Abstract
Speciation rates among extant lineages of organisms vary extensively, but our understanding of the causes of this variation and, therefore, the processes of speciation is still remarkably incomplete. Both theoretical and empirical studies have indicated that sexual selection is important in speciation, but earlier discussions have focused almost exclusively on the potential role of female mate choice. Recent findings of postmating reproductive conflicts of interest between the sexes suggest a quite different route to speciation. Such conflicts may lead to perpetual antagonistic coevolution between males and females and may thus generate rapid evolutionary divergence of traits involved in reproduction. Here, we assess this hypothesis by contrasting pairs of related groups of insect species differing in the opportunity for postmating sexual conflict. Groups where females mate with many males exhibited speciation rates four times as high as in related groups where females mate only once. Our results not only highlight the general importance of postmating sexual selection in speciation, but also support the recent suggestion that sexual conflict is a key engine of speciation.
Sexual selection has been shown to be a key component in the process of speciation (1–6), but reproductive competition may contribute to the evolution of reproductive isolation in several ways (3). Recent research has suggested that differences in the evolutionary interests of males and females may provide an important route to speciation (7–16). Because conflicts of interest between interacting loci residing within the same genome, but favoring different sexes can result in very rapid antagonistic coevolution (9–10, 16), such conflicts may be capable of generating reproductive isolation (14). Sexual conflict over the postmating interests of males and females is virtually ubiquitous, and stems from competition between males over the fertilization of eggs (17). Males stand to gain from adaptations that increase sperm competition success (7) and female short-term egg production (18), as well as from those that act to decrease female remating rate (9), even if these benefits are achieved at the expense of female fitness (19). Many of these postmating conflicts are mediated by various components transferred to the female in the male seminal fluid and by female receptivity to these substances (9, 20). Whenever female interests are compromised by males, the female reproductive system will evolve to depress these costs, in turn creating perpetual or episodic postmating sexual selection (by sperm competition and/or cryptic female choice) for novel male adaptations by biasing postmating fertilization success among males toward males most able to manipulate female reproduction in their own interests. Such sexually antagonistic adaptations will generate rapid coevolution between male and female reproductive physiology (9) and morphology (8), eventually resulting in reproductive isolation between allopatric populations (11, 14). Here we assess the general importance of such postmating sexual conflict for the rate of speciation, by comparing extant species richness in pairs of related clades of insects differing in the opportunity for postmating sexual conflict.
To test whether the intensity of postmating sexual conflict covaries with speciation rates, we analyzed a series of paired phylogenetic contrasts (21). In each contrast, we compared the number of described extant species in a clade where females typically mate with many different males (polyandry) with the number in a closely related clade where females typically mate with only one male (monandry). In polyandrous species, the ejaculates of several males will compete over fertilizations within the female, and male traits that aid in this reproductive competition will be favored even if they convey costs to females (7, 14, 19, 22). There will thus be ample opportunity for postmating sexual conflict (see above) and therefore also for antagonistic coevolution between the sexes under polyandry (9–10, 12, 16). In monandrous species, in contrast, male ejaculates will not compete and the reproductive success of a male will instead increase with any postmating elevation of his mate's fitness. Male and female interests are hence identical after the mating under monandry (16), provided that no parental care of offspring occurs (23). Postmating sexual conflict, as well as any resulting postmating sexual selection, will thus be absent or minimal in monandrous species. Because the two clades in a given contrast share a common ancestor, the relative number of extant species in these clades will reflect differences in speciation rate. Hence, if postmating sexual conflict plays a significant role in insect speciation, we expect clades with opportunity for such conflicts (i.e., polyandrous) to be more speciose.
