Summary
Increasing evidence points to a heritable contribution to the development of lymphoma. The goal of this study was to determine the rate of familial lymphoproliferative malignancy among consecutive lymphoma patients presenting to a tertiary care center and to enroll families with multiple affected first degree relatives on a data and tissue collection study. Beginning in 2004 all new patients presenting to the Dana-Farber Cancer Institute with non-Hodgkin’s or Hodgkin’s lymphoma or chronic lymphocytic leukemia (CLL) were asked to complete a one-page IRB-approved self-administered family history questionnaire. 55.4% of 1948 evaluable patients reported a 1st degree relative with a malignancy, highest among CLL probands. Lymphoid malignancies were particularly common, with 9.4% of all probands reporting a 1st degree relative with a related LPD. This frequency was again highest for CLL, at 13.3% of CLL probands, compared to 8.8% of NHL probands and 5.9% of HL probands (p=0.002). The prevalence of CLL was significantly increased in parents of CLL probands (p < 0.05), and a greater risk of NHL was seen in fathers of NHL probands than in mothers (p=0.026). We conclude that familial aggregation of lymphoproliferative disorders is common among newly diagnosed patients, varies significantly by diagnosis and contributes meaningfully to the population disease burden.
Keywords: familial lymphoma, familial CLL, heritable, lifetime risk
Introduction
Lymphomas comprise a heterogeneous group of malignancies arising from lymphoid tissues, but with variable clinical features and of unclear cause. The incidence of many lymphomas increased from the 1970s to the mid 1990s but has since leveled off (Groves et al, 2000). Although some risk factors for lymphoma development have been identified, including autoimmune disease (Ekstrom Smedby et al, 2008;Mikuls et al, 2006;Salloum et al, 1996), certain infections like EBV and HIV (Hjalgrim et al, 2003;Kirk et al, 2001), and occupational exposure to pesticides (Garry et al, 1996;Nanni et al, 1996;Schroeder et al, 2001;Waterhouse et al, 1995), the cause in most cases remains obscure. Similarly, although CLL risk has been tentatively associated with exposure to agricultural chemicals (Blair et al, 2007) or a history of pneumonia or sinusitis (Landgren et al, 2007a;Landgren et al, 2007b), most individuals do not have these risk factors.
Several lines of evidence suggest that inherited genetic risk factors contribute to the development of lymphomas. First, several rare genetic immunodeficiency disorders including Wiskott-Aldrich syndrome and ataxia telangiectasia are associated with significantly increased risks of lymphoma (Gatti and Good 1971;Zaizov-Marx et al, 2003). Second, if an individual has developed a non-Hodgkin’s or Hodgkin’s lymphoma, a monozygotic twin of that individual has a greater risk of also developing the disease than a dizygotic twin (Lichtenstein et al, 2000;Verkasalo et al, 1999). In addition, multiple epidemiologic studies have associated a family history of lymphoma with 1.5 -4X relative risks of developing lymphoma, with minor variations by type (Altieri et al, 2005;Chang et al, 2005;Wang et al, 2007). A number of recent association studies have also associated lymphoma risk with heritable polymorphisms in genes involved in immunity and inflammation (Cerhan et al, 2007;Lan et al, 2006;Rothman et al, 2006), DNA repair (Hill et al, 2006;Kerridge et al, 2002;Rudd et al, 2006;Sarmanova et al, 2001) and genes involved in folate metabolism (Lim et al, 2007;Skibola et al, 2004).
The most recent pooled case-control analysis of the InterLymph Consortium confirmed an increased familial risk across the spectrum of lymphomas and found higher risks in particular for those with affected siblings or affected male relatives (Wang et al, 2007). The largest familial risk appears to be associated with CLL, based on an analysis of the Swedish Family Cancer database which found a 7.5X relative risk for CLL in a first degree relative of a CLL patient, as compared to a 1.45X risk for NHL and 2.35X risk for HL (Goldin et al, 2004). In this study no difference in risk was found between parents or siblings or related to sex of the relative (Goldin et al, 2004), although earlier studies in CLL alone had suggested a higher familial risk in females, whether proband cases or relatives (Cannon-Albright et al, 1994;Linet et al, 1989).
Given these findings, significant effort has been devoted to attempting to identify genes or loci involved in the heritability of CLL and lymphoma. A number of small linkage studies were completed initially in CLL (Goldin et al, 2003;Ng et al, 2006), culminating recently in the identification of a region on chromosome 13 (13q21) as a possible site of susceptibility in familial CLL (Ng et al, 2007). Unfortunately, a systematic search for mutations in protein-coding genes in that region found none (Ng et al, 2007). Studying the minimal region of deletion at 13q14 has identified the role of miR15 and miR16 in the pathogenesis of sporadic CLL, but their involvement if any in familial CLL remains to be determined (Calin et al, 2002;Calin et al, 2005). Other recent genome-wide linkage searches have identified different possible susceptibility loci, including 2q21, 6p22 and 18q21 (Sellick et al, 2007;Sellick et al, 2005). Recently a polymorphism in the death-associated protein kinase 1 (DAPK1) gene was associated with familial CLL in a single family, but this allele has not been found again in a larger number of cases (Raval et al, 2007). Thus to date the search for genes involved in a significant percentage of familial lymphoproliferative disease has been disappointing.
