Abstract
Butirosin is unique among the naturally occurring aminoglycosides, having a substituted amino group at position 1 (N1) of the 2-deoxystreptamine ring with an (S)-4-amino-2-hydroxybutyrate (AHB) group. While bacterial resistance to aminoglycosides can be ascribed chiefly to drug inactivation by plasmid-encoded aminoglycoside-modifying enzymes, the presence of an AHB group protects the aminoglycoside from binding to many resistance enzymes, and hence, the antibiotic retains its bactericidal properties. Consequently, several semisynthetic N1-substituted aminoglycosides, such as amikacin, isepamicin, and netilmicin, were developed. Unfortunately, butirosin, amikacin, and isepamicin are not resistant to inactivation by 3′-aminoglycoside O-phosphotransferase type IIIa [APH(3′)-IIIa]. We report here the crystal structure of APH(3′)-IIIa in complex with an ATP analog, AMPPNP [adenosine 5′-(β,γ-imido)triphosphate], and butirosin A to 2.4-Å resolution. The structure shows that butirosin A binds to the enzyme in a manner analogous to other 4,5-disubstituted aminoglycosides, and the flexible antibiotic-binding loop is key to the accommodation of structurally diverse substrates. Based on the crystal structure, we have also constructed a model of APH(3′)-IIIa in complex with amikacin, a commonly used semisynthetic N1-substituted 4,6-disubstituted aminoglycoside. Together, these results suggest a strategy to further derivatize the AHB group in order to generate new aminoglycoside derivatives that can elude inactivation by resistance enzymes while maintaining their ability to bind to the ribosomal A site.
Aminoglycosides encompass a vast group of molecules with important antibiotic activities. They are often prescribed for the treatment of serious nosocomial infections and, in some cases, protozoan infections (6). Moreover, their potential as antiviral agents has also been under intense investigation in recent years (33, 47, 50). The high efficacy of aminoglycosides as antibacterial agents is in part due to their bactericidal capability. Most aminoglycosides kill the bacteria by targeting the A site of the 16S rRNA (40). Upon binding of the drug, key nucleotides in the decoding region of the ribosome undergo conformational changes that promote the interactions between mRNA and near-cognate or noncognate tRNA, leading to errors in protein translation (45). Unfortunately, the extensive use of aminoglycosides has undermined their effectiveness, due to the emergence of resistance in pathogens. Resistance to aminoglycosides can predominantly be attributed to covalent modification of the drug catalyzed by plasmid-encoded aminoglycoside-modifying enzymes, especially the O-phosphotransferases, which generally give high levels of resistance (16). Covalent modification diminishes the antibiotics' ability to bind to their targets, and therefore, they can no longer exert their bactericidal effects (34).
Butirosins are unique among the naturally occurring aminoglycosides due to the (S)-4-amino-2-hydroxybutyrate (AHB) side chain moiety substituted at the amino group at position 1 (N1) of the 2-deoxystreptamine ring. While most aminoglycosides routinely lose their potency due to inactivation by an aminoglycoside-modifying enzyme, butirosin is able to elude many inactivating enzymes, thus retaining its bactericidal capabilities (51). This observation prompted the development of semisynthetic N1-substituted aminoglycoside antibiotics such as amikacin and arbekacin (kanamycin A and dibekacin derivated at N1 by AHB, respectively) (23, 26, 30, 31), isepamicin (gentamicin B substituted with 4-amino-2-hydroxypropionyl at N1) (41), and netilmicin (sisomicin with an ethyl group introduced at N1) (54). These compounds have been shown to be clinically useful against some aminoglycoside-resistant strains, such as methicillin-resistant Staphylococcus aureus (29). It is thought that the AHB and other acyl side chains at the 1-amino position hinder binding to the aminoglycoside-modifying enzymes without affecting binding to the ribosome A site. Recently, the capability of amikacin binding to the ribosome was illustrated by Kondo et al. in the crystal structure of an RNA fragment containing the A site in complex with amikacin (28).
Nonetheless, many of these N1-substituted aminoglycosides are not immune to inactivation by all aminoglycoside kinases. For example, butirosin, amikacin, and isepamicin can be inactivated by 3′-aminoglycoside O-phosphotransferase type IIIa [APH(3′)-IIIa] via the addition of a phosphate group. APH(3′)-IIIa is found in many gram-positive bacteria, such as enterococci and staphylococci, and is perhaps the most-studied aminoglycoside resistance factor, as it has an unusually broad substrate profile (37).
