Abstract
Mitochondrial DNA (mtDNA) is organized in protein-DNA macrocomplexes called nucleoids. Average nucleoids contain 2–8 mtDNA molecules, which are organized by the histone-like mitochondrial transcription factor A. Besides well-characterized constituents, such as single-stranded binding protein or polymerase γ (Polγ), various other proteins with ill-defined functions have been identified. We report for the first time that mammalian nucleoids contain essential enzymes of an integral antioxidant system. Intact nucleoids were isolated with sucrose density gradients from rat and bovine heart as well as human Jurkat cells. Manganese superoxide dismutase (SOD2) was detected by Western blot in the nucleoid fractions. DNA, mitochondrial glutathione peroxidase (GPx1), and Polγ were coimmunoprecipitated with SOD2 from nucleoid fractions, which suggests that an antioxidant system composed of SOD2 and GPx1 are integral constituents of nucleoids. Interestingly, in cultured bovine endothelial cells the association of SOD2 with mtDNA was absent. Using a sandwich filter-binding assay, direct association of SOD2 by salt-sensitive ionic forces with a chemically synthesized mtDNA fragment was demonstrated. Increasing salt concentrations during nucleoid isolation on sucrose density gradients disrupted the association of SOD2 with mitochondrial nucleoids. Our biochemical data reveal that nucleoids contain an integral antioxidant system that may protect mtDNA from superoxide-induced oxidative damage.—Kienhöfer, J., Häussler, D. J. F., Ruckelshausen, F., Muessig, E., Weber, K., Pimentel, D., Ullrich, V., Bürkle, A., Bachschmid, M. M. Association of mitochondrial antioxidant enzymes with mitochondrial DNA as integral nucleoid constituents.
Keywords: SOD2, nucleoid complex, aging, smooth muscle cell, TFAM, Polγ
Tissues with high metabolic activity, such as the myocardium or brain, rely on a perfectly functioning mitochondrial respiratory chain for energy supply, which is characterized by coupled oxidative phosphorylation and low generation of reactive oxygen species (ROS). Mutations or deletions of the mitochondrial genome (1,2,3,4) or posttranslational oxidative modifications of respiratory chain proteins (5,6,7,8,9) can cause mitochondrial dysfunction (5, 10,11,12,13,14,15,16,17). It is hypothesized that these alterations may increase electron leakage within the respiratory chain, thus enhancing mitochondrial ROS formation (18,19,20,21,22). Consequently ATP production decreases, and ROS interfere with cellular redox regulation (23, 24). One example of this is limiting the bioavailability of nitric oxide (·NO) in the cardiovascular system.
The intron-free vertebrate mitochondrial DNA (mtDNA) (16.5 kbp) encodes 13 proteins of the respiratory chain complexes I, III, and IV as well as the ATP synthase (complex V), 2 structural ribosomal RNAs of mitochondrial ribosomes, and 22 tRNAs. The organization of mtDNA is commonly regarded as a free plasmid-like molecule located in the mitochondrial matrix. However, early reports from the 1960s based on electron microscopy showed that mtDNA is structured as protein-DNA macrocomplexes called nucleoids (25, 26). Earlier studies used cesium salt gradients to isolate mtDNA, which caused disruption of the nucleoids so that their existence remained undiscovered for a long time. Present methods use gentle lysis of isolated mitochondria followed by sequential gradient centrifugation, allowing isolation of intact nucleoids (27). As a result, various new proteins involved in metabolic processes, DNA repair, or scaffolding have been identified as part of the nucleoid structure (27,28,29,30,31,32).
Nucleoids are ubiquitously distributed among plants, fungi, and animals (26, 29, 33,34,35,36,37,38) and play an important role in regulating replication/translation, maintenance, repair, and recombination of mtDNA. In general, nucleoids harbor 2–8 mtDNA copies, and several hundred of these complexes exist in a cell (36). Nucleoids are either membrane associated (39) or located in the mitochondrial matrix.
Mitochondrial single-stranded DNA binding protein (mtSSB), mitochondrial polymerase γ (Polγ), and the mitochondrial transcription factor A (TFAM) are major constituents of nucleoids and are important for mtDNA organization (26, 40,41,42). TFAM seems to have a histone-like function based on 2 high-mobility group (HMG) boxes, which are characteristic of a group of chromosomal proteins in the nucleus. Besides acting as a transcription factor, TFAM also organizes nucleoids by folding and wrapping mtDNA (43,44,45,46,47,48,49,50). The exclusive localization to mtDNA makes TFAM an ideal marker for mitochondrial nucleoids. Interestingly, the calculated packing density of mitochondrial nucleoids is comparable to the packing density of bacterial nucleoids (51), and it seems that TFAM is the major component in coordinating the packing of the mtDNA (52).
Faithful mtDNA replication is essential to ensure oxidative phosphorylation and preserves cell function during development and regeneration. Several maternally inherited human diseases (53), such as diabetes mellitus, deafness syndrome, mitochondrial encephalomyopathy, lactic acidosis, stroke-like syndrome (2, 54, 55), and Kearns-Sayre syndrome (2, 54), highlight the importance of mtDNA. Point mutations or deletions were shown to correlate with an increased rate of apoptosis, free radical formation, and energy depletion, leading to impairment of tissue function (54).
The effects of mtDNA mutations were further demonstrated in transgenic mice with a proofreading-deficient Polγ (mtDNA-mutator mouse). The phenotype of these animals showed typical signs of apoptosis (6, 56), which caused premature aging characterized by kyphosis (curvature of the spine), cardiac hypertrophy, and osteoporosis. A recent study showed that a transgenic mouse expressing cardiac-targeted, mutated human Polγ (57) developed early aging symptoms combined with enhanced ROS formation and severe cardiomyopathy, similar to observations in the mtDNA-mutator mouse. These data support the common view that mutated mtDNA is a major contributor to aging and various disorders (58,59,60,61,62).
Based on the organization of mtDNA into nucleoids and their proximity to the ROS-generating respiratory chain, nucleoids may have an integrated antioxidant system to protect mtDNA from oxidative damage. This hypothesis is further substantiated by the notion that Escherichia coli manganese superoxide dismutase (SOD2) binds to and may protect bacterial DNA, whereas the iron-containing bacterial isoform lacks this association (63). Mitochondria contain highly efficient enzymes to detoxify ROS, such as SOD2, glutathione peroxidase (GPx1), and members of the thioredoxin superfamily that may be included in the nucleoid structure.
Here we present biochemical evidence that nucleoid complexes isolated from rat or bovine heart or Jurkat cells include SOD2. This provides the first evidence that an antioxidant system in direct association with mtDNA may protect against ROS-mediated damage generated by the respiratory chain. Alteration of the nucleoid-associated antioxidant system may have great impact in development of chronic diseases and in the aging process.
MATERIALS AND METHODS
All chemicals were of analytical grade and obtained from Sigma-Aldrich (St. Louis, MO, USA), Fluka (Buchs, Switzerland), or Merck (Darmstadt, Germany).
Animals
Animal treatment was in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the U.S. National Institutes of Health and was granted by the Ethics Committee of the University Konstanz. Rats were sacrificed, and the heart was removed for mitochondria isolation. Bovine hearts and bovine aortas for the primary culture were obtained from the local slaughterhouse.
Cell culture
Human Jurkat T cell lymphoma cells (clone E6) were grown at 37°C in RPMI 1640 medium (Biochrom, Berlin, Germany), with 1% l-glutamine, 10% bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin (Biochrom).
Primary cultures of bovine aortic endothelial cells and human smooth muscle cells were prepared and grown as described previously (64, 65). Human veins were obtained during bypass surgery at the Heart Center Bodensee (Kreuzlingen, Switzerland). In accordance with the Declaration of Helsinki, consent of the ethics committee was obtained, and a written consent was given by the patients.
