.05, **P<0.01, and ***P<0.001. P values for single-agents reflect bortezomib or PCI-24781 vs control; P values for the combination reflects bortezomib/PCI-24781 vs single-agent concentrations (expect for NF-kB1, where P of <0.001 reflects bortezomib/PCI-24781 combination vs bortezomib, while combination vs PCI-24781 P is <0.05). PCI, PCI-24781; Bort, bortezomib.
Discussion
We show that the broad spectrum hydroxamic acid-based HDACi, PCI-24781, induced time- and concentration-dependent apoptosis in a HL cell line, several NHL cell lines, and in primary CLL/SLL cells. PCI-24781 had an IC50 of <1μM in the NHL lines and <1.5μM for L428 cells, both clinically achievable concentrations (10). Apoptosis occurred through ΔΨm, ROS generation, and caspase activation in all cell lines. We observed that PCI-24781 alone induced a four-fold increase in ROS. Furthermore, apoptosis induced by PCI-24781 was ROS-dependent, as cell death was abrogated when cells were pretreated with the anti-oxidant agent, catalase.
We also observed synergistic apoptosis in NHL cells when bortezomib was combined with PCI-24781. Combination studies of novel agents are important, in part to overcome clinical resistance to single-agent therapy in disease subsets where response is more limited, such as with bortezomib for diffuse large B-cell lymphoma (30) or HL (31). Further, this work extends and offers mechanistic insights into the previous work in other tumor types regarding the ROS-dependent synergy between HDACi and bortezomib (16-20). The mode of apoptosis induction by the PCI-24781/bortezomib combination involved activation of both extrinsic (caspase-8) and intrinsic (caspase-9) caspase pathways. Compared to either agent alone, PCI-24781 and bortezomib together led to highly increased levels of cleaved caspase-8, caspase-9, caspase-3, and PARP. The upregulation of several members of the TNF receptor superfamily may lead to the activation of the extrinsic pathway, while the activation of the intrinsic pathway via caspase-9 is consistent with the relatively early ΔΨm that is observed here. Moreover, cell death was caspase-dependent as shown with pan-caspase inhibition, which inhibited apoptosis induced by PCI-24781 alone and combined with bortezomib.
The activation of NF-kB is known to play a critical role in the oncogenesis of lymphoid malignancies (35, 36). Treatment with PCI-24781 alone led to downregulation of several components of the proteasome complex as well as many NF-kB target genes. While the combination of PCI-24781 and bortezomib resulted in further down-regulation of several NF-kB target genes including c-Myc (at protein and mRNA levels), myc-regulated genes, as well as the two catalytic subunits of IKK (IKKα and IKKβ) (Table 1 and Figure 6A). More directly, the DNA-binding activity was also decreased following treatment of cells with these compounds as shown by the gel shift assay. The canonical NF-kB pathway appeared to more dominant here as the PCI-24781/bortezomib combination primarily affected the p65/p50 complex through reduction of IKK activity and p50 expression (and its precursor, NF-kB1), which led to decreased nuclear translocation and decreased binding of NF-kB to its target promoters. HDACi that increase acetylation of proteins require an intact NF-kB signaling pathway to induce cell cycle arrest in human myeloid leukemia cells (37).
We also present here the first reported gene expression profiling data using the combination of an HDACi and bortezomib. A potential explanation of PCI-24781-induced cell death involves direct upregulation of pro-oxidant genes (with resultant NF-kB inhibition and ROS production) and through the effects of direct inhibition of NF-kB and associated changes in anti-oxidant genes. Following PCI-24781 +/- bortezomib, oxidative stress markers were upregulated, while anti-oxidant genes were downregulated. The oxidative stress marker HMOX-1, which was upregulated in this study, can inhibit NF-kB activation by preventing its translocation into the nucleus and inhibiting the degradation of IkB (32, 33); indeed HMOX-1 has been shown to enhance bortezomib-induced cell death in leukemic cells (34). Further, we found that PCI-24781 downregulated the expression of many anti-oxidant genes including TXN2 and TNXRD2. Activation of NF-kB is known to play an important role in the oxidative stress response of tumor cells (38) in part through the regulation of anti-oxidant genes suggesting here that downregulation of thioredoxin-2 and other anti-oxidant genes, inhibition of NF-kB, and induction of ROS could all act together to explain the mechanism of activity of PCI-24781 in lymphoma. It is also interesting that following PCI-24781 treatment, gene expression data showed downregualtion of c-FLIP and the pro-survival BIRC family members including survivin and apollon, which may all prevent cytochrome c release and caspase activation. Further studies are needed to confirm the detailed mechanism of caspase activation in response to PCI-24781 and/or bortezomib in lymphoma.