Materials and Methods
We searched for potential phylogenetic contrasts (21) in previous reviews (24) and comparative studies (25–28), as well as in reference databases and on the World-Wide Web. Three criteria had to be met for inclusion of a clade/contrast in our analysis. First, reliable and accordant data from several sources on female mating frequencies had to be available. These data typically consisted of female spermatophore/ejaculate counts in natural populations and/or detailed field/laboratory studies of mating behavior. Second, the phylogeny of the clades included in a given contrast had to be well established. Third, only pairs of closely related clades with a documented difference in female mating frequencies (polyandrous versus monandrous mating systems) were included among our contrasts. Since the vast majority of insects are polyandrous (24), each of our contrasts originated by a documented monandrous clade. If the sister clade to this monandrous clade was found to be polyandrous according to our first inclusion criterion, the polyandrous sister clade was used in the comparison. If not, we searched for a documented polyandrous clade as closely related as possible to the focal monandrous clade, and used this clade to construct the contrast. If no closely related polyandrous clade existed, the contrast was discarded. Our selection of phylogenetic contrasts is based on a large number of published references as well as on personal contacts with a large number of expert colleagues. A complete list of these sources can be found in the supplementary material at www.pnas.org.
To avoid potential effects of nonrandom inclusion of contrasts, our selection of contrasts was temporally structured so that all potential cases were first screened and assessed according to the inclusion criteria above. Decisions to include or exclude potential contrasts in our analyses were based solely on these three criteria. The retrieval of species number data (the total number of described species in a clade) was without exception performed subsequent to these decisions, and in no case was a contrast excluded after species number data had been collected. Thus, the selection of contrasts for our analyses was naïve with regards to species number data, and can thus not be nonrandom with respect to the speciosity of different clades.
Because our knowledge of the reproductive biology of well-studied orders (e.g., Lepidoptera and Diptera) far exceeds that of less well-studied orders (e.g., Trichoptera and Neuroptera), our selection was nonrandom with regards to the orders represented. However, the fact that well-studied orders are overrepresented in our data does not in itself present a problem unless these orders differ profoundly from other insects, in which case the generality of our conclusions would be more limited. Our analyses indicate that this is not the case (see below).
Species were classified as monandrous only if the majority (>50%) of mated females had been found to mate only once in their lifetime. Our data set includes larger clades (i.e., families and certain genera) where mating frequency data are not available for all species. Such clades were included only if all available data gave concordant evidence for one of the two mating systems (polyandry or monandry), if these data were deemed reliable, and if all species within the clade were homogeneous with regards to their general reproductive biology. Despite these restrictive criteria, a limited occurrence of polyandrous species in clades classified as monandrous (and vice versa) is possible whenever mating frequency data are not at hand for all species in the clade. Similarly, our definition of monandry allows a limited occurrence of polyandrous females also in some species classified as monandrous. Even though the monandrous and polyandrous clades exhibited a marked difference in the average frequency of female remating in all contrasts included in our analyses, such misclassifications could potentially introduce bias in our analyses. It is important to note, however, that this would contribute only to an underestimation of the true effect of absence of postmating sexual conflict on species richness (25) and would thus render our tests more conservative (i.e., would elevate the type II statistical error rate).
Results
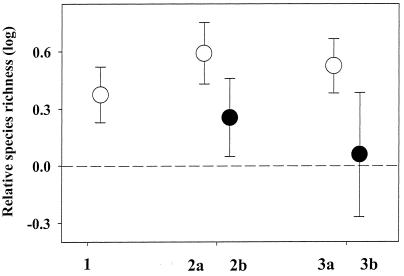
We were able to identify 25 phylogenetic contrasts, representing five different orders, all of which were independent in the sense that no clade was represented in more than one contrast (Table 1). The mean relative species richness, defined as the number of described species in the polyandrous clade divided by that in the monandrous clade, across all phylogenetic contrasts was R̂ = 3.98. The distribution of log-transformed relative species richness R did not differ significantly from normality (Lilliefors test, P = 0.405), and the null hypothesis of H0: log R̂ = 0 was thus tested, and rejected, by a t test (t = 2.45, df = 24, Pα/2 = 0.011). A nonparametric and more conservative analogue of this test, a sign test of H0: R̂ = 1, confirmed this result (Pα/2 = 0.022). Nine of the contrasts involve true sister groups (i.e., the clades share a common and unique ancestor) and 16 involve groups that are more distantly related. Because of the added variance of phylogenetic “noise” in contrasts involving more distantly related groups, the variance of these two types of contrasts might differ (21). However, neither the variance nor the magnitude of log-transformed relative species richness depended significantly on whether the clades in a contrast were sister groups or more distantly related (Levene's test for homogeneity of variances, P = 0.354; t = 1.12, df = 23, P = 0.275) or whether the contrast involved a within- or a between-family comparison (t = 1.53, df = 23, P = 0.139; Levene's test for homogeneity of variances, P = 0.469) (see Fig. 1). Restricting the test of the null hypothesis of H0: log R̂ = 0 to include only sister group contrasts did not alter our conclusions (t = 3.54, df = 8, Pα/2 = 0.004). Further, there were no significant differences between the five orders in the magnitude of relative species richness (ANOVA, F4,20 = 0.864, P = 0.503).