Considerable knowledge of the accommodation of the structurally diverse substrates by APH(3′)-IIIa has been gained in recent years. Crystal structures of APH(3′)-IIIa in the apo, nucleotide-bound, and ternary complexes, with both the nucleotide and the aminoglycoside antibiotic, have been determined (9, 18, 24). The ternary complexes confirmed the importance of electrostatic interactions in substrate binding and revealed that discrete pockets and a flexible binding loop are used to accommodate aminoglycosides of different shapes and sizes (18). To date, the conformations of several aminoglycosides, including those that are substituted at N1 such as amikacin, isepamicin, and butirosin, bound to APH(3′)-IIIa, have been studied by nuclear magnetic resonance (NMR) spectroscopy (12-14, 48). These studies suggest that different aminoglycosides may assume markedly different conformations and that there is little similarity between the binding properties of 4,5- and 4,6-disubstituted aminoglycosides. Moreover, a comparison of the aminoglycoside conformations observed in the ternary crystal structures of APH(3′)-IIIa and those obtained from NMR experiments shows limited resemblance. Most importantly, none of the NMR studies have indicated how the substitution at N1 is accommodated by the kinase. Here, we report the crystal structure of APH(3′)-IIIa in complex with AMPPNP [adenosine 5′-(β,γ-imido)triphosphate; a nonhydrolyzable ATP analog] and butirosin A, as well as a computationally deduced model for APH(3′)-IIIa bound with amikacin, a prototypical semisynthetic N1-substituted aminoglycoside. These results improve our understanding of the effectiveness of APH(3′)-IIIa as a resistant enzyme and provide information for the development of new derivatives.
MATERIALS AND METHODS
Crystallization and data collection.
APH(3′)-IIIa was obtained based on the procedure described by McKay et al. (37). Butirosin A-bound APH(3′)-IIIa was crystallized from a solution containing 15 mg/ml of protein, 2.5 mM of AMPPNP (Sigma-Aldrich, St. Louis, MO), 1.5 mM of butirosin A (Sigma-Aldrich, St. Louis, MO), and 2 mM MgCl2 using the hanging-drop technique. After incubation of 1 week at 22°C, small, needle-shaped crystals that were approximately 0.25 mm long appeared under one condition, consisting of 40% (vol/vol) polyethylene glycol 600, 0.1 M sodium acetate (pH 4.5), and 0.2 M magnesium chloride. Additive screens I to III (Hampton Research) were used to enhance crystal quality. Upon the inclusion of one of the additives, 3% (wt/vol) d-(+)-sucrose, the size and shape of the crystals were considerably improved. The resultant crystals were egg shaped and measured approximately 0.40 mm in length and 0.25 mm at the widest part in the middle. These crystals belonged to space group P42212, with unit cell dimensions of a = b = 80.1 Å and c = 110 Å, and contained one APH(3′)-IIIa molecule in each asymmetric unit. Resolution data (2.4 Å) were collected from a single crystal under cryogenic conditions at beamline X8C of the National Synchrotron Light Source, equipped with an ADSC Quantum charge-coupled-device detector (λ = 1.072 Å). Intensities were integrated and scaled using the HKL program suite (44), giving statistics summarized in Table 1.
TABLE 1.
Diffraction data and refinement statistics
| Statistic(s) | Value(s) for APH (3′)-IIIa-AMPPNP- butirosin A |
|---|---|
| Data collection statistics | |
| Space group | P42212 |
| Unit cell parameters | a = b = 80.1 Å, |
| c = 111 Å; | |
| α = β = γ = 90° | |
| Resolution limit (Å) | 2.4 |
| No. of reflections observed (unique) | 201,410 (14,576) |
| Data redundancy (outer shell) | 14.1 (13.1) |
| Completeness (outer shell) (%) | 99.1 (94.8) |
| Rsym (outer shell) (%) | 4.4 (13.7) |
| Mean I/σ(I) (outer shell) | 44.1 (19.4) |
| Model refinement statisticsa | |
| No. of reflections | |
| Working set | 14,475 |
| Test set | 1,473 |
| Total no. of atoms | 2,297 |
| No. of protein atoms | 2,170 |
| No. of Mg2+ atoms | 2 |
| No. of cofactor atoms | 31 |
| No. of substrate atoms | 38 |
| No. of solvent atoms | 85 |
| Rcryst/Rfree | 0.217/0.261 |
| RMSD | |
| Bond length (Å) | 0.007 |
| Bond angle (°) | 1.286 |
Ten percent of randomly selected reflections were designated as test reflections for use in R cross-validation.
Structure determination and refinement.