Isolation of mitochondria from tissues
Rat and bovine hearts were homogenized at 4°C in mitochondria isolation buffer [250 mM sucrose; 10 mM Hepes, pH 7.4; 1mM EGTA; 0.5% (w/v) fatty acid-free BSA; 1 mM glutathione] in a Dounce homogenizer (25 strokes). Tissue homogenates were centrifuged at 750 g for 10 min. The supernatant containing mitochondria was collected and further centrifuged at 7500 g for 10 min. After the second centrifugation step, the mitochondria were enriched in the pellet. The pellet was resuspended in 10 mM Hepes, pH 7.4, and 250 mM sucrose, and both centrifugation steps (10 min 750 g and 10 min 7500 g) were repeated. Thereafter, the mitochondrial pellet was further purified by isopycnic gradient centrifugation as described previously (27).
Isolation of mitochondria from cell culture
Mitochondria were prepared from cultured Jurkat cells (3 l medium) (Biochrom) and from primary bovine endothelial cells. Mitochondria of bovine endothelial cells were isolated from a total of 120 10-cm cell culture dishes. Cells were collected by centrifugation at 200 g, and mitochondria were prepared as described (27).
Biochemical isolation of intact mitochondrial nucleoids
Intact nucleoids were isolated according to a modified method (Fig. 1A) published by Garrido et al. (27). In addition to the standard gradient buffer, 20 or 200 mM NaCl was added to study the association-dissociation equilibrium of nucleoid protein constituents to mtDNA.
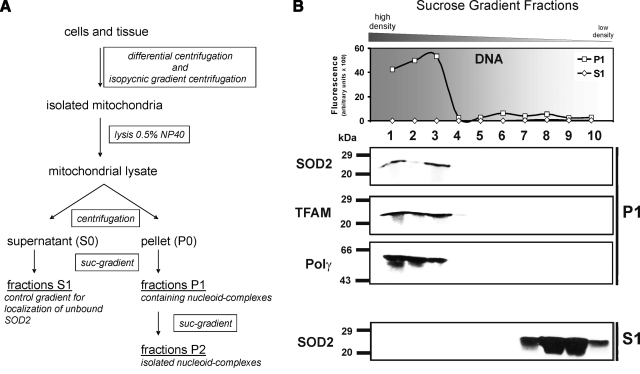
Figure 1.
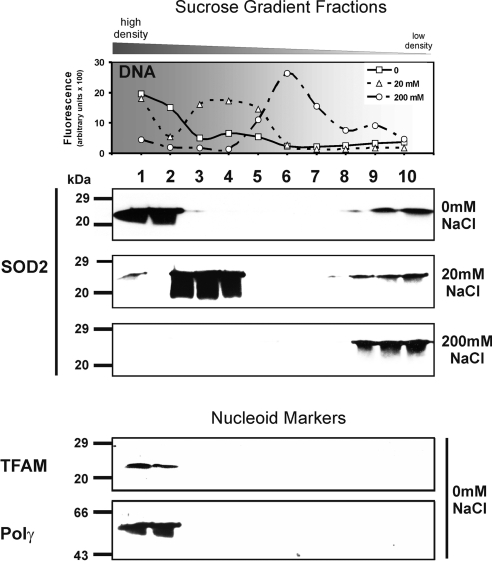
Isolation of human nucleoids from Jurkat cells. A) Tissue or cells were disrupted with a Dounce homogenizer in isoosmotic mitochondrial isolation buffer. Mitochondria were isolated with differential centrifugation followed by an isopycnic gradient centrifugation step as described in methods. Pure mitochondria were lysed with the nonionic detergent Nonidet P-40 and fractionated into supernatant (S0) and pellet (P0) by centrifugation. Pure nucleoids were isolated from the pellet by 2 sequential sucrose step gradients (P1 and P2; 75–20% sucrose). B) Nucleoids were purified from Jurkat mitochondria by a step gradient, and the P1 fractions were analyzed for their DNA (SYBR Green fluorescence)/protein distribution (Western blot analysis). MtDNA was concentrated in P1 fractions 1–3 (top panel). To identify nucleoid-containing fractions, Western blotting against TFAM and Polγ was performed. Nucleoid markers and SOD2 were found in the mtDNA-containing fractions (middle panel). Some unbound SOD2 was found in the low-density S fractions (bottom panel). Data shown are representative of 8 independent experiments.
After the centrifugation step the sucrose gradient (20–75%) was fractionated into 1 ml portions from the bottom to the top. Samples were dialyzed against NE2 buffer (27) at 4°C overnight and subjected to SDS-PAGE analysis. The mtDNA content in the fractions was measured by SYBR Green™ (Invitrogen, Carlsbad, CA, USA) fluorescence in a 96-well plate. Samples were analyzed with a Spectra Fluor fluorescence reader (Tecan, Crailsheim, Germany) at excitation 485 nm and emission 535 nm.
Characterization of the sucrose gradient fractions
Samples of gradient fractions were subjected to standard SDS-PAGE on 12% polyacrylamide gels and Western on nitrocellulose membranes (Schleicher & Schuell BioScience, Dassel, Germany). Primary antibodies and dilutions used included polyclonal rabbit serum directed against human TFAM (kindly provided by Rudolf J. Wiesner, University of Heidelberg, Germany), 1:5000; polyclonal SOD2 antiserum (Stressgene Bioreagents, Victoria, BC, Canada), 1:10,000; polyclonal Polγ antiserum (Acris, Hiddenhausen, Germany), 1:5000; polyclonal glutathione peroxidase I antiserum (LabFrontier, Seoul, South Korea), 1:2000; monoclonal cytochrome c (BD Biosciences, Erembodegem, Belgium), 1:3000; monoclonal fumarate hydratase (Abcam, Cambridge, MA, USA), 1:200; and polyclonal histone H1 antisera (Santa Cruz Biotechnology, Heidelberg, Germany).
Activity of malate dehydrogenase
The activity of the trichloroacetic acid cycle enzyme malate dehydrogenase (MDH) was determined photometrically by measuring the malate-dependent NAD+ turnover as described elsewhere (66, 67). For each nucleoid preparation, a representative fraction was selected. In case of S1 preparations, fraction 9 was selected, and for the P1/P2 preparations, fraction 2 was selected. For each assay, 5 μl (S1 preparations) or 50 μl (P preparations) of the isolated nucleoids was used.
SOD activity assay
SOD activity was measured with the SOD assay kit (Sigma-Aldrich) according to the manufacturer’s instructions.
Coimmunoprecipitation
Protein A was swelled in IP buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 4 mM EDTA; 0.25% gelatin) for 3 h. Sucrose gradient fractions were mixed with freshly prepared protein A. Antibodies were added and allowed to precipitate the antigen and to bind to protein A overnight at 4°C. The samples were rinsed with IP buffer (centrifugation at 4000 g for 5 min), SDS sample buffer was added, and proteins were separated by SDS-PAGE and analyzed by Western blotting with an antibody against SOD2 (Stressgene Bioreagents), 1:10,000. Antibodies for immunoprecipitation were used as follows: monoclonal DNA (Progen, Heidelberg, Germany), 1:20; polyclonal SOD2 (Stressgen Bioreagents), 1:300; polyclonal glutathione peroxidase I (LabFrontier), 1:200; and polyclonal Polγ (Acris), 1:200.