Table 1.
Paired phylogenetic contrasts
| Order | Polyandrous family | Polyandrous clade | No. of species | Monandrous family | Monandrous clade | No. of species |
|---|---|---|---|---|---|---|
| Coleoptera | Anobiidae | Ernobius spp. | 53 | Anobiidae | Xestobium spp. | 10 |
| Dermestidae | Dermestes spp. | 73 | Dermestidae | Trogoderma spp. | 120 | |
| Elateridae | Agriotes spp. | 228 | Elateridae | Selatosomus spp. | 74 | |
| Diptera | Muscidae | Coenosia spp. | 353 | Anthomyiidae | Delia spp. | 289 |
| Cecidomyiidae | Rhopalomyia spp. | 157 | Cecidomyiidae | Mayetiola spp. | 30 | |
| Chironomidae | Chironomus spp. | >300 | Chironomidae | Pontomyia spp. | 4 | |
| Chironomidae | Stictochironomus spp. | 34 | Chironomidae | Clunio spp. | 18 | |
| Drosophilidae | Total for family | 3,400 | Culicidae | Total for family | 3,500 | |
| Dryomyzidae | Total for family | 20 | Calliphoridae | Total for family | >1,000 | |
| Tephritidae | Anastrepha spp. | 196 | Tephritidae | Bactrocera spp. | 486 | |
| Sciaridae | Total for family | 1,750 | Bibionidae | Total for family | 660 | |
| Scatophagidae | Scatophaga spp. | 55 | Muscidae | Musca spp. | 63 | |
| Ephemeroptera | Siphlonuridae | Siphlonurus spp. | 37 | Caenidae | Caenis spp. | 115 |
| Homoptera | Psyllidae | Cacopsylla spp. | >100 | Diaspididae | Aonidiella spp. | 30 |
| Lepidoptera | Noctuidae | Total for family | 21,000 | Psychidae | Total for family | 600 |
| Tortricidae | Choristoneura spp. | 37 | Tortricidae | Epiphyas spp. | 40 | |
| Nymphalidae | Eueides spp. (aliphera clade) | 7 | Nymphalidae | Eueides spp. (vibilia clade) | 5 | |
| Nymphalidae | Heliconius spp. (silvaniform clade) | 15 | Nymphalidae | Heliconius spp. (sara/sapho clade) | 7 | |
| Nymphalidae | Polygonia/Kaniska/Roddia spp. | 18 | Nymphalidae | Nymphalis spp. | 6 | |
| Nymphalidae | Acraea spp. | 240 | Nymphalidae | Cethosia spp. | 13 | |
| Pieridae | Dixeia spp. | 15 | Pieridae | Ascia spp. | 14 | |
| Pieridae | Colias/Zerene spp. | 77 | Pieridae | Phoebis spp. | 16 | |
| Pieridae | Euchloe spp. | 15 | Pieridae | Anthocaris spp. | 14 | |
| Pieridae | Eurema spp. | 85 | Pieridae | Gonepteryx spp. | 6 | |
| Pieridae | Dismorphia spp. | 86 | Pieridae | Leptidea spp. | 8 |
Figure 1.
The ratio of species richness in polyandrous clades to that in monandrous clades across the phylogenetic contrasts. Given is mean (±SE) log-transformed relative species richness of the following: 1, all 25 phylogenetic contrasts; 2a, contrasts involving only true sister taxa; 2b, contrasts not involving sister taxa; 3a, contrasts involving within-family comparisons; and 3b, those involving between-family comparisons. Dashed line indicates the null hypothesis of equal number of species in the two types of clades.