The structure of APH(3′)-IIIa with AMPPNP and butirosin A was solved by molecular replacement using AMoRe (42), as implemented in the CCP4 suite of programs (11). The structure of APH(3′)-IIIa-ADP-kanamycin (18), excluding the ligands and solvent molecules, was used as the search model. After several cycles of refinement using the CNS software (8), AMPPNP, Mg2+, and butirosin A molecules were placed in the active site based on difference electron density maps (2Fo − Fc and Fo − Fc). Ideal stereochemistry applied to butirosin A during subsequent refinement was based on that of neomycin B and values from the energy-minimized conformation obtained from the molecular mechanics program MM2 (1, 10), implemented in Chem3D (CambridgeSoft). Examination of initial electron density maps showed that the aminoglycoside-binding loop (residues 150 to 165) and the hinge region (residues 101 to 106) required remodeling. Based on difference electron density maps as well as simulated annealing omit maps, these sections were rebuilt using the program O (25). Successive cycles of refinement were alternated with manual intervention, and the addition of solvent molecules was continued until no significant improvement in the model statistics was observed. Final refinement statistics are given in Table 1.
Modeling of APH(3′)-IIIa ternary complex with amikacin.
The initial conformation of amikacin was taken from the crystal structure of the antibiotic bound to an oligonucleotide containing the bacterial ribosome A site (Protein Data Bank ID: 2G5Q) (28). Amikacin was then modeled into the aminoglycoside-binding pocket of APH(3′)-IIIa, according to the butirosin A ternary complex reported here, by superposing the 2-deoxystreptamine and the prime rings of the aminoglycosides. AMPPNP and the magnesium ions from the butirosin A ternary complex were included in the amikacin model during the simulation.
Molecular dynamics simulations were performed with the GROMACS (version 3.3.1) suite of programs (5, 32), using the GROMACS 87 force field. The coordinates of AMPPNP and amikacin were submitted to the Dundee PRODRG2 server (52) to generate GROMACS topology files and to add polar hydrogens. The topology file of amikacin was adjusted in order to institute restraints on the conformation of the ring structures of the aminoglycoside. A truncated octahedron periodic water box was made and was solvated using the simple point charge water model. Twenty-one sodium counterions were added to neutralize the overall charge. After energy minimization using the steepest descent method, the system was subjected to a 20-ps position-restrained dynamics simulation. The protein atom positions were restrained, as prescribed by the linear constraint algorithm (22), while the solvent and ligands were allowed to relax about the protein. Particle-mesh Ewald electrostatics (15, 17), in combination with the temperature and pressure coupling methods of Berendsen et al. (4), were used. Next, a 100-ps molecular dynamics simulation was carried out under the same conditions, except the restraints on the atomic positions were removed. An average structure was calculated and then further refined using a steepest descent and conjugate gradient energy minimization. The resultant coordinates can be found in the supplemental material.
Protein structure accession number.
Coordinates have been deposited in the Protein Data Bank under accession number 3H8P.
RESULTS AND DISCUSSION
Overall structure of butirosin A-bound APH(3′)-IIIa.
To date, seven crystal structures of APH(3′)-IIIa have been determined. They include the AMPPNP and butirosin A-bound structure reported here, the apo form (9), the ADP- and AMPPNP-bound structures (9, 24), the ternary complexes with ADP and kanamycin A or neomycin B (18), and the inactive form of the enzyme inhibited by an ankyrin repeat protein (27). These structures confirm the overall rigidity of APH(3′)-IIIa, as no gross domain movements can be detected upon the binding of natural ligands in the active site. However, four segments have been consistently observed to display differing conformations in the various APH(3′)-IIIa structures. These segments are residues 21 to 26, 100 to 112, 147 to 170, and 226 to 238. A detailed description has been reported previously (9, 18). In brief, segments encompassing residues 100 to 112 and 226 to 238 are inherently flexible, whereas the conformation of the other two segments is dependent on the nature of the bound ligand. Residues 21 to 26 are associated with nucleotide binding and residues 147 to 170 are associated with aminoglycoside binding.
Nucleotide binding.
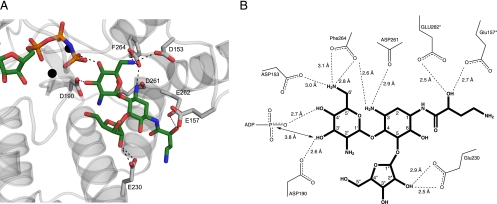
Unlike the kanamycin A and neomycin B structures, which have a bound nucleotide in the form of ADP, the butirosin A ternary complex was crystallized with an ATP analogue, AMPPNP. The AMPPNP molecule can be placed unambiguously into the electron density found in the cleft between the N and C termini of the APH(3′)-IIIa-butirosin A complex in a location and orientation identical to those of the other nucleotide-bound APH(3′)-IIIa complexes (Fig. 1). In addition, residues 21 to 26, which form a lid over the phosphate moieties of the