Sandwich slot blot filter binding assay
The assay described by Czerwinski et al. (68) was modified as follows. Nitrocellulose (NC; Bio-Rad, Hercules, CA, USA) and nylon (Osmonics, Westborough, MA, USA) filters were precut to fit in the Bio-Dot SF (Bio-Rad) slot blot apparatus. NC filters were presoaked for 10 min in 0.4 M KOH (reduction of DNA adsorption) and neutralized. Nylon and NC filters were then equilibrated for 30 min in binding buffer (5 mM potassium phosphate buffer, 0.3% glycerol, 400 μM MgCl2). Binding assays were performed with fixed DNA concentration of 0.5 nM and varying SOD2 concentrations in binding buffer with 50 μg/ml BSA on ice for 60 min. Recombinant SOD2 from E. coli was obtained from Sigma-Aldrich. According to the manufacturer, the protein was purified from E. coli by heat precipitation (60°C), streptomycin and ammonium sulfate precipitation, and 2 subsequent ion-exchange chromatography steps on CM-52 and DEAE, respectively, according to the method published by Keele et al. (69). The purity of the protein was 80%, determined by SDS-PAGE with 2 major unidentified contaminants at ∼35 and ∼50 kDa. Binding assays with SOD1 (bovine, EC 1.15.1.1; Sigma-Aldrich) were performed under the same conditions.
Equilibrated membranes were flushed with 200 μl of binding buffer in the slot blot apparatus, and reaction mixtures were loaded and rinsed with 200 μl of binding buffer. DNA bound to nylon membranes was UV cross-linked (3× auto cross-link) with a UV Stratalinker 2400 (Stratagene, Cedar Creek, CA, USA). The NC filter was blocked with Roti®-Block (Carl Roth, Karlsruhe, Germany) for 25 min, incubated with the streptavidin-coupled IRDYE® 800CW (Li-Cor, Lincoln, NE, USA) and washed 3 times with TBS-T (10 mM Tris-HCl, pH 8.0; 100 mM NaCl; 0.1% Tween 20), whereas the nylon filter was washed in PBS containing 0.1% SDS for 30 min and incubated with the same dye. The membranes were rinsed with TBS-T. Biotinylated DNA was visualized with an infrared scanner (Li-Cor) and quantified with the Odyssey application software (ver. 2.1.112; Li-Cor).
Confocal microscopy of human smooth muscle cells
Human smooth muscle cells were seeded on 8-well glass chamber slides (Nalgen Nunc, Naperville, IL, USA) at a density of 1–1.5 × 105 cells/well to reach 50–60% of confluence on the day of the experiment. Cells were fixed in 4% paraformaldehyde/PBS for 10 min at room temperature. After washing in PBS, cells were permeabilized, and nonspecific binding of the first antibody was blocked with 0.1% Triton X-100 and 1% BSA in PBS for 30 min at room temperature. After a washing step, primary antibodies were incubated overnight at 4°C in a humidified chamber. The polyclonal SOD2 antibody (Stressgen Bioreagents) was 1:100, and the monoclonal DNA (PROGEN) antibody was 1:10 diluted in PBS. After 3 × 5 min washing in PBS, the secondary antibodies (labeled with Alexa488 or Alexa546) were applied for 60 min at room temperature. Secondary antibodies (MoBiTec, Göttingen, Germany) were diluted 1:300 in PBS. Immunostained cells were mounted in glycerol/PBS (3:1).
Metabolic BrdU labeling was performed with the BrdU Immunofluorescence Assay Kit (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s instructions. Immunostained cells were imaged with a Zeiss LSM510 Meta confocal microscope (Carl Zeiss, Oberkochen, Germany).
RESULTS
SOD2 is present in nucleoids
The method by Garrido et al. (27) was modified as indicated in Fig. 1A and yielded sufficiently pure nucleoids (P2 fraction) from cultured cells or tissue by serial sucrose step gradient centrifugation and fractionation. The nucleoid purity was confirmed by molecular markers for nucleoids, such as TFAM (52) and Polγ (29), and assayed for evidence of contamination by mitochondrial matrix (fumarate hydratase and malate dehydrogenase) and inner membrane proteins (cytochrome c) or nuclear DNA (histone H1; data not shown).
Mitochondria were isolated from human Jurkat cells, lysed, and divided by centrifugation into a pellet (P0) and supernatant (S0) fraction that were both further separated on a sucrose step gradient (Fig. 1B). All gradient fractions (P1 and S1) were screened for mtDNA with SYBR Green fluorescent detection. Only the fast sedimenting P1 fractions 1–3 contained mtDNA (Fig. 1B, top panel, square symbol), indicating a protein-DNA macromolecular complex. These fractions were not contaminated by mitochondrial matrix proteins, which was confirmed by the tricarboxylic acid cycle enzyme fumarate hydratase (FH) (Supplemental Fig. S2A). FH was present only in the upper P1 fractions 8–10, but not in the nucleoid containing P1 fractions 1–3. As a second control for matrix contaminations, MDH activity was assessed (Supplemental Table S1). The P1 fraction 2 was 0.43% and for P2 fraction 2, 0.29% of the MDH activity in S1 fraction 9. These reductions in MDH activity of >99.5% in both P fractions, together with the results on FH, strongly indicate that contaminations of nucleoid fractions by mitochondrial matrix proteins were insignificant.
The DNA content in the S1 fractions of the supernatant (Fig. 1B, top panel, rhombus symbol) was below the detection limit, but proteins of the mitochondrial matrix such as FH (Supplemental Fig. S2A) and inner membrane such as cytochrome c (data not shown) as well as unbound SOD2 were detected in the slow sedimenting S1 fractions 7–10 (Fig. 1B, bottom panel).
The P1 fraction with the highest DNA content (Fraction 3, Fig. 1B, top panel, square symbol) was treated with the restriction endonuclease HindIII to verify mtDNA and exclude nuclear DNA contamination. We detected defined mtDNA fragments of 10.2 and 5.5 kBp (Supplemental Fig. S1, lane 1), whereas intact mtDNA was found at 16.5 kBp (Supplemental Fig. S1, lane 2).
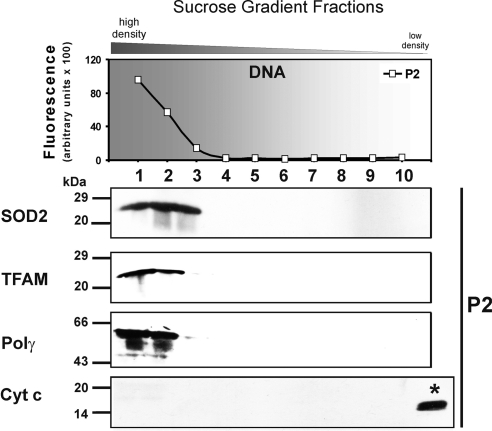
SOD2, TFAM, and Polγ were detected in the fast-sedimenting P1 fractions 1–3 of the pellet only (Fig. 1B, middle panel; Supplemental Fig. S2A), which suggests that SOD2 is a protein constituent of nucleoids. To further purify and exclude contamination of nucleoids with matrix proteins, P1 fractions 1–3 were pooled, treated again with lysis buffer, and separated on a second sucrose step gradient (P2). Once more, SOD2 appeared in P2 fractions 1–3 only, signifying a direct association with mtDNA (Fig. 2; see also Supplemental Fig. S2B). The fractions of the second gradient were free of contamination with soluble matrix or inner membrane proteins, as proven by the absence of cytochrome c (Fig. 2), FH (Supplemental Fig. S2B), and MDH (Supplemental Table S1). Thus, a significant proportion of the otherwise matrix-located SOD2 appears to be mtDNA associated when compared to the nucleoid markers TFAM and Polγ. Furthermore, SOD2 activity correlates with the Western blot results on SOD2 protein, and 60% (Supplemental Table S1) of its activity compared to the S1 fraction 9 remained in the nucleoid P1 and P2 fractions 2, whereas contaminating MDH activity was <0.5% of the activity in S1 fraction 9.
Figure 2.