Despite this significant research effort focused on possible mechanisms of heritability in lymphoproliferative disorders, the implications for clinical practice of small relative risks identified in epidemiologic studies have been obscure. How often will physicians see patients with LPDs who also have a close relative with the same or another LPD? What should the physician tell a curious patient who wonders about familial associations in lymphoma? In order to help us better understand the answers to these clinically relevant questions, we initiated systematic screening of all patients with lymphoproliferative disorders seen at DFCI, beginning in 2004, in order to determine the clinical significance of familial predisposition and the resulting impact on family members. Since the frequency of other malignancies in first degree relatives of probands with lymphomas was also unknown, we collected data on solid tumors as well.
Patients and Methods
Selection of Patients
Systematic screening of consecutive new patients seen in the Dana-Farber Cancer Institute Lymphoma Program, which includes CLL, began in November 2004 and is ongoing through the present. The data presented here cover the period November 2004 to February 2007. Eligible patients included those with any non-Hodgkin’s lymphoma, Hodgkin’s lymphoma or CLL/SLL. A one page IRB-approved screening questionnaire was mailed to each patient prior to the visit and supplied again at the clinic, to be completed prior to their visit. The questionnaire asked about any history of malignancy in parents, siblings or children, and about any history of lymphoproliferative disorder in any other blood relative. Those patients who reported a lymphoproliferative disorder in a first degree relative were invited to participate in a more detailed study which included a more extensive family history questionnaire, medical history, confirmation of the presence of a pathologically-proven LPD in the proband and relative, and germline and tumor banking. The family history questionnaires employed in this study may be viewed at http://research3.dfci.harvard.edu/jbrown/.
During this period, 2012 patients with lymphoproliferative disorders were scheduled to be seen, of whom 64 were ineligible, due to adoption or unknown status of parental cancers, leading to 1948 evaluable and screened patients. The questionnaire return rate was 71%. For the remaining patients, physician notes were reviewed. Of these 1948, 183 (9.4%; 95% CI 8.1–10.8%) reported a first degree relative (alive or dead) with a LPD. Of that group 62 have been registered to the more detailed tissue banking study. Many have not been registered because the affected relatives are deceased and lack available records or pathology.
This protocol was approved by the Dana-Farber Cancer Institute Institutional Review Board, and informed consent was obtained from all patients enrolled on the more detailed study. Those patients who did not wish to return the screening questionnaire could choose not to, without follow-up from study staff; alternatively patients could return the questionnaire but decline follow-up contact.
Questionnaire Interpretation
Proband diagnoses were confirmed by DFCI physicians and independent hematopathology review at the Brigham and Women’s Hospital. For analysis purposes, subgroups were created that included all NHLs, HL (which included nodular lymphocyte predominant HL), and CLL/SLL. For those patients who reported a WHO or REAL diagnosis in a relative, that diagnosis was recorded. For those who reported Hodgkin’s lymphoma, or non-Hodgkin’s lymphoma not otherwise specified, those diagnoses were noted as such. “Lymphoma not otherwise specified” was taken as non-Hodgkin’s lymphoma for the purposes of the analysis (Glaser et al, 2007). “Leukemia not otherwise specified” was excluded due to the ambiguity with acute leukemias or chronic myelogenous leukemia. “Leukemia not otherwise specified” was noted in 26 first degree relatives, 33 second degree relatives and 12 third degree relatives.
For solid malignancies, common primary malignancies were noted as such. If a single individual was listed as having both a common primary malignancy and a common site of metastasis for that malignancy (for example, lung with bone or liver), they were coded as lung cancer and not considered to have separate bone or liver cancer.
Statistical Analysis
Frequency tables have been analyzed by the Chi-Square test. Unless otherwise noted, the nominal p values given are for the three group comparison (CLL, NHL, HL). 95% confidence intervals are exact binomial confidence intervals. The nominal p values for the comparison of reported solid tumor rates among parents of CLL probands and NHL probands were determined using the Fisher exact test. The p value for the comparison of maternal to paternal rates of NHL was corrected for multiple comparisons using the method of Bonferroni.
Results
Patient Characteristics
The screened population consisted of 62% non-Hodgkin’s lymphoma patients, 23% CLL/SLL patients and 15% Hodgkin’s lymphoma patients, as shown in Table 1. The breakdown of NHL subtypes is also shown in Table 1. The median age of screened NHL patients was 58 (range 16–98), comparable to the median age of screened CLL patients, which was also 58 (range 28–90). Hodgkin’s probands were significantly younger, with a median age of 36 (range 18–85). The NHL and CLL probands are younger than the median age at diagnosis in the SEER population (67 and 72, respectively), possibly reflecting referral bias, although the age of the Hodgkin’s probands is comparable to SEER (38). The age of the probands is significant, because the data from the NHL and CLL groups are directly comparable, whereas the HL group is likely to have fewer evaluable first degree relatives and fewer first degree relatives affected with any cancer, due to the younger age of the probands.
Table I.