Finally, PCI-24781 mediated a prominent arrest of the lymphoma cells in the G0/G1 phase of the cell cycle, resulting in a marked decrease in S phase. Blockade of the cell cycle was accompanied by increased expression of p21, a cyclin-dependent kinase (CDK) inhibitor that plays an important role in cell cycle arrest during the G1 or G2 phase (39). Prominent increases were also observed in other CDK inhibitors, including CDKN1B, CDKN1C, and CDKN2B (Table 1). Concordant decreases in many cyclins and CDKs, most prominently CDK4 and cyclin A2, likely contributed to the dramatic increase in G1 arrest and subsequent apoptosis in these lymphoma cell lines. As expected with an HDACi, there was also an accumulation of acetylated histone H3 and H4 with PCI-24781 treatment in these cells, which was synergistically increased by adding bortezomib; however, we and others have shown that histone acetylation is not directly correlated with sensitivity (11, 40). It is likely however, that the enhanced accumulation of p21 is due to increased histone acetylation in these cells (27). Further, histone acetylation (and its surrogate for compounds that inhibit HDAC6, tubulin acetylation) has proved to be an important and sensitive pharmacodynamic marker of HDAC activity in clinical studies, including the PCI-24781 studies currently underway.
In summary, our results show that the pan-HDACi, PCI-24781, when used alone exhibited concentration-dependent apoptosis in a HL cell line, several NHL cell lines, and primary CLL/SLL cells. Furthermore, the combination of PCI-24781 with bortezomib was strongly synergistic in all NHL cell lines. Apoptosis induced by PCI-24781 and by the combination were ROS- and caspase-dependent. Disruption of the cellular redox state with production of ROS and downregulation of oxidative stress response genes may represent an important mechanism underlying lymphoma cell death in response to PCI-24781 +/- bortezomib. We showed that the NF-kB pathway, which is a major regulator of the oxidative stress response in cells, was inhibited by PCI-24781 and further when combined with bortezomib. Finally, we showed that acetylation of histones and re-expression of p21 was greatly enhanced when bortezomib was added to PCI-24781. Thus, the combination of PCI-24781/bortezomib triggered cell death through interacting mechanisms including ROS generation, mitochondrial disruption, proteasome and NF-kB inhibition, cell cycle arrest, and caspase activation. Taken together our data suggest that PCI-24781 has potential therapeutic value in HL and NHL as a single-agent and in combination with bortezomib.
Supplementary Material
Acknowledgements
We thank the members of flow cytometry core facility of the Robert H. Lurie Comprehensive Cancer Center of Northwestern University, Chicago, for assistance with the FACS analysis.
Supported in part from grants from the National Cancer Institute (AME, K23 CA109613-A1).
Footnotes
Statement of translational relevance
Histone deacetylase (HDAC) inhibitors affect genes that result in growth inhibition, differentiation, and apoptosis of cancer cells. The HDAC inhibitor, Zolinza (vorinostat) was recently FDA approved as monotherapy for relapsed/refractory cutaneous T-cell lymphoma. Further pre-clinical study and clinical application of new-generation HDAC inhibitors in other subtypes of lymphoma is warranted. In the present study, we investigated the apoptotic potential and related molecular mechanisms of the new HADC inhibitor, PCI-24781. We showed that PCI-24781, at clinically achievable concentrations, induced concentration-dependent apoptosis in several non-Hodgkin lymphoma cell lines, a Hodgkin lymphoma cell line, and in primary lymphoid leukemia (CLL/SLL) cells. Cell death was ROS- and caspase-dependent and was associated with activation of NF-kB pathways. In addition, synergistic cell death was achieved when PCI-24781 was combined with the proteasome inhibitor, bortezomib. These data support the clinical use of PCI-24781 as a single-agent or combined with bortezomib for the treatment of patients with relapsed/refractory lymphoma.