Isolation of pure nucleoids. The DNA-containing P1 fractions (1,2,3) were collected and further purified on a second sucrose step gradient (75–20%), resulting in P2 fractions (as shown in Fig. 1A). MtDNA-containing fractions were identified with SYBR Green and detected in P2 fractions 1–3 (top panel). As positive control for the nucleoids fractions were probed for TFAM and Polγ by Western blot analysis. Nucleoid markers and SOD2 appeared in the mtDNA fractions. P2 fractions were also analyzed with an antibody against cytochrome c (*positive control) as a marker for inner membrane contamination. Data shown are representative of 8 independent experiments.
SOD2 coimmunoprecipitates with mtDNA and glutathione peroxidase
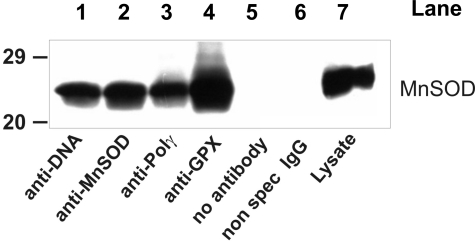
Large quantities of nucleoids were isolated from bovine heart mitochondria, and sucrose gradient fractions were analyzed with Western blots. The distribution of SOD2 and association with mtDNA were very similar to the results in Jurkat cells (Supplemental Fig. S3), which suggests that among mammals SOD2 integration into mtDNA complexes may be species encompassing. Immunoprecipitation of DNA, Polγ, glutathione peroxidase (GPx1), and SOD2 from isolated bovine nucleoids and Western blot detection for SOD2 confirmed the direct interaction of SOD2 with mtDNA or nucleoid constituents (Fig. 3).
Figure 3.
Immunoprecipitation of bovine heart nucleids. Heart mitochondrial nucleoids were immunoprecipitated with antibodies as indicated (lanes 1–4). Analysis of the immunoprecipitates by Western blotting against SOD2 showed that the SOD2 is included in the nucleoid complex. SOD2 and GPx1 are constituents of a putative nucleoid antioxidant system. Nonspecific binding and cross-reactivity were excluded by appropriate controls (lanes 5 and 6; unspecific IgG and without antibody). Data shown are representative of 3 independent experiments.
The functional role of SOD2 in trapping superoxide proximal to mtDNA and preventing oxidative lesions suggests that GPx1 may be a component of the nucleoid structure to eliminate the SOD product, H2O2. GPx1 coimmunoprecipitated with mtDNA (Fig. 3) and Western blot analysis of sucrose fractions (results not shown) confirmed the presence of GPx1 in nucleoids.
SOD2 binds directly with DNA
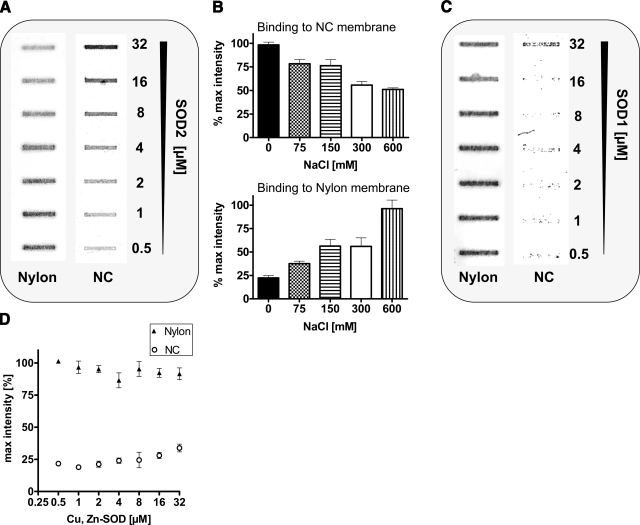
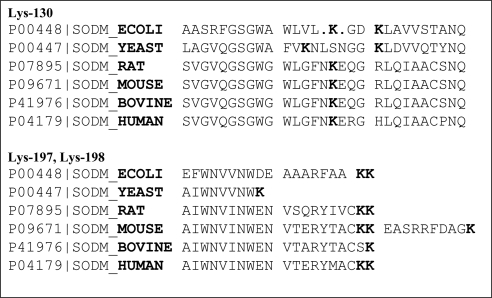
Interaction of SOD2 with DNA-bound proteins such as TFAM or binding of SOD2 to mtDNA could not be concluded from the previous experiments. Therefore, a sandwich slot blot filter assay (70) was established to investigate the direct interaction of recombinant SOD2 with mtDNA (Fig. 4A). A random sequence (34 bp) from human mtDNA (1150–1174) was synthesized as a biotin-labeled reverse and forward primer and annealed into a double strand. DNA was incubated in binding buffer with commercially available recombinant E. coli SOD2 from Sigma-Aldrich (commercial sources of human SOD2 are truncated or contain a C-terminal 6xHis-tag, and attempts to overexpress human SOD2 in bacteria failed because of the formation of inclusion bodies) and filtered through a sandwich of NC and nylon membranes. Nevertheless, the bacterial protein still shares 60% sequence homology to the bovine sequence and 45% to the human sequence. The protein purity of the SOD2 preparation was ≈80%, determined by SDS-PAGE. Proteins and protein-DNA complexes were adsorbed on the NC membrane, whereas free DNA was adsorbed on the nylon membrane. Biotin-labeled DNA was detected by fluorescent streptavidin. Figure 4A, B clearly demonstrates that SOD2 directly binds to DNA. Overexpression of a C-terminal EGFP-tagged SOD2 in HEK-cells translocated to the mitochondria, but association with mtDNA was absent (data not shown). Analogous experiments with SOD1 did not show any interaction with mtDNA (Fig. 4C, D). This result confirms previously published findings (63, 68), which demonstrated SOD2-DNA interaction by filter (68) and gel shift assays (63). The C-terminal α-helical lysines (K197, K198 of human MnSOD) in conjunction with K130 in a loop region are likely DNA-protein interaction sites (71) and may form a structurally conserved DNA binding domain (see Fig. 8; Supplemental Fig. S4). In contrast, bovine SOD1 is comprised of β-sheets, contains also a C-terminal Lys, but lacks entirely DNA association and has no homology to bovine SOD2.
Figure 4.
Slot blot filter assay to prove interaction of mtDNA and recombinant SOD2. A) Sandwich slot blot filter assay demonstrates the concentration-dependent interaction of recombinant E. coli SOD2 (0.5–32 μM) with a biotinylated sequence from human mtDNA (0.5 nM; 34-mer). SOD2-DNA complexes bind to NC, whereas unbound DNA binds to the nylon filter. Bound DNA was visualized by a Streptavidin-IR dye (LiCor; IRDYE 800 CW) and quantified by densitometry. B) The interaction of SOD2 with biotinylated DNA was concentration dependently disrupted by increasing salt concentrations (NaCl: 75–600 mM). Data shown are representative of 3 independent experiments. C) Sandwich slot blot filter assay with bovine SOD1 to demonstrate that binding of SOD to DNA is specific for SOD2 only. Increasing concentrations of SOD1 were incubated with biotinylated DNA, slot blotted onto the filter sandwich (nitrocellulose on top for proteins and nylon on the bottom for DNA), stained with IR-fluorescent dye labeled streptavidin, and analyzed with densitometry. Data shown are representatives of 3 independent experiments. D) Densitomertic analysis of 3 independent experiments.
Figure 5.
Isolation of rat heart nucleoids with increasing salt concentrations. Nucleoids were purified from rat heart. Top panel: MtDNA containing P1 fractions were quantified with SYBR Green. Nucleoids sedimented more slowly with increasing salt concentrations (20 and 200 mM NaCl), indicating that ionic forces are involved in the association of SOD2 with nucleoids. Middle panel: Under standard conditions (0 mM NaCl), the nucleoids migrated to the bottom of the gradient (fractions 1–3). At 20 mM NaCl, most of the SOD2 was still associated with mtDNA (fractions 4–5), but according to the mtDNA distribution the nucleoid complex started to dissociate at low salt (nucleoids were also identified by TFAM and Polγ; data not shown). At high salt (200 mM NaCl), attachment of the SOD2 was completely disrupted. Bottom panel: As markers for the nucleoid complexes in 0 mM fractions, we used antibodies against TFAM and Polγ. SOD2, TFAM, Polγ, and mtDNA were present in the same fractions. Data shown are representative of 3 independent experiments.