Diagnoses of Screened Probands
| Diagnosis | N | % of Total | |
|---|---|---|---|
| CLL/SLL | 452 | 23% | |
| Non-Hodgkin’s Lymphoma | 1210 | 62% | % of NHLs: |
| Follicular | 383 | 19.7% | 32% |
| DLBCL | 292 | 15.0% | 24% |
| Waldenstrom’s | 133 | 6.8% | 11% |
| Marginal zone | 119 | 6.1% | 9.8% |
| T-NHLs | 93 | 4.8% | 7.7% |
| Mantle | 78 | 4.0% | 6.4% |
| Other | 112 | 5.7% | 9.3% |
| Hodgkin’s lymphoma | 286 | 15% | |
| Total | 1948 |
Frequency of Affected Relatives with Malignancy
Of the entire screened population, 55.4% reported a first degree relative with cancer. This frequency differed significantly by diagnosis, with 63.7% (95% CI 59.1–68.2%) of CLL patients reporting at least one affected first degree relative, compared to 56.1% (95% CI 53.3–58.9%) of NHL patients and 39.2% (95% CI 33.5–45.1%) of HL patients (p < 0.0001 for the three group comparison; p = 0.04 for CLL compared to NHL). If 2nd degree relatives with cancer are considered in addition, 65.0% of all probands reported a relative with malignancy: 70.1% (95% CI 65.7–74.3%) of CLL probands, which was significantly greater than NHL probands at 64% (95% CI 61.3–66.8%) and HL probands at 61.2% (95% CI 55.3–66.9%) (p=0.02 for the three group comparison and p=0.02 for CLL compared to NHL). It is important to note that these differences are significant between CLL and NHL, where the probands are of similar age, not just between HL and CLL/NHL.
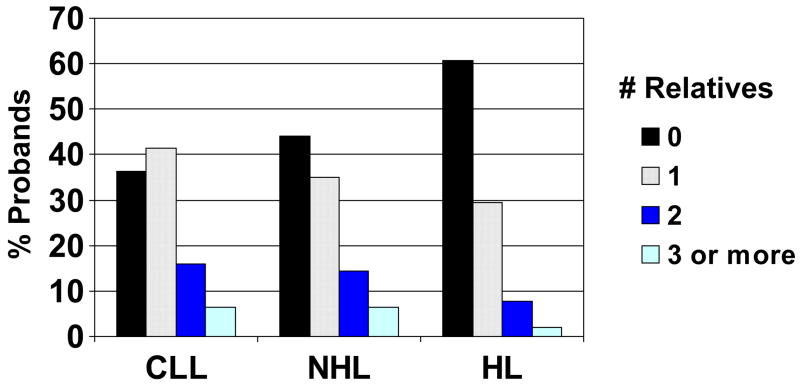
The median number of first degree relatives affected with cancer was one for all groups, among probands reporting any first degree relatives with cancer, but the distribution shifted significantly by diagnosis as shown in Figure 1, with more CLL patients reporting at least one affected relative and fewer HL patients doing so. A total of 382 probands reported 505 siblings with cancer; 21.9% of CLL probands reported a sibling with cancer, as compared to 21.1% of NHL probands and 9.8% of HL probands. This difference is likely driven by the younger age of the HL probands. However, for parents, at least one parent with cancer was reported by 53.1% (95% CI 48.4–57.8%) of CLL patients, 46.2% (95% CI 43.4–49.1%) of NHL patients, and 34.3% of HL patients (p = 0.000003 for the three group comparison). This difference between CLL and NHL patients alone is also statistically significant (p=0.03), indicating a difference not attributable to the age of the probands, which is comparable.
Figure 1.
Number of first-degree relatives with cancer by proband diagnosis (p < 0.0001). Proband type is shown on the x axis.
Frequency of Affected Relatives with Lymphoproliferative Malignancy
Of these 1948, 183 reported a first degree relative (alive or dead) with a LPD. Among all probands 9.4% reported a 1st degree relative with a lymphoproliferative disorder, and 13.6% a 1st or 2nd degree relative with a lymphoproliferative disorder (Table 2). These rates differed significantly by diagnosis, with 13.3% (95% CI 10.3–16.8%) of CLL probands reporting a 1st degree relative with a LPD, 8.8% (95% CI 7.2–10.5%) of NHL probands, and 5.9% (95% CI 3.5–9.3%) of HL probands (p= 0.002). The number of families with two or more affected first degree relatives with LPDs was small, and included 8 with CLL (1.8%), 10 with NHL (0.83%) and none with HL (0%).
Table II.
Lymphoproliferative Malignancy Among 1st and 2nd Degree Relatives
| Any 1st Degree Relative with LPD | Any 1st or 2nd Degree Relative with LPD | |
|---|---|---|
| Percentage of Probands Reporting: | ||
| All | 9.4% | 13.6% |
| By Proband Diagnosis: | ||
| NHL | 8.8% | 12.6% |
| HL | 5.9% | 11.9% |
| CLL | 13.3% | 17.5% |
This difference in frequency of affected relatives by proband diagnosis was true also for the proportion of those with affected 1st or 2nd degree relatives, with 17.5% (95% CI 14.1–21.3%) of CLL probands, 12.6% (95% CI 10.7–14.6%)) of NHL probands and 11.9% (95% CI 8.4–16.2%) of HL probands reporting an affected 1st or 2nd degree relative (p= 0.026). The number of probands with two or more relatives of any type affected with LPDs broke down as follows: CLL (N=19, 4.2%, including 4 with 3 affected relatives (0.9%)); HL (N=4, 1.4%, including 2 with 3 affected relatives (0.7%)); NHL (N=31, 2.5%, including 8 with 3 affected relatives (0.7%)).