A few key biological characteristics of groups of organisms are known to be associated with extant species richness, the most general of which are trophic ecology, range of geographic distribution, and latitude (3, 21, 29). To assess whether our conclusions could be affected by potentially confounding effects, we tested for the influence of these factors. The polyandrous and monandrous clades shared a similar trophic ecology in 20 of our 25 contrasts (see supplementary material at www.pnas.org). The mean relative species richness did not differ significantly between contrasts involving clades with similar and dissimilar trophic ecologies (Kolmogorov–Smirnov two-sample test, P = 0.117). We nevertheless tested for the effect of mating system on species richness, using only contrasts in which major effects of trophic ecology on relative species richness cannot be present, namely contrasts in which both clades share a similar trophic ecology. The null hypothesis of a similar number of species in polyandrous and monandrous clades (H0: log R̂ = 0) was strongly rejected (t = 3.58, df = 19, Pα/2 < 0.001) when this restricted data set was used. Thus, our conclusions not only were robust against the potentially confounding effects of differences between clades in trophic ecology but were actually strengthened when such effects were removed.
We assessed distributional range by recording the number of biogeographic regions occupied by each clade in our data set (see supplementary material at www.pnas.org). For each contrast, we then formed a ratio between the number of regions occupied by the polyandrous clade to that of the monandrous clade. This measure of relative distribution (D) thus measure the difference in geographic range between the polyandrous and the monandrous clade in each contrast. A value higher than 1 indicates that the polyandrous clade is more widely distributed than the monandrous clade, and a value lower than 1 describes the reverse situation. As expected, the relative distribution D was positively correlated with relative species richness R across all contrasts (Spearman rank correlation, r = 0.527, df = 23, Pα/2 = 0.003). This result simply confirms that the more widely distributed one clade is relative to the other, the more species it tends to contain relative to the other. We then tested for potentially confounding effects of distributional range in two different ways. First, the average distributional ranges of the two types of clades were compared. Polyandrous clades occupied on average 3.88 (SD = 1.88) and monandrous clades occupied on average 3.84 (SD = 2.01) biogeographic regions. The number of biogeographic regions occupied by the two types of clades was not significantly different (Wilcoxon signed rank test, P = 0.867; paired t test, P = 0.929), suggesting that our results were not caused by indirect effects of differences in distribution. Second, and more importantly, we tested whether the predicted relative species richness (R) was equal to or larger than unity when the two clades in a contrast occupied the same number of biogeographic regions. We did this, using the whole data set, in the following way. To yield predicted relative species richness, we performed a linear regression with log-transformed relative species richness as the dependent variable and log-transformed relative distribution as the independent variable (log R = 0.343 + 1.000 log D; Pα/2 = 0.016). The predicted intercept (c) in this regression would be zero under a null hypothesis stating that polyandrous and monandrous clades are equally species rich, i.e., polyandrous and monandrous clades should contain a similar number of species when occupying a similar number of biogeographic regions. This null hypothesis (H0: c = 0; HA: c > 0) was tested and rejected by a t test (c = 0.343, t = 2.54, df = 23, Pα/2 = 0.009), demonstrating that the elevation of the regression was significantly higher than predicted under the null hypothesis. Restricting this test to include only contrasts where both clades share a similar trophic ecology (see above) further increased the confidence of our conclusion (c = 0.482, t = 3.54, df = 18, Pα/2 = 0.001). These intercepts are statistically analogous to, and not significantly different from, the log-transformed mean relative species richness presented above.