Figure 6.
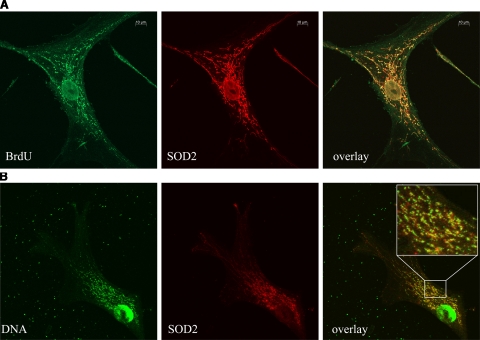
Confocal images of human vascular smooth muscle cells. MtDNA is organized as protein-DNA complexes (nucleoids) visible as punctate staining within the mitochondrial network. A) Confocal microscopy with antibodies recognizing SOD2 (red) and DNA-incorporated BrdU (metabolic BrdU labeling, green). Yellow represents colocalized SOD2 with mtDNA (overlay). B) Confocal microscopy with antibodies against SOD2 (red) and DNA (green). Yellow represents colocalized SOD2 with mtDNA (overlay). Colocalization of SOD2 with mtDNA confirms (yellow) that a significant proportion of SOD2 is integrated in nucleoids. Furthermore, nucleoid heterogeneity in size and composition is illustrated.
Figure 7.
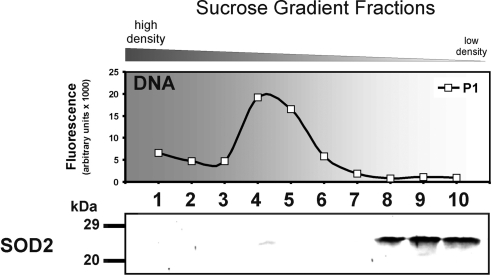
Lack of SOD2 association with nucleoids isolated from bovine aortic endothelial cells. Top panel: MtDNA containing P1 fractions were quantified with SYBR Green. MtDNA was concentrated in P1 fractions 1–6. Bottom panel: SOD2 was only present in the low-density P1 fractions 8–10 and not in the mtDNA-containing fractions. Notably the mtDNA content is significantly higher than in the other preparation. Data shown are representative of 4 independent experiments.
Figure 8.
Highly conserved Lys residues at the C-terminus of SOD2. The C-terminal α-helical lysines (K197, K198 of human SOD2) in conjunction with K130 in a loop region are likely DNA interaction sites. We hypothesize that they form a structurally conserved DNA binding domain of SOD2.
MtDNA and SOD2 interact by ionic forces
Ionic binding forces involved in SOD2-mtDNA interaction were manipulated by increasing salt concentration (Fig. 4B) in buffers used for the filter binding assay or for mitochondrial lysis. High salt concentration dependently disrupted the DNA-SOD2 complexes (Fig. 4B) detected with the filter binding assay.
To further substantiate our hypothesis of SOD2-mtDNA as a general principle among mammalian species mitochondria were obtained in larger quantities from rat heart (Supplemental Fig. S2C, D). Nucleoids showed a comparable distribution and protein composition in the sucrose gradient as observed for Jurkat cell nucleoids (Fig. 2) as well as bovine heart nucleoids (Supplemental Fig. S3).
Mitochondrial preparations were incubated with 0 (standard condition), 20, or 200 mM NaCl, and rat heart nucleoids were isolated as described, yielding P1 fractions (Fig. 5). As a control for the presence of the nucleoids, the 0 mM NaCl fractions were probed for the presence of TFAM and Polγ (Fig. 5, bottom panel; Supplemental Fig. S2C, D).
Compared to those preparations that only contained 20 mM Tris-HCl buffer (0 mM NaCl; compare Fig. 1B) the salt treatment caused mtDNA to shift into slow-sedimenting fractions (Fig. 5, top panel), which suggests dissociation of proteins from nucleoid complexes. SOD2 remained bound to mtDNA at 20 mM NaCl and was copurified together with an obviously lighter mtDNA in the P1 fractions 3–5. At 200 mM NaCl, the mtDNA found in P1 fractions 5–7 was devoid of SOD2, which accumulated in P1 fractions 8–10 (Fig. 5, middle panel). This proves a dissociation of the mtDNA-SOD2 complexes at high salt, indicating a largely ionic interaction between the 2 macromolecules.
Contamination by mitochondrial matrix proteins was assessed by the presence of FH (Supplemental Fig. S2C, D) or MDH (Supplemental Table S1) in the P fractions, similar to the Jurkat preparation. FH was undetectable in the nucleoid-containing fractions, and MDH activity in P1 and P2 fractions 2 compared to S1 fraction 9 was reduced by >99.7% (Supplemental Table S1). In contrast, SOD2 activity was 69% compared to S1 fraction in P1 and 63% for P2, respectively.
mtDNA colocalizes with SOD2 in human smooth muscle cells
Human smooth muscle cell DNA was metabolically labeled by BrdU incorporation and stained with an antibody against BrdU or with an antibody recognizing DNA. MtDNA is visible as punctae that may represent a single nucleoid (Fig. 6). The mitochondrial network is clearly visible by the SOD2 staining. An overlay demonstrates a high degree of colocalization of SOD2 and mtDNA in mitochondria located around the nucleus. Peripheral mitochondria show a clear separation of SOD2 and mtDNA (shown by separated green and red dots). These findings are in accordance with the results of the nucleoid isolation, where we found SOD2 associated with the mtDNA but also unbound SOD2 (Fig. 1B; S1 fractions).
Lack of association of SOD2 with mtDNA in bovine endothelial cells
Nucleoids were isolated from bovine endothelial cells as described above. Surprisingly, there was no association of SOD2 with mtDNA found. Furthermore, different nucleoid densities were noticed as compared to other preparations (Fig. 7). Nucleoids appeared in P1 fractions 4–6 and not in P1 fractions 1–3 as observed for rat heart nucleoids (Fig. 5) or bovine heart nucleoids (Supplemental Fig. S3). This partially resembles the mtDNA distribution pattern of the isolation of nucleoids from mitochondria lysed with 20 mM NaCl (Fig. 5, top panel), which suggests a difference in the composition of endothelial cell nucleoids. Western blots of these fractions confirmed the lack of SOD2 association with mtDNA. SOD2 was found in the slow sedimenting P1 fractions 8–10 (Fig. 7) only.
DISCUSSION
Our data provide biochemical evidence that some portion of the enzyme is associated with the mitochondrial nucleoid structure and binds directly to mtDNA. This new finding is based on the detection of SOD2 in sucrose gradient isolated nucleoids by Western blots and coimmunoprecipitation of SOD2 with antibodies against DNA and Polγ from nucleoid fractions. It applies for mitochondria isolated from several sources, such as bovine and rat heart, Jurkat cells, and human vascular smooth muscle cells. Because the matrix protein FH does not copurify with the nucleoids and there is a marked reduction of MDH activity in the nucleoid fractions, it is unlikely that association of SOD2 with mtDNA is due to contamination of the nucleoid fraction.
Pathophysiological importance of the mitochondrial SOD2
Under physiological conditions, the mitochondrial respiratory chain is a major source of superoxide radical, converting ∼0.1% of oxygen into ROS. Therefore, the mitochondrial restricted SOD2 is essential for cell survival as evidenced by the lethality of Sod2−/− mice (72). These animals die within a few days after birth and exhibit a variety of phenotypes, including neurodegeneration, cardiovascular abnormalities, and extensive mitochondrial dysfunction and damage. Several pathophysiological conditions, such as inflammation or chronic hypertension, are associated with a counter-regulatory increase in SOD2 to cope with increased ROS formation (73).