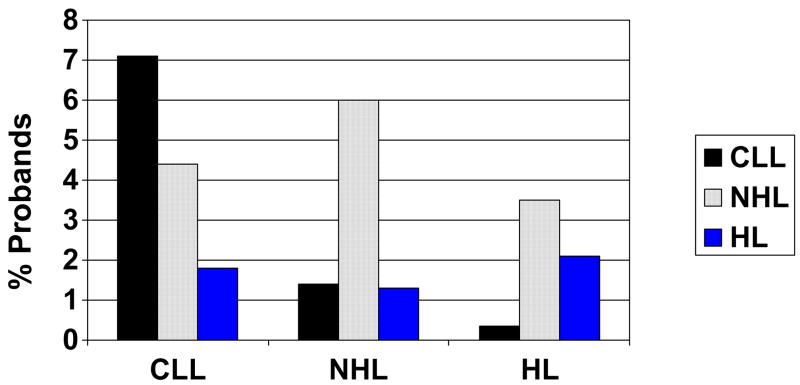
The distribution of lymphoproliferative disorders among 1st degree relatives is shown in Figure 2. Although for both CLL and NHL the same LPD is most commonly seen in the affected first degree relative, nonetheless there are significant rates of other LPDs. For Hodgkin’s lymphoma probands, both HL and NHL are seen at high rates in first degree relatives of the screened population.
Figure 2.
Percentage of probands reporting a given lymphoproliferative disorder among their first degree relatives (p < 0.0001). Proband type is shown on the x axis.
Rates of Malignancy in Parents of Probands
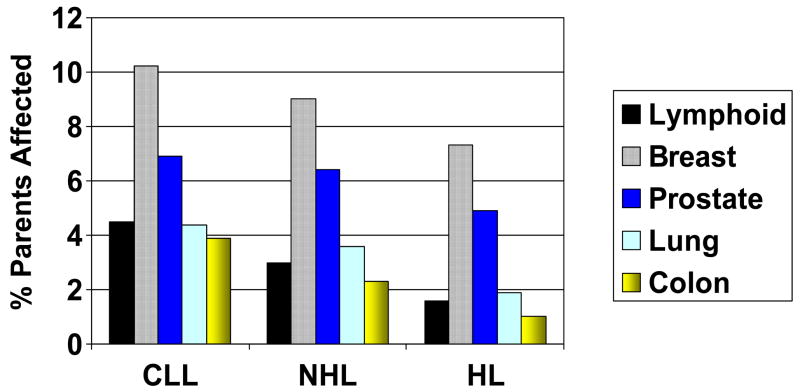
Reported prevalences of common solid tumors and lymphoid malignancies were determined in the parents of probands, as shown in Figure 3. Lymphoid malignancies appear relatively enriched, particularly in CLL probands, whose parents have higher rates of lymphoid malignancies than lung or colon cancer. Among NHL and HL parents, lymphoid malignancies are more commonly reported than colon cancer.
Figure 3.
Reported prevalence of solid and lymphoid malignancy among parents of affected probands, again separated by proband diagnosis (shown on the x axis).
Although these reported prevalences of parental malignancy are not true lifetime estimates, since many of these individuals are still alive and at risk, nonetheless a comparison to US SEER estimated complete prevalence counts as of 1/1/2004 was performed. These results suggested systematic undercounting of solid tumors (breast, colon, lung, prostate; data not shown), likely related to the lack of lifetime data and reduced sensitivity of self-reporting of familial solid tumors among probands with hematopoietic malignancy (Chang et al, 2006). We therefore compared the reported prevalences of these solid tumors in parents of CLL probands to the reported prevalences in parents of NHL probands. Although no significant difference was observed in the rates of breast cancer, prostate cancer or lung cancer, a higher rate of colon cancer was reported in parents of CLL probands (3.9%) than in parents of NHL probands (2.3%) (p = 0.016).
We also calculated the reported prevalences of LPDs in parents of probands with CLL and NHL (Table 3), in which the sensitivity of reporting is better (Chang et al, 2006;Glaser et al, 2007). These data show that the reported prevalences of CLL in parents of CLL probands, even without full lifetime data, are increased compared to SEER lifetime estimates (p < 0.05) and increased compared to parents of NHL probands (p = 0.001 for mothers; 0.007 for fathers). Among parents of NHL probands, the rates are greater for a mother with CLL or a father with NHL, but the confidence intervals do overlap with SEER; the importance of this is unclear given the relatively small number of events and shorter period of observation. A significant difference was observed, however, between the rates of NHL in fathers and mothers of NHL probands, with a greater risk in the fathers (p = 0.026). The numbers of parents with HL in either group was too small to permit adequate analysis.
Table III.
Selected Estimates of LPD Prevalence in Parents of Probands
| Estimated Lifetime Risk in Parents: | CLL Probands DFCI | SEER Lifetime Risk | NHL Probands DFCI |
|---|---|---|---|
| Mother: CLL | 3.1 (1.7–5.1) | 0.36 (.35-.37) | 0.74 (0.3–1.4) |
| Father: CLL | 2.0 (0.9–3.7) | 0.56 (.54-.57) | 0.50 (0.2–1.1) |
| Mother: NHL | 1.1 (0.3–2.6) | 1.87 (1.85–1.9) | 1.24 (0.7–2.0) |
| Father: NHL | 1.55 (0.6–3.2) | 2.19 (2.16–2.22) | 2.81 (2.0–3.9) |
| Mother: HL | 0.66 (0.1–1.9) | 0.2 (0.2–0.21) | 0.33 (0.09–0.8) |
| Father: HL | 0.22 (0.006–1.2) | 0.24 (0.23–0.25) | 0.5 (0.2–1.1) |
Statistically significant findings in this table are underlined.