Some comparative data suggest that the rate of speciation increases toward the equator, which potentially may contribute to latitudinal diversity gradients (29). We assessed the potential influence of latitude by assigning each biogeographic region a score, based on whether it includes equatorial areas (score = 1) or not (score = 2). We first calculated a latitudinal index for each clade, defined as the average score across all regions occupied by that clade. We then formed a ratio between the latitudinal indices of the poly- to the monandrous clade in each contrast. This ratio was negatively correlated with relative species richness R across all contrasts (Spearman rank correlation, r = −0.602, df = 23, Pα = 0.001), showing that polyandrous clades were relatively more species rich when inhabiting relatively more equatorial areas. We tested for potentially confounding effects of latitude in two different ways. First, we compared the average latitudinal index of the two types of clades. The latitudinal index did not differ between poly- (mean = 1.41, SD = 0.25) and monandrous (mean = 1.42, SD = 0.27) clades (Wilcoxon signed rank test, P = 0.794; paired t test, P = 0.909), suggesting that our main results were not caused by indirect effects of differences in distribution. Second, by analogy with distributional range, we tested whether the predicted relative species richness (R) was equal to or larger than unity when the two clades in a contrast were equally equatorial in their distribution (see above for statistical methods). The null hypothesis of an equal number of species when equally equatorial in their distribution (for log-transformed data; H0: c = 0; HA: c > 0) was tested and rejected by a t test (c = 0.351, t = 2.62, df = 23, Pα/2 = 0.008). As was the case for distributional range, restricting this test to include only contrasts where both clades share a similar trophic ecology further increased the confidence of our conclusion (c = 0.489, t = 3.64, df = 18, Pα/2 = 0.001).
These analyses confirm that speciation in insects indeed seems to be more rapid in more widely distributed clades (21) and in clades inhabiting more equatorial areas (29). However, our main conclusion not only was robust against the potentially confounding effects of differences between poly- and monandrous clades in distribution but was, if anything, strengthened when such effects were controlled for.
Discussion
Our analyses showed that clades with an opportunity for postmating sexual conflict and postmating sexual selection exhibit a strongly elevated level of speciation compared with groups without such opportunity, and this overall conclusion was not affected by taxonomic construction of comparisons or by the ecology and distribution of the groups involved. The magnitude of the detected effect is remarkably large, given that extant species richness represents the net difference between past speciation and extinction. Because extinction rates should if anything be higher under more intense sexual conflict (13, 19, 30), our estimate of a 4-fold overall difference in speciation rate is most certainly a conservative one. Our results not only establish the general importance of sexual selection and sexual conflict in speciation, but also yield specific insights into the processes involved in this effect.
When two partially diverged populations come into secondary contact, selection against hybridization may drive the evolution of reproductive isolation. The general importance of such reinforcement, however, remains one of the greatest controversies in research on speciation (13, 31), and it is clear that reinforcement is not responsible for the pattern detected in our study for the following reasons. In monandrous species, each female will mate with either a con- or a heterospecific male and will thus produce either only conspecific offspring or only hybrids. This contrasts with polyandrous species, in which each female may mate with both con- and heterospecific males and may thus produce a certain proportion of hybrid offspring. For any given degree of hybrid inviability, this results in a lower variance in female fitness (Vwf) and a lower opportunity for selection among females (If = Vwf) against hybridization under polyandry. While the intensity of selection against hybridization (i.e., the covariance between hybridization avoidance and female fitness) may not differ greatly in the two types of clades, it should, if anything, be more intense under monandry compared with polyandry. Reinforcement should thus, if at all contributing to differences in species richness between poly- and monandrous clades, generate a trend opposite to that observed here.
Earlier discussions of the role of sexual selection in speciation have focused almost exclusively on evolutionary divergence of female mating preferences (1–6, 31). However, diverging premating female mate choice is highly unlikely to contribute in any significant way to the pattern documented here. First, unlike the bird species that have been the subjects of earlier comparative studies (32–35), the insect species included in our study exhibit few sexual ornaments indicative of intense female mate choice. Second, sexual selection resulting from premating female choice may not in general be weaker under monandry (a single male may mate with many females also in monandrous species), and variance in male mating success can be high in both polyandrous and monandrous species (35, 36). Third, and most importantly, the most influential and critical difference between monandrous and polyandrous clades is without doubt the absence of postmating sexual conflict and postmating sexual selection under monandry (16, 20, 25).