The common polymorphism in the mitochondrial leader sequence, which supposedly decreases the mitochondrial import of SOD2, is associated with the development of various forms of cancer, such as breast or prostate carcinoma (74). In summary, SOD2 is essential to protect mitochondrial physiology and the mitochondrial genome from oxidative damage.
Does SOD2 protect mtDNA from oxidative damage?
For many of the known nucleoid-associated proteins, with the exception of TFAM, mtSSB, and Polγ, no clear function has been assigned (30, 34, 75). For SOD2 it is obvious to suggest that a proximity of SOD2 to mtDNA could prevent superoxide toxicity, in which protein-bound FeIII and a subsequent Fenton reaction with H2O2 yields ·OH radicals or a ferryl ion. Hydrogen peroxide should therefore be deleterious as well and requires an enzymatic system for detoxification. This was confirmed by coimmunoprecipitation of SOD2 with an antibody against glutathione peroxidase 1 and by Western blot analysis of sucrose density gradient fractions. The emerging concept of a functional antioxidant system established in the nucleoid structure is supported by the notion that free mtDNA is more vulnerable to X-ray or H2O2-induced damage than mtDNA organized in nucleoids (76). One may argue that dense packing alone may already result in such protection, but it was found that in E. coli SOD2 is directly associated with DNA and could protect against superoxide, whereas bacterial FeSOD showed no association and provided no protection (63). The evolutionary explanation for the bacterial origin of mitochondria is another argument for similarities in the association of SOD2 in the bacterial and mitochondrial genome.
Indirect proof of an increased antioxidant capability of mtDNA associated vs. free matrix-located SOD2 may come from a comparison between endothelial and smooth muscle cells. The latter were reported to be more resistant toward oxidative stress-induced mtDNA lesions than endothelial cells (77). It may be speculated that the rapidly proliferating mitochondrial network in the endothelium could be the cause for the absence of an association of SOD2 with mtDNA.
How does SOD2 interact with mtDNA?
We conclude from data obtained by a filter-binding assay that direct binding of SOD2 to mtDNA can occur without participation of a scaffolding protein such as TFAM. Increasing the ionic strength by addition of sodium chloride during the isolation procedure dissociated SOD2 from isolated nucleoids or prevented binding of SOD2 to a synthetic DNA fragment in the filter-binding assay. TFAM of isolated nucleoids still remained bound to mtDNA (refer also to refs. 26, 27, 34) under these conditions, which suggests that the 2 proteins bind with different affinities to mtDNA. For TFAM the existence of 2 HMG boxes (78) in the sequence may provide a stronger interaction with mtDNA, which would agree with its suggested scaffolding properties. The instability of SOD2 binding at high salt again favors ionic binding forces, which could be explained by the presence of several C-terminally located positively charged lysines and their interaction with the negatively charged phosphate backbone of mtDNA. Corresponding lysines in SOD2 are conserved from E. coli to mammals (Fig. 8) (63, 68, 79). However, interaction with other proteins in the nucleoid structure cannot be excluded at the current stage but will be addressed in future investigations with truncation mutants and side-directed mutagenesis of the Lys residues.
Is the association of proteins to mtDNA metabolically controlled?
Several nucleoid populations with different associated proteins likely exist (29, 51); however, the physiological function remains unclear. As herein a lack of SOD2 association in endothelial cells is reported, one may postulate that a dynamic metabolically or stress-driven process of association and dissociation occurs. For the process of mtDNA replication, transcription, and repair, a separation of mtDNA attached proteins has to be postulated, as assumed for immature Xenopus oocytes compared to mature ones (80). Similar to the histones in nuclear DNA, a process of acetylation/deacetylation could control (81) the attachment of nucleoid proteins to mtDNA, thus modulating nucleoid density for maintenance and replication/transcription. Also metabolic demands may be associated in the control of the nucleoid density and integrated proteins.
The mammalian mitochondrial genome solely codes components of the respiratory chain. Therefore, an increase in ATP demand and oxidative metabolism requires the induction of proteins of the respiratory chain. The D-loop conformation of mtDNA, which is an indicator of enhanced oxidative metabolism and transcriptional activity, is more abundant after exercising the skeletal muscle (82). This indicates that mtDNA may exist in different conformations that are regulated by metabolic demand.
CONCLUSIONS
The relationship between SOD2 and mtDNA may exist to prevent mitochondrial dysfunction and allow for protection in a number of different tissues and disease processes. We demonstrated that mtDNA is associated with an antioxidant system in mammalian cells with SOD2 binding directly to mtDNA. The implications for this complex in physiological and pathophysiologic processes have not been elucidated but likely play an important role in diseases linked to chronic oxidation, such as diabetes or aging.
Supplementary Material
Acknowledgments
We thank Dr. Elisa Ferrando-May for assistance with the confocal microscope, Dr. Rudolf J. Wiesner (University of Heidelberg, Heidelberg, Germany) for supplying the anti-TFAM antibody, Dr. W. Hofer for assisting with the MDH activity assays, Rebecca Zee and Alicia Evangelista for preparing the manuscript, and Dr. Richard A. Cohen for his support. This work was supported by grant BU 698/6-1 of the Deutsche Forschungsgemeinschaft (DFG). D.J.F.H. and M.M.B. were supported by the Whitaker Cardiovascular Institute and NIH grants R01 AG027080-04 and P01 HL081738-03. D.P. was supported by NIH grant K08HL071563.