The rates of CLL reported in both mothers and fathers of CLL probands are statistically greater than the rates reported in SEER (p < 0.05).
The rate of NHL in fathers of NHL probands is greater than the rate of NHL in mothers of NHL probands (p = 0.026).
Confirmation of Reported Diagnoses
Since many of the patients with self-reported familial LPDs were enrolled on our more detailed cohort tissue banking study, we were able to determine the accuracy of reported lymphoma diagnoses. To date, of 284 enrolled probands and affected family members, 229 have pathology reports available at present, and 96% of those reports confirm the reported diagnosis. Of the remaining 10 pathology reports, 9 confirmed lymphoma of a different subtype (3.9% misclassification rate), with only one failing to document lymphoma. Thus the accuracy of reported diagnoses for the probands and their 1st degree relatives was quite high in this patient population.
Discussion
This study systematically evaluates the frequency of familial lymphoproliferative disorders in a consecutive population of affected patients. Our results confirm the clinical experience that led to the initiation of this study, namely that a significant fraction of affected patients have a relative who also has a LPD. Approximately 1 in 6 CLL patients and 1 in 8 lymphoma patients have a 1st or 2nd degree relative with a related lymphoproliferative disorder. Although awareness of familial predisposition is increasingly common in CLL, this finding in lymphoma is not as widely known.
Our findings are consonant with the increased relative risks of LPDs suggested for first degree relatives in prior epidemiologic studies. Since no truly appropriate comparison group exists for our data, it is difficult to correlate our results directly with these relative risk estimates. Similarly, it has been difficult to translate those relative risks into easily understandable clinical expectations, which is a strength of our data.
Substantial evidence suggests that familial aggregation of lymphoproliferative disorders has a significant genetic component. The early literature in these diseases contains reports of families that have virtually Mendelian inheritance of LPDs, and it is well known that a number of single gene immunodeficiency diseases are associated with high rates of lymphomas. The challenge in the case of lymphoproliferative diseases is defining the phenotype(s) that we expect within families. The high rates of any cancer observed in first degree relatives in this study suggest a greater susceptibility to cancer in general. The variation in cancer and LPD rates by diagnosis, greater for CLL than NHL and HL, further suggests that the underlying biological predisposition may vary among the diseases. The differences observed between CLL and NHL indicate that this finding is real and not merely an artifact of different proband ages. At the same time, however, the clear crossover of LPDs among families regardless of proband diagnosis strongly suggests that similar genes may predispose to any of these diseases.
Patterns of inheritance may provide some clues to pathogenesis. It is of interest that we observed a higher rate of NHL in fathers of NHL probands than in mothers; this effect is larger than would be expected based on the relative incidence of NHL in men and women. A similar association was recently reported by the InterLymph Consortium (Wang et al, 2007). In contrast, among parents of CLL probands, the relative increase in risk compared to SEER was greater for mothers than fathers, which may reflect the lower baseline risk for mothers. Similarly, Mauro et al. have reported that a family history of hematologic malignancy is more common among women than men with CLL(Mauro et al, 2006). These phenotypes, including any differences between CLL and NHL, will need to be clarified in family studies, including our ongoing cohort study of affected families, and through enhanced understanding of the underlying biology.
The major possible weakness of our study is its reliance on self-reported family history data. Self-reported data on familial cancer has the potential for both misstating the frequency of disease and misclassifying disease. For example, we observed probable underreporting of common solid tumors in parents compared to SEER frequencies. In a previous case-control study of self-reported family history in lymphoma patients, the sensitivity of reporting of common solid tumors ranged from 50–70% (Chang et al, 2006), which is not dissimilar from our observations. Reports of the accuracy of lymphoma reporting in similar studies has varied but is significantly better; the above study reported a sensitivity of 85% with 89% specificity for LPDs. A separate study of familial Hodgkin’s lymphoma reported 100% validation of affected first degree relatives, but dropped to 81% if more distant relatives were included (Glaser et al, 2007). In our study, the accuracy of reported diagnoses was quite high, with all but one enrolled individual documented to have lymphoma, and only 3.9% of those misclassified by type. These data confirm the validity of our reported frequencies.
This study has focused on a selected patient population seen at a tertiary care referral center. Because of that the observed rates of LPDs in this population may not be fully generalizable to community practice. Nonetheless the observation of a consecutive population of lymphoma patients with this high frequency of familial LPDs is significant, and further study of these patients will prove critical to elucidating the genetic basis of familial LPDs. The genes involved may well prove to be those in which polymorphisms have been associated with lymphoma risk: cytokines, DNA repair, folate metabolism. Systematic genomics approaches applied to large numbers of families are most likely to clarify this question in the coming years.