Postmating sexual selection has received very little attention in discussions of speciation, because it has been believed that this type of selection is stabilizing and therefore lacks the ability to generate rapid evolutionary divergence in reproductive characters (37, 38). Males have been thought to simply optimize their competitive ability within what has been believed to be a relatively static and passive environment, constituted by the female reproductive tract. Recent research, however, has demonstrated that postmating sexual conflict can impel divergent postmating sexual selection among males (9, 16), by sperm competition (12, 39) and/or by cryptic female choice (ref 20; G. Gavrilets, G.A., and U.F., unpublished work), and that the female reproductive tract is neither evolutionarily static nor passive (9, 20, 40, 41). Understanding the evolution of the female reproductive tract is critical, because this sets the stage for sperm competition and dictates cryptic female preferences (20). While mechanisms other than sexual conflict (notably “good-genes” and “Fisherian run-away” selection) could generate divergent evolution of female reproductive tract morphology and physiology (20, 42–44), and thus also lead to increased rates of speciation under polyandry, these scenarios are associated with theoretical problems (refs. 11, 45, 46; G. Gavrilets, G.A., and U.F., unpublished work) and are currently not supported by experimental data (9, 41).
Studies of conspecific sperm precedence in insects have confirmed that postmating sexual selection, most likely propelled by sexually antagonistic coevolution (38, 39), rapidly generates postmating reproductive barriers between closely related species (39, 47–49). Increased rates of speciation should thus be an evolutionary corollary of postmating sexual conflict (11, 14). Our study supports this suggestion. Sexual conflict indeed seems to be a key “engine of speciation.”
Supplementary Material
Acknowledgments
This study was made possible by the generous help of a large number of experts on the biology and systematics of various groups of insects (see supplementary material, available either at www.pnas.org or from the author upon request). We are deeply grateful to all of these colleagues. R. Alexander, J. Andrés, P. Eklöv, L. Persson, L. Rowe, and three anonymous reviewers provided constructive comments on the manuscript. Financial support was provided by the Swedish Natural Science Research Council.
References
- 1.Lande R. Proc Natl Acad Sci USA. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dieckman U, Doebeli M. Nature (London) 1999;400:354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- 3.West-Eberhard M J. Quart Rev Biol. 1983;58:155–183. [Google Scholar]
- 4.Seehausen O, van Alphen J J M, Witte F. Science. 1997;277:1808–1811. [Google Scholar]
- 5.Hoy R R, Hoikkala A, Kaneshiro K. Science. 1988;240:217–219. doi: 10.1126/science.3127882. [DOI] [PubMed] [Google Scholar]
- 6.McMillan W O, Jiggins C D, Mallet J. Proc Natl Acad Sci USA. 1997;94:8628–8633. doi: 10.1073/pnas.94.16.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman T, Liddle L F, Kalb J M, Wolfner M F, Partridge L. Nature (London) 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- 8.Arnqvist G, Rowe L. Proc R Soc London Ser B. 1995;261:123–127. [Google Scholar]
- 9.Rice W R. Nature (London) 1996;381:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 10.Rice W R. Proc Natl Acad Sci USA. 1998;95:6217–6221. doi: 10.1073/pnas.95.11.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander R D, Marshall D C, Cooley J R. In: The Evolution of Mating Systems in Insects and Arachnids. Choe J C, Crespi B J, editors. Cambridge, U.K.: Cambridge Univ. Press; 1996. pp. 4–31. [Google Scholar]
- 12.Chapman T, Partridge L. Nature (London) 1996;381:189–190. doi: 10.1038/381189a0. [DOI] [PubMed] [Google Scholar]