References
- Lee H C, Wei Y H. Oxidative stress, mitochondrial DNA mutation, and apoptosis in aging. Exp Biol Med (Maywood) 2007;232:592–606. [PubMed] [Google Scholar]
- Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- Silva J P, Larsson N G. Manipulation of mitochondrial DNA gene expression in the mouse. Biochim Biophys Acta. 2002;1555:106–110. doi: 10.1016/s0005-2728(02)00263-3. [DOI] [PubMed] [Google Scholar]
- Jeng J Y, Yeh T S, Lee J W, Lin S H, Fong T H, Hsieh R H. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem. 2008;103:347–357. doi: 10.1002/jcb.21625. [DOI] [PubMed] [Google Scholar]
- Lee H C, Wei Y H. Role of mitochondria in human aging. J Biomed Sci. 1997;4:319–326. doi: 10.1007/BF02258357. [DOI] [PubMed] [Google Scholar]
- Kujoth G C, Hiona A, Pugh T D, Someya S, Panzer K, Wohlgemuth S E, Hofer T, Seo A Y, Sullivan R, Jobling W A, Morrow J D, Van Remmen H, Sedivy J M, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla T A. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Davies K J. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- Bohr V A, Stevnsner T, de Souza-Pinto N C. Mitochondrial DNA repair of oxidative damage in mammalian cells. Gene. 2002;286:127–134. doi: 10.1016/s0378-1119(01)00813-7. [DOI] [PubMed] [Google Scholar]
- Richter C. Oxidative damage to mitochondrial DNA and its relationship to ageing. Int J Biochem Cell Biol. 1995;27:647–653. doi: 10.1016/1357-2725(95)00025-k. [DOI] [PubMed] [Google Scholar]
- Wei Y H, Lu C Y, Wei C Y, Ma Y S, Lee H C. Oxidative stress in human aging and mitochondrial disease-consequences of defective mitochondrial respiration and impaired antioxidant enzyme system. Chin J Physiol. 2001;44:1–11. [PubMed] [Google Scholar]
- Schapira A H. Mitochondrial dysfunction in neurodegenerative disorders. Biochim Biophys Acta. 1998;1366:225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- Ide T, Tsutsui H, Kinugawa S, Utsumi H, Kang D, Hattori N, Uchida K, Arimura K, Egashira K, Takeshita A. Mitochondrial electron transport complex I is a potential source of oxygen free radicals in the failing myocardium. Circ Res. 1999;85:357–363. doi: 10.1161/01.res.85.4.357. [DOI] [PubMed] [Google Scholar]
- Madamanchi N R, Runge M S. Mitochondrial dysfunction in atherosclerosis. Circ Res. 2007;100:460–473. doi: 10.1161/01.RES.0000258450.44413.96. [DOI] [PubMed] [Google Scholar]
- Leuner K, Hauptmann S, Abdel-Kader R, Scherping I, Keil U, Strosznajder J B, Eckert A, Muller W E. Mitochondrial dysfunction: the first domino in brain aging and Alzheimer’s disease? Antioxid Redox Signal. 2007;9:1659–1675. doi: 10.1089/ars.2007.1763. [DOI] [PubMed] [Google Scholar]
- Aime S, Digilio G, Bruno E, Mainero V, Baroni S, Fasano M. Modulation of the antioxidant activity of HO* scavengers by albumin binding: a 19F-NMR study. Biochem Biophys Res Commun. 2003;307:962–966. doi: 10.1016/s0006-291x(03)01307-x. [DOI] [PubMed] [Google Scholar]
- Ekstrand M I, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, Thams S, Bergstrand A, Hansson F S, Trifunovic A, Hoffer B, Cullheim S, Mohammed A H, Olson L, Larsson N G. Progressive Parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc Natl Acad Sci U S A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Larsson N G. Mitochondrial dysfunction as a cause of ageing. J Intern Med. 2008;263:167–178. doi: 10.1111/j.1365-2796.2007.01905.x. [DOI] [PubMed] [Google Scholar]
- Casteilla L, Rigoulet M, Penicaud L. Mitochondrial ROS metabolism: modulation by uncoupling proteins. IUBMB Life. 2001;52:181–188. doi: 10.1080/15216540152845984. [DOI] [PubMed] [Google Scholar]
- Wallace D C. Mouse models for mitochondrial disease. Am J Med Genet. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- Esposito L A, Melov S, Panov A, Cottrell B A, Wallace D C. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci U S A. 1999;96:4820–4825. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikova V G, Vinogradov A D. Generation of superoxide by the mitochondrial complex I. Biochim Biophys Acta. 2006;1757:553–561. doi: 10.1016/j.bbabio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Bovina C, D'Aurelio M, Fato R, Formiggini G, Genova M L, Giuliano G, Merlo Pich M, Paolucci U, Parenti Castelli G, Ventura B. Role of mitochondria in oxidative stress and aging. Ann N Y Acad Sci. 2002;959:199–213. doi: 10.1111/j.1749-6632.2002.tb02094.x. [DOI] [PubMed] [Google Scholar]
- Bachschmid M, Schildknecht S, Ullrich V. Redox regulation of vascular prostanoid synthesis by the nitric oxide-superoxide system. Biochem Biophys Res Commun. 2005;338:536–542. doi: 10.1016/j.bbrc.2005.08.157. [DOI] [PubMed] [Google Scholar]
- Frein D, Schildknecht S, Bachschmid M, Ullrich V. Redox regulation: a new challenge for pharmacology. Biochem Pharmacol. 2005;70:811–823. doi: 10.1016/j.bcp.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Nass M M, Nass S, Afzelius B A. The general occurrence of mitochondrial DNA. Exp Cell Res. 1965;37:516–539. doi: 10.1016/0014-4827(65)90204-1. [DOI] [PubMed] [Google Scholar]
- Van Tuyle G C, McPherson M L. A compact form of rat liver mitochondrial DNA stabilized by bound proteins. J Biol Chem. 1979;254:6044–6053. [PubMed] [Google Scholar]
- Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek A M, Spelbrink J N. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Mao C C, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale A B, Fearnley I M, Harbour M, Robinson A J, Reichelt S, Spelbrink J N, Walker J E, Holt I J. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bogenhagen D F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- Chen X J, Wang X, Kaufman B A, Butow R A. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- Hobbs A E, Srinivasan M, McCaffery J M, Jensen R E. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa I, Fumoto S, Kuroiwa T, Sando N. Characterization of DNA-binding proteins involved in the assembly of mitochondrial nucleoids in the yeast Saccharomyces cerevisiae. Plant Cell Physiol. 1995;36:1179–1188. [PubMed] [Google Scholar]
- Miyakawa I, Okazaki-Higashi C, Higashi T, Furutani Y, Sando N. Isolation and characterization of mitochondrial nucleoids from the yeast Pichia jadinii. Plant Cell Physiol. 1996;37:816–824. doi: 10.1093/oxfordjournals.pcp.a029017. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D F, Wang Y, Shen E L, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Shaw J M, Okamoto K. Mmm1p spans both the outer and inner mitochondrial membranes and contains distinct domains for targeting and foci formation. J Biol Chem. 2003;278:48997–49005. doi: 10.1074/jbc.M308436200. [DOI] [PubMed] [Google Scholar]
- Legros F, Malka F, Frachon P, Lombes A, Rojo M. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- Miyakawa I, Aoi H, Sando N, Kuroiwa T. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J Cell Sci. 1984;66:21–38. doi: 10.1242/jcs.66.1.21. [DOI] [PubMed] [Google Scholar]
- Miyakawa I, Sando N, Kawano S, Nakamura S, Kuroiwa T. Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J Cell Sci. 1987;88:431–439. doi: 10.1242/jcs.88.4.431. [DOI] [PubMed] [Google Scholar]
- Nass M M. Mitochondrial DNA: advances, problems, and goals. Science. 1969;165:25–35. doi: 10.1126/science.165.3888.25. [DOI] [PubMed] [Google Scholar]
- Van Tuyle G C, Pavco P A. The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J Cell Biol. 1985;100:251–257. doi: 10.1083/jcb.100.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke G D, Pavco P A, Ledwith B J, Van Tuyle G C. Structural and functional studies of the rat mitochondrial single strand DNA binding protein P16. Arch Biochem Biophys. 1990;282:116–124. doi: 10.1016/0003-9861(90)90094-f. [DOI] [PubMed] [Google Scholar]
- Pavco P A, Van Tuyle G C. Purification and general properties of the DNA-binding protein (P16) from rat liver mitochondria. J Cell Biol. 1985;100:258–264. doi: 10.1083/jcb.100.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R P, Lisowsky T, Parisi M A, Clayton D A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- Kang D, Hamasaki N. Mitochondrial transcription factor A in the maintenance of mitochondrial DNA: overview of its multiple roles. Ann N Y Acad Sci. 2005;1042:101–108. doi: 10.1196/annals.1338.010. [DOI] [PubMed] [Google Scholar]