Acknowledgments
This work was supported in part by NIH grants K23 CA115682 to JRB and the Okonow-Lipton Fund. ASF is supported in part by NIH grant 2P01CA092625. The authors are indebted to Donnette Bailey and Sharinda Roper, the New Patient Coordinators of the Lymphoma Program, and to the staff of the Dana 1 hematologic malignancies clinic, for their assistance with the distribution and collection of the family history questionnaires. We also wish to thank Elke Raderschall PhD for her assistance with sample processing on the more detailed cohort study.
Footnotes
This study has been presented in part at the Annual Meeting of the American Society of Hematology in December 2006 and at the IW-CLL Meeting in September 2007.
References
- Altieri A, Bermejo JL, Hemminki K. Familial risk for non-Hodgkin lymphoma and other lymphoproliferative malignancies by histopathologic subtype: the Swedish Family-Cancer Database. Blood. 2005;106:668–672. doi: 10.1182/blood-2005-01-0140. [DOI] [PubMed] [Google Scholar]
- Blair A, Purdue MP, Weisenburger DD, Baris D. Chemical exposures and risk of chronic lymphocytic leukaemia. British Journal of Haematology. 2007;139:753–761. doi: 10.1111/j.1365-2141.2007.06874.x. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. New England Journal of Medicine. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Cannon-Albright LA, Thomas A, Goldgar DE, Gholami K, Rowe K, Jacobsen M, McWhorter WP, Skolnick MH. Familiality of cancer in Utah. Cancer Research. 1994;54:2378–2385. [PubMed] [Google Scholar]
- Cerhan JR, Ansell SM, Fredericksen ZS, Kay NE, Liebow M, Call TG, Dogan A, Cunningham JM, Wang AH, Liu-Mares W, Macon WR, Jelinek D, Witzig TE, Habermann TM, Slager SL. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110:4455–4463. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang ET, Smedby KE, Hjalgrim H, Glimelius B, Adami HO. Reliability of self-reported family history of cancer in a large case-control study of lymphoma. Journal of the National Cancer Institute. 2006;98:61–68. doi: 10.1093/jnci/djj005. [DOI] [PubMed] [Google Scholar]
- Chang ET, Smedby KE, Hjalgrim H, Porwit-MacDonald A, Roos G, Glimelius B, Adami HO. Family history of hematopoietic malignancy and risk of lymphoma. Journal of the National Cancer Institute. 2005;97:1466–1474. doi: 10.1093/jnci/dji293. [DOI] [PubMed] [Google Scholar]
- Ekstrom Smedby K, Vajdic CM, Falster M, Engels EA, Martinez-Maza O, Turner J, Hjalgrim H, Vineis P, Seniori Costantini A, Bracci PM, Holly EA, Willett E, Spinelli JJ, La Vecchia C, Zheng T, Becker N, De Sanjose S, Chiu BC, Dal Maso L, Cocco P, Maynadie M, Foretova L, Staines A, Brennan P, Davis S, Severson R, Cerhan JR, Breen EC, Birmann B, Grulich AE, Cozen W. Autoimmune disorders and risk of non-Hodgkin lymphoma subtypes: a pooled analysis within the InterLymph Consortium. Blood. 2008;111:4029–38. doi: 10.1182/blood-2007-10-119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry VF, Tarone RE, Long L, Griffith J, Kelly JT, Burroughs B. Pesticide Appliers with Mixed Pesticide Exposure: G-Banded Analysis and Possible Relationship to Non-Hodgkin’s Lymphoma. Cancer Epidemiology, Biomarkers & Prevention. 1996;5:11–16. [PubMed] [Google Scholar]
- Gatti RA, Good RA. Occurrence of Malignancy in Immunodeficiency Diseases. Cancer. 1971;28:89–98. doi: 10.1002/1097-0142(197107)28:1<89::aid-cncr2820280117>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Glaser SL, Chang ET, Horning SJ, Clarke CA. Understanding the validity of self-reported positive family history of lymphoma in extended families to facilitate genetic epidemiology and clinical practice. Leukemia and Lymphoma. 2007;48:1110–1118. doi: 10.1080/10428190701302434. [DOI] [PubMed] [Google Scholar]
- Goldin LR, Ishibe N, Sgambati M, Marti GE, Fontaine L, Lee MP, Kelley JM, Scherpbier T, Buetow KH, Caporaso NE. A genome scan of 18 families with chronic lymphocytic leukaemia. British Journal of Haematology. 2003;121:866–873. doi: 10.1046/j.1365-2141.2003.04372.x. [DOI] [PubMed] [Google Scholar]
- Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104:1850–1854. doi: 10.1182/blood-2004-01-0341. [DOI] [PubMed] [Google Scholar]
- Groves FD, Linet MS, Travis LB, Devesa SS. Cancer Surveillance Series: Non-Hodgkin’s Lymphoma Incidence by Histologic Subtype in the United States From 1978 Through 1995. Journal of the National Cancer Institute. 2000;92:1240–1251. doi: 10.1093/jnci/92.15.1240. [DOI] [PubMed] [Google Scholar]