- 13.Parker G A, Partridge L. Phil Trans R Soc London Ser B. 1998;353:261–274. doi: 10.1098/rstb.1998.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice W R. In: Endless Forms: Species and Speciation. Howard D J, Berlocher S H, editors. New York: Oxford Univ. Press; 1998. pp. 261–270. [Google Scholar]
- 15.Gavrilets S. Nature (London) 2000;403:886–889. doi: 10.1038/35002564. [DOI] [PubMed] [Google Scholar]
- 16.Holland B, Rice W R. Proc Natl Acad Sci USA. 1999;96:5083–5088. doi: 10.1073/pnas.96.9.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stockley P. Trends Ecol Evol. 1997;12:154–159. doi: 10.1016/s0169-5347(97)01000-8. [DOI] [PubMed] [Google Scholar]
- 18.Chapman T, Miyatake T, Smith H K, Partridge L. Proc R Soc London Ser B. 1998;265:1879–1894. doi: 10.1098/rspb.1998.0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice W R. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 20.Eberhard W G. Female Control: Sexual Selection by Cryptic Female Choice. Princeton, NJ: Princeton Univ. Press; 1996. [Google Scholar]
- 21.Barraclough T G, Vogler A P, Harvey P H. Phil Trans R Soc London Ser B. 1998;353:241–249. [Google Scholar]
- 22.Rice W R. Science. 1992;256:1436–1439. doi: 10.1126/science.1604317. [DOI] [PubMed] [Google Scholar]
- 23.Trivers R L. In: Sexual Selection and the Descent of Man. Campbell B, editor. Chicago: Aladine; 1972. pp. 136–179. [Google Scholar]
- 24.Thornhill R, Alcock J. The Evolution of Insect Mating Systems. Cambridge, MA: Harvard Univ. Press; 1983. [Google Scholar]
- 25.Arnqvist G. Nature (London) 1998;393:784–786. [Google Scholar]
- 26.Ridley M. Anim Behav. 1989;37:535–545. [Google Scholar]
- 27.Ridley M. Funct Ecol. 1990;4:75–84. [Google Scholar]
- 28.Ridley M. Biol Rev. 1988;63:509–549. [Google Scholar]
- 29.Cardillo M. Proc R Soc London Ser B. 1999;266:1221–1225. [Google Scholar]
- 30.Tanaka Y. J Theor Biol. 1996;180:197–206. doi: 10.1006/jtbi.1996.0096. [DOI] [PubMed] [Google Scholar]
- 31.Coyne J A, Orr H A. Phil Trans R Soc London Ser B. 1998;353:287–305. doi: 10.1098/rstb.1998.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barraclough T G, Harvey P H, Nee S. Proc R Soc London Ser B. 1995;259:211–215. [Google Scholar]
- 33.Møller A P, Cuervo J J. Evolution. 1998;52:859–869. doi: 10.1111/j.1558-5646.1998.tb03710.x. [DOI] [PubMed] [Google Scholar]
- 34.Mitra S, Landel H, Pruett Jones S. Auk. 1996;113:544–551. [Google Scholar]
- 35.Owens I P F, Hartley I R. Proc R Soc London Ser B. 1998;265:397–407. [Google Scholar]
- 36.Shellman-Reeve J S. Proc R Soc London Ser B. 1999;266:137–144. [Google Scholar]
- 37.Howard D J, Reece M, Gregory P G, Chu J, Cain M L. In: Endless Forms: Species and Speciation. Howard D J, Berlocher S H, editors. New York: Oxford Univ. Press; 1998. pp. 279–288. [Google Scholar]
- 38.Howard D J. Annu Rev Ecol Syst. 1999;30:109–132. [Google Scholar]
- 39.Price C S C. Nature (London) 1997;388:663–666. doi: 10.1038/41753. [DOI] [PubMed] [Google Scholar]
- 40.Wilson N, Tubman S C, Eady P E, Robertson G W. Proc R Soc London Ser B. 1997;264:1491–1495. [Google Scholar]
- 41.Clark A G, Begun D J, Prout T. Science. 1999;283:217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- 42.Eberhard W G. Am Nat. 1993;142:564–571. doi: 10.1086/285556. [DOI] [PubMed] [Google Scholar]
- 43.Eberhard W G. In: Sperm Competition and Sexual Selection. Birkhead T R, Møller A P, editors. London: Academic; 1998. pp. 91–116. [Google Scholar]
- 44.Keller L, Reeve H K. Adv Study Behav. 1995;24:291–315. [Google Scholar]
- 45.Kirkpatrick M, Ryan M J. Nature (London) 1991;350:33–38. [Google Scholar]
- 46.Arnqvist G. Biol J Linn Soc. 1997;60:365–379. [Google Scholar]
- 47.Nakano S. Kontyû. 1985;53:112–119. [Google Scholar]
- 48.Wade M J, Patterson H, Chang N W, Johnson N A. Heredity. 1994;72:163–167. doi: 10.1038/hdy.1994.23. [DOI] [PubMed] [Google Scholar]
- 49.Gregory P G, Howard D J. Evolution. 1997;48:705–710. doi: 10.1111/j.1558-5646.1994.tb01355.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.