- Kang D, Kim S H, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Kanki T, Nakayama H, Sasaki N, Takio K, Alam T I, Hamasaki N, Kang D. Mitochondrial nucleoid and transcription factor A. Ann N Y Acad Sci. 2004;1011:61–68. doi: 10.1007/978-3-662-41088-2_7. [DOI] [PubMed] [Google Scholar]
- Garstka H L, Facke M, Escribano J R, Wiesner R J. Stoichiometry of mitochondrial transcripts and regulation of gene expression by mitochondrial transcription factor A. Biochem Biophys Res Commun. 1994;200:619–626. doi: 10.1006/bbrc.1994.1493. [DOI] [PubMed] [Google Scholar]
- Garstka H L, Schmitt W E, Schultz J, Sogl B, Silakowski B, Perez-Martos A, Montoya J, Wiesner R J. Import of mitochondrial transcription factor A (TFAM) into rat liver mitochondria stimulates transcription of mitochondrial DNA. Nucleic Acids Res. 2003;31:5039–5047. doi: 10.1093/nar/gkg717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensler S, Weber K, Schmitt W E, Perez-Martos A, Enriquez J A, Montoya J, Wiesner R J. Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res. 2001;29:3657–3663. doi: 10.1093/nar/29.17.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniura-Weber K, Goffart S, Garstka H L, Montoya J, Wiesner R J. Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 2004;32:6015–6027. doi: 10.1093/nar/gkh921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D F, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- Kaufman B A, Durisic N, Mativetsky J M, Costantino S, Hancock M A, Grutter P, Shoubridge E A. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J. Overview on visceral manifestations of mitochondrial disorders. Neth J Med. 2006;64:61–71. [PubMed] [Google Scholar]
- Anan R, Nakagawa M, Miyata M, Higuchi I, Nakao S, Suehara M, Osame M, Tanaka H. Cardiac involvement in mitochondrial diseases. A study on 17 patients with documented mitochondrial DNA defects. Circulation. 1995;91:955–961. doi: 10.1161/01.cir.91.4.955. [DOI] [PubMed] [Google Scholar]
- Koga Y, Akita Y, Junko N, Yatsuga S, Povalko N, Fukiyama R, Ishii M, Matsuishi T. Endothelial dysfunction in MELAS improved by l-arginine supplementation. Neurology. 2006;66:1766–1769. doi: 10.1212/01.wnl.0000220197.36849.1e. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink J N, Rovio A T, Bruder C E, Bohlooly Y M, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs H T, Larsson N G. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Lewis W, Day B J, Kohler J J, Hosseini S H, Chan S S, Green E C, Haase C P, Keebaugh E S, Long R, Ludaway T, Russ R, Steltzer J, Tioleco N, Santoianni R, Copeland W C. Decreased mtDNA, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiewacz H D, Hamann A. DNA reorganization and biological aging: a review. Biochemistry (Mosc) 1997;62:1275–1284. [PubMed] [Google Scholar]
- Ballinger S W, Patterson C, Knight-Lozano C A, Burow D L, Conklin C A, Hu Z, Reuf J, Horaist C, Lebovitz R, Hunter G C, McIntyre K, Runge M S. Mitochondrial integrity and function in atherogenesis. Circulation. 2002;106:544–549. doi: 10.1161/01.cir.0000023921.93743.89. [DOI] [PubMed] [Google Scholar]
- Brown K A, Didion S P, Andresen J J, Faraci F M. Effect of aging, MnSOD deficiency, and genetic background on endothelial function: evidence for MnSOD haploinsufficiency. Arterioscler Thromb Vasc Biol. 2007;27:1941–1946. doi: 10.1161/ATVBAHA.107.146852. [DOI] [PubMed] [Google Scholar]
- Van der Loo B, Bachschmid M, Skepper J N, Labugger R, Schildknecht S, Hahn R, Mussig E, Gygi D, Luscher T F. Age-associated cellular relocation of Sod 1 as a self-defense is a futile mechanism to prevent vascular aging. Biochem Biophys Res Commun. 2006;344:972–980. doi: 10.1016/j.bbrc.2006.03.224. [DOI] [PubMed] [Google Scholar]
- Van Houten B, Woshner V, Santos J H. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 2006;5:145–152. doi: 10.1016/j.dnarep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Steinman H M, Weinstein L, Brenowitz M. The manganese superoxide dismutase of Escherichia coli K-12 associates with DNA. J Biol Chem. 1994;269:28629–28634. [PubMed] [Google Scholar]
- Schildknecht S, Daiber A, Ghisla S, Cohen R A, Bachschmid M M. Acetaminophen inhibits prostanoid synthesis by scavenging the PGHS-activator peroxynitrite. FASEB J. 2008;22:215–224. doi: 10.1096/fj.06-8015com. [DOI] [PubMed] [Google Scholar]
- Schildknecht S, Bachschmid M, Weber K, Maass D, Ullrich V. Endotoxin elicits nitric oxide release in rat but prostacyclin synthesis in human and bovine vascular smooth muscle cells. Biochem Biophys Res Commun. 2005;327:43–48. doi: 10.1016/j.bbrc.2004.11.132. [DOI] [PubMed] [Google Scholar]
- Spamer C, Pette D. Activities of malate dehydrogenase, 3-hydroxyacyl-CoA dehydrogenase and fructose-1,6-diphosphatase with regard to metabolic subpopulations of fast- and slow-twitch fibres in rabbit muscles. Histochemistry. 1979;60:9–19. doi: 10.1007/BF00495725. [DOI] [PubMed] [Google Scholar]
- Nemeth P M, Pette D, Vrbova G. Comparison of enzyme activities among single muscle fibres within defined motor units. J Physiol. 1981;311:489–495. doi: 10.1113/jphysiol.1981.sp013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerwinski J D, Hovan S C, Mascotti D P. Quantitative nonisotopic nitrocellulose filter binding assays: bacterial manganese superoxide dismutase-DNA interactions. Anal Biochem. 2005;336:300–304. doi: 10.1016/j.ab.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Keele B B, Jr, McCord J M, Fridovich I. Superoxide dismutase from Escherichia coli B: a new manganese-containing enzyme. J Biol Chem. 1970;245:6176–6181. [PubMed] [Google Scholar]
- Wong I, Lohman T M. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci U S A. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe N M, Thornton J M. Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J Mol Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- Melov S, Schneider J A, Day B J, Hinerfeld D, Coskun P, Mirra S S, Crapo J D, Wallace D C. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat Genet. 1998;18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- Faraci F M, Didion S P. Vascular protection: superoxide dismutase isoforms in the vessel wall. Arterioscler Thromb Vasc Biol. 2004;24:1367–1373. doi: 10.1161/01.ATV.0000133604.20182.cf. [DOI] [PubMed] [Google Scholar]
- Oberley L W, Buettner G R. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979;39:1141–1149. [PubMed] [Google Scholar]
- Shadel G S. Mitochondrial DNA, aconitase ‘wraps’ it up. Trends Biochem Sci. 2005;30:294–296. doi: 10.1016/j.tibs.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Guliaeva N A, Kuznetsova E A, Gaziev A I. Proteins associated with mitochondrial DNA protect it against the action of X-rays and hydrogen peroxide. Biofizika. 2006;51:692–697. [PubMed] [Google Scholar]
- Ballinger S W, Patterson C, Yan C N, Doan R, Burow D L, Young C G, Yakes F M, Van Houten B, Ballinger C A, Freeman B A, Runge M S. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res. 2000;86:960–966. doi: 10.1161/01.res.86.9.960. [DOI] [PubMed] [Google Scholar]
- Parisi M A, Clayton D A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- Hopkin K A, Papazian M A, Steinman H M. Functional differences between manganese and iron superoxide dismutases in Escherichia coli K-12. J Biol Chem. 1992;267:24253–24258. [PubMed] [Google Scholar]
- Shen E L, Bogenhagen D F. Developmentally-regulated packaging of mitochondrial DNA by the HMG-box protein mtTFA during Xenopus oogenesis. Nucleic Acids Res. 2001;29:2822–2828. doi: 10.1093/nar/29.13.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinardo M M, Musicco C, Fracasso F, Milella F, Gadaleta M N, Gadaleta G, Cantatore P. Acetylation and level of mitochondrial transcription factor A in several organs of young and old rats. Biochem Biophys Res Commun. 2003;301:187–191. doi: 10.1016/s0006-291x(02)03008-5. [DOI] [PubMed] [Google Scholar]
- Annex B H, Williams R S. Mitochondrial DNA structure and expression in specialized subtypes of mammalian striated muscle. Mol Cell Biol. 1990;10:5671–5678. doi: 10.1128/mcb.10.11.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.