- Hill DA, Wang SS, Cerhan JR, Davis S, Cozen W, Severson RK, Hartge P, Wacholder S, Yeager M, Chanock SJ, Rothman N. Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood. 2006;108:3161–3167. doi: 10.1182/blood-2005-01-026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjalgrim H, Askling J, Rostgaard K, Hamilton-Dutoit S, Frisch M, Zhang JS, Madsen M, Rosdahl N, Konradsen HB, Storm HH, Melbye M. Characteristics of Hodgkin’s Lymphoma after Infectious Mononucleosis. The New England Journal of Medicine. 2003;349:1324–1332. doi: 10.1056/NEJMoa023141. [DOI] [PubMed] [Google Scholar]
- Kerridge I, Lincz L, Scorgie F, Hickey D, Granter N, Spencer A. Association between Xenobiotic Gene Polymorphisms and non-Hodgkin’s Lymphoma Risk. British Journal of Hematology. 2002;118:477–481. doi: 10.1046/j.1365-2141.2002.03606.x. [DOI] [PubMed] [Google Scholar]
- Kirk O, Pedersen C, Cozzi-Lepri A, Antunes F, Miller V, Gatell JM, Katlama C, Lazzarin A, Skinhoj P, Barton SE. Non-Hodgkin lymphoma in HIV-infected patients in the era of highly active antiretroviral therapy. Blood. 2001;98:3406–3412. doi: 10.1182/blood.v98.12.3406. [DOI] [PubMed] [Google Scholar]
- Lan Q, Zheng T, Rothman N, Zhang Y, Wang SS, Shen M, Berndt SI, Zahm SH, Holford TR, Leaderer B, Yeager M, Welch R, Boyle P, Zhang B, Zou K, Zhu Y, Chanock S. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–4108. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgren O, Gridley G, Check D, Caporaso NE, Morris Brown L. Acquired immune-related and inflammatory conditions and subsequent chronic lymphocytic leukaemia. British Journal of Haematology. 2007a;139:791–798. doi: 10.1111/j.1365-2141.2007.06859.x. [DOI] [PubMed] [Google Scholar]
- Landgren O, Rapkin JS, Caporaso NE, Mellemkjaer L, Gridley G, Goldin LR, Engels EA. Respiratory tract infections and subsequent risk of chronic lymphocytic leukemia. Blood. 2007b;109:2198–2201. doi: 10.1182/blood-2006-08-044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and Heritable Factors in the Causation of Cancer. The New England Journal of Medicine. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Lim U, Wang SS, Hartge P, Cozen W, Kelemen LE, Chanock S, Davis S, Blair A, Schenk M, Rothman N, Lan Q. Gene-nutrient interactions among determinants of folate and one-carbon metabolism on the risk of non-Hodgkin lymphoma: NCI-SEER case-control study. Blood. 2007;109:3050–3059. doi: 10.1182/blood-2006-07-034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linet MS, Van Natta ML, Brookmeyer R, Khoury MJ, McCaffrey LD, Humphrey RL, Szklo M. Familial cancer history and chronic lymphocytic leukemia. A case-control study. American Journal of Epidemiology. 1989;130:655–664. doi: 10.1093/oxfordjournals.aje.a115387. [DOI] [PubMed] [Google Scholar]
- Mauro FR, Giammartini E, Gentile M, Sperduti I, Valle V, Pizzuti A, Guarini A, Giannarelli D, Foa R. Clinical features and outcome of familial chronic lymphocytic leukemia. Haematologica. 2006;91:1117–1120. [PubMed] [Google Scholar]
- Mikuls TR, Endo JO, Puumala SE, Aoun PA, Black NA, O’Dell JR, Stoner JA, Boilesen EC, Bast MA, Bergman DA, Ristow KM, Ooi M, Armitage JO, Habermann TM. Prospective study of survival outcomes in Non-Hodgkin’s lymphoma patients with rheumatoid arthritis. Journal of Clinical Oncology. 2006;24:1597–1602. doi: 10.1200/JCO.2005.04.6227. [DOI] [PubMed] [Google Scholar]
- Nanni O, Amadori D, Lugaresi C, Falcini F, Scarpi E, Saragoni A, Buiatti E. Chronic Lymphocytic Leukemias and Non-Hodgkin’s Lymphomas by Histological Type in Farming-Animal Breeding Workers: A Population Case-Control Study Based on A Priori Exposure Matrices. Occupational and Environmental Medicine. 1996;53:652–657. doi: 10.1136/oem.53.10.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D, Marti GE, Fontaine L, Toro JR, Caporaso N, Goldin LR. High-density mapping and follow-up studies on chromosomal regions 1, 3, 6, 12, 13 and 17 in 28 families with chronic lymphocytic leukaemia. British Journal of Haematology. 2006;133:59–61. doi: 10.1111/j.1365-2141.2006.05972.x. [DOI] [PubMed] [Google Scholar]
- Ng D, Toure O, Wei MH, Arthur DC, Abbasi F, Fontaine L, Marti GE, Fraumeni JF, Jr, Goldin LR, Caporaso N, Toro JR. Identification of a novel chromosome region, 13q21.33–q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood. 2007;109:916–925. doi: 10.1182/blood-2006-03-011825. [DOI] [PubMed] [Google Scholar]
- Raval A, Tanner SM, Byrd JC, Angerman EB, Perko JD, Chen SS, Hackanson B, Grever MR, Lucas DM, Matkovic JJ, Lin TS, Kipps TJ, Murray F, Weisenburger D, Sanger W, Lynch J, Watson P, Jansen M, Yoshinaga Y, Rosenquist R, de Jong PJ, Coggill P, Beck S, Lynch H, de la Chapelle A, Plass C. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman N, Skibola CF, Wang SS, Morgan G, Lan Q, Smith MT, Spinelli JJ, Willett E, De Sanjose S, Cocco P, Berndt SI, Brennan P, Brooks-Wilson A, Wacholder S, Becker N, Hartge P, Zheng T, Roman E, Holly EA, Boffetta P, Armstrong B, Cozen W, Linet M, Bosch FX, Ennas MG, Holford TR, Gallagher RP, Rollinson S, Bracci PM, Cerhan JR, Whitby D, Moore PS, Leaderer B, Lai A, Spink C, Davis S, Bosch R, Scarpa A, Zhang Y, Severson RK, Yeager M, Chanock S, Nieters A. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncology. 2006;7:27–38. doi: 10.1016/S1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
- Rudd MF, Sellick GS, Webb EL, Catovsky D, Houlston RS. Variants in the ATM-BRCA2-CHEK2 axis predispose to chronic lymphocytic leukemia. Blood. 2006;108:638–644. doi: 10.1182/blood-2005-12-5022. [DOI] [PubMed] [Google Scholar]
- Salloum E, Cooper DL, Howe G, Lacy J, Tallini G, Crouch J, Schultz M, Murren J. Spontaneous regression of lymphoproliferative disorders in patients treated with methotrexate for rheumatoid arthritis and other rheumatic diseases. Journal of Clinical Oncology. 1996;14:1943–1949. doi: 10.1200/JCO.1996.14.6.1943. [DOI] [PubMed] [Google Scholar]
- Sarmanova J, Benesova K, Gut I, Nedelcheva-Kristensen V, Tynkova L, Soucek P. Genetic Polymorphisms of Biotransformation Enzymes in Patients with Hodgkin’s and non-Hodgkin’s Lymphomas. Human Molecular Genetics. 2001;10:1265–1273. doi: 10.1093/hmg/10.12.1265. [DOI] [PubMed] [Google Scholar]
- Schroeder JC, Olshan AF, Baric R, Dent GA, Weinberg CR, Yount B, Cerhan JR, Lynch CF, Schuman LM, Tolbert PE, Rothman N, Cantor KP, Blair A. Agricultural Risk Factors for t(14;18) Subtypes of Non-Hodgkin’s Lymphoma. Epidemiology. 2001;12:701–709. doi: 10.1097/00001648-200111000-00020. [DOI] [PubMed] [Google Scholar]
- Sellick GS, Goldin LR, Wild RW, Slager SL, Ressenti L, Strom SS, Dyer MJ, Mauro FR, Marti GE, Fuller S, Lyttelton M, Kipps TJ, Keating MJ, Call TG, Catovsky D, Caporaso N, Houlston RS. A high-density SNP genome-wide linkage search of 206 families identifies susceptibility loci for chronic lymphocytic leukemia. Blood. 2007;110:3326–3333. doi: 10.1182/blood-2007-05-091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick GS, Webb EL, Allinson R, Matutes E, Dyer MJ, Jonsson V, Langerak AW, Mauro FR, Fuller S, Wiley J, Lyttelton M, Callea V, Yuille M, Catovsky D, Houlston RS. A high-density SNP genomewide linkage scan for chronic lymphocytic leukemia-susceptibility loci. American Journal of Human Genetics. 2005;77:420–429. doi: 10.1086/444472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola CF, Forrest MS, Coppede F, Agana L, Hubbard A, Smith MT, Bracci PM, Holly EA. Polymorphisms and haplotypes in folate-metabolizing genes and risk of non-Hodgkin lymphoma. Blood. 2004;104:2155–2162. doi: 10.1182/blood-2004-02-0557. [DOI] [PubMed] [Google Scholar]
- Verkasalo PK, Kaprio J, Koskenvuo M, Pukkala E. Genetic Predisposition, Environment and Cancer Incidence: A Nationwide Twin Study in Finland, 1976–1995. International Journal of Cancer. 1999;83:743–749. doi: 10.1002/(sici)1097-0215(19991210)83:6<743::aid-ijc8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Wang SS, Slager SL, Brennan P, Holly EA, De Sanjose S, Bernstein L, Boffetta P, Cerhan JR, Maynadie M, Spinelli JJ, Chiu BC, Cocco PL, Mensah F, Zhang Y, Nieters A, Dal Maso L, Bracci PM, Costantini AS, Vineis P, Severson RK, Roman E, Cozen W, Weisenburger D, Davis S, Franceschi S, La Vecchia C, Foretova L, Becker N, Staines A, Vornanen M, Zheng T, Hartge P. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10 211 cases and 11 905 controls from the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;109:3479–3488. doi: 10.1182/blood-2006-06-031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse D, Carman WJ, Schottenfeld D, Gridley G, McLean S. Cancer Incidence in the Rural Community of Tecumseh, Michigan. Cancer. 1995;77:763–770. doi: 10.1002/cncr.1996.2820770402. [DOI] [PubMed] [Google Scholar]
- Zaizov-Marx R, Liberzon E, Zilberstein J, Stark B, Gavriel H, Avrahami G, Cohen IJ, Goshen Y, Yaniv I, Avigad S. The Role of ATM Gene Molecular Variants in Childhood Sporadic Lymphoma. Blood. 2003;102:897a. [Google Scholar]