References
- 1.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 2.Laird PW. Cancer epigenetics. Human molecular genetics. 2005;14(Spec No 1):R65–76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 3.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 4.Rosato RR, Grant S. Histone deacetylase inhibitors: insights into mechanisms of lethality. Expert Opin Ther Targets. 2005;9:809–24. doi: 10.1517/14728222.9.4.809. [DOI] [PubMed] [Google Scholar]
- 5.Ungerstedt JS, Sowa Y, Xu WS, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:673–8. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–52. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 7.Ruefli AA, Ausserlechner MJ, Bernhard D, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10833–8. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W, Ngo L, Perez G, Dokmanovic M, Marks PA. Intrinsic apoptotic and thioredoxin pathways in human prostate cancer cell response to histone deacetylase inhibitor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15540–5. doi: 10.1073/pnas.0607518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosato RR, Almenara JA, Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer research. 2003;63:3637–45. [PubMed] [Google Scholar]
- 10.Adimoolam S, Sirisawad M, Chen J, Thiemann P, Ford JM, Buggy JJ. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proc Natl Acad Sci U S A. 2007;104:19482–7. doi: 10.1073/pnas.0707828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buggy JJ, Cao ZA, Bass KE, et al. CRA-024781: a novel synthetic inhibitor of histone deacetylase enzymes with antitumor activity in vitro and in vivo. Mol Cancer Ther. 2006;5:1309–17. doi: 10.1158/1535-7163.MCT-05-0442. [DOI] [PubMed] [Google Scholar]
- 12.Undevia LJ SD, Schilsky RL, Loury D, Balasubramanian S, Mani C, Sirisawad M, Buggy JJ, Miller RA, Ratain MJ. Phase I study of the safety, pharmacokinetics (PK) and pharmacodynamics (PD) of the histone deacetylase inhibitor (HDACi) PCI-24781. J Clin Oncol. 2008 May 20;(suppl):2008. abstr 14514. [Google Scholar]
- 13.Perez-Galan P, Roue G, Villamor N, Montserrat E, Campo E, Colomer D. The proteasome inhibitor bortezomib induces apoptosis in mantle-cell lymphoma through generation of ROS and Noxa activation independent of p53 status. Blood. 2006;107:257–64. doi: 10.1182/blood-2005-05-2091. [DOI] [PubMed] [Google Scholar]
- 14.Yu C, Rahmani M, Dent P, Grant S. The hierarchical relationship between MAPK signaling and ROS generation in human leukemia cells undergoing apoptosis in response to the proteasome inhibitor Bortezomib. Experimental cell research. 2004;295:555–66. doi: 10.1016/j.yexcr.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Ling YH, Liebes L, Zou Y, Perez-Soler R. Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to Bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. The Journal of biological chemistry. 2003;278:33714–23. doi: 10.1074/jbc.M302559200. [DOI] [PubMed] [Google Scholar]
- 16.Adachi M, Zhang Y, Zhao X, et al. Synergistic effect of histone deacetylase inhibitors FK228 and m-carboxycinnamic acid bis-hydroxamide with proteasome inhibitors PSI and PS-341 against gastrointestinal adenocarcinoma cells. Clin Cancer Res. 2004;10:3853–62. doi: 10.1158/1078-0432.CCR-03-0806. [DOI] [PubMed] [Google Scholar]
- 17.Feng R, Oton A, Mapara MY, Anderson G, Belani C, Lentzsch S. The histone deacetylase inhibitor, PXD101, potentiates bortezomib-induced anti-multiple myeloma effect by induction of oxidative stress and DNA damage. Br J Haematol. 2007;139:385–97. doi: 10.1111/j.1365-2141.2007.06772.x. [DOI] [PubMed] [Google Scholar]
- 18.Heider U, von Metzler I, Kaiser M, et al. Synergistic interaction of the histone deacetylase inhibitor SAHA with the proteasome inhibitor bortezomib in mantle cell lymphoma. European journal of haematology. 2008;80:133–42. doi: 10.1111/j.1600-0609.2007.00995.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller CP, Ban K, Dujka ME, et al. NPI-0052, a novel proteasome inhibitor, induces caspase-8 and ROS-dependent apoptosis alone and in combination with HDAC inhibitors in leukemia cells. Blood. 2007;110:267–77. doi: 10.1182/blood-2006-03-013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai Y, Chen S, Kramer LB, Funk VL, Dent P, Grant S. Interactions between bortezomib and romidepsin and belinostat in chronic lymphocytic leukemia cells. Clin Cancer Res. 2008;14:549–58. doi: 10.1158/1078-0432.CCR-07-1934. [DOI] [PubMed] [Google Scholar]
- 21.Evens AM, Lecane P, Magda D, et al. Motexafin gadolinium generates reactive oxygen species and induces apoptosis in sensitive and highly resistant multiple myeloma cells. Blood. 2005;105:1265–73. doi: 10.1182/blood-2004-03-0964. [DOI] [PubMed] [Google Scholar]
- 22.Chandra J, Tracy J, Loegering D, et al. Adaphostin-induced oxidative stress overcomes BCR/ABL mutation-dependent and -independent imatinib resistance. Blood. 2006;107:2501–6. doi: 10.1182/blood-2005-07-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao ZA, Bass KE, Balasubramanian S, et al. CRA-026440: a potent, broad-spectrum, hydroxamic histone deacetylase inhibitor with antiproliferative and antiangiogenic activity in vitro and in vivo. Molecular cancer therapeutics. 2006;5:1693–701. doi: 10.1158/1535-7163.MCT-06-0042. [DOI] [PubMed] [Google Scholar]
- 24.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 25.Zamzami N, Marchetti P, Castedo M, et al. Inhibitors of permeability transition interfere with the disruption of the mitochondrial transmembrane potential during apoptosis. FEBS letters. 1996;384:53–7. doi: 10.1016/0014-5793(96)00280-3. [DOI] [PubMed] [Google Scholar]
- 26.Henry-Mowatt J, Dive C, Martinou JC, James D. Role of mitochondrial membrane permeabilization in apoptosis and cancer. Oncogene. 2004;23:2850–60. doi: 10.1038/sj.onc.1207534. [DOI] [PubMed] [Google Scholar]
- 27.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10014–9. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blagosklonny MV, Robey R, Sackett DL, et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. olecular cancer therapeutics. 2002;1:937–41. [PubMed] [Google Scholar]
- 29.Chai F, Evdokiou A, Young GP, Zalewski PD. Involvement of p21(Waf1/Cip1) and its cleavage by DEVD-caspase during apoptosis of colorectal cancer cells induced by butyrate. Carcinogenesis. 2000;21:7–14. doi: 10.1093/carcin/21.1.7. [DOI] [PubMed] [Google Scholar]
- 30.Goy A, Younes A, McLaughlin P, et al. Phase II study of proteasome inhibitor bortezomib in relapsed or refractory B-cell non-Hodgkin’s lymphoma. J Clin Oncol. 2005;23:667–75. doi: 10.1200/JCO.2005.03.108. [DOI] [PubMed] [Google Scholar]
- 31.Blum KA, Johnson JL, Niedzwiecki D, Canellos GP, Cheson BD, Bartlett NL. Single agent bortezomib in the treatment of relapsed and refractory Hodgkin lymphoma: cancer and leukemia Group B protocol 50206. Leuk Lymphoma. 2007;48:1313–9. doi: 10.1080/10428190701411458. [DOI] [PubMed] [Google Scholar]
- 32.Oh GS, Pae HO, Lee BS, et al. Hydrogen sulfide inhibits nitric oxide production and nuclear factor-kappaB via heme oxygenase-1 expression in RAW264.7 macrophages stimulated with lipopolysaccharide. Free Radic Biol Med. 2006;41:106–19. doi: 10.1016/j.freeradbiomed.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 33.Chaea HJ, Kim HR, Kang YJ, et al. Heme oxygenase-1 induction by (S)-enantiomer of YS-51 (YS-51S), a synthetic isoquinoline alkaloid, inhibits nitric oxide production and nuclear factor-kappaB translocation in ROS 17/2.8 cells activated with inflammatory stimulants. Int Immunopharmacol. 2007;7:1559–68. doi: 10.1016/j.intimp.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 34.Hamamura RS, Ohyashiki JH, Kurashina R, et al. Induction of heme oxygenase-1 by cobalt protoporphyrin enhances the antitumour effect of bortezomib in adult T-cell leukaemia cells. Br J Cancer. 2007;97:1099–105. doi: 10.1038/sj.bjc.6604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nature reviews. 2002;2:301–10. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 36.Rao PH, Houldsworth J, Dyomina K, et al. Chromosomal and gene amplification in diffuse large B-cell lymphoma. Blood. 1998;92:234–40. [PubMed] [Google Scholar]
- 37.Dai Y, Rahmani M, Grant S. An intact NF-kappaB pathway is required for histone deacetylase inhibitor-induced G1 arrest and maturation in U937 human myeloid leukemia cells. Cell cycle (Georgetown, Tex. 2003;2:467–72. [PubMed] [Google Scholar]
- 38.Bubici C, Papa S, Dean K, Franzoso G. Mutual cross-talk between reactive oxygen species and nuclear factor-kappa B: molecular basis and biological significance. Oncogene. 2006;25:6731–48. doi: 10.1038/sj.onc.1209936. [DOI] [PubMed] [Google Scholar]
- 39.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–15. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 40.Dejligbjerg M, Grauslund M, Christensen IJ, Tjornelund J, Buhl Jensen P, Sehested M. Identification of predictive biomarkers for the histone deacetylase inhibitor belinostat in a panel of human cancer cell lines. Cancer Biomark. 2008;4:101–9. doi: 10.3233/cbm-2008-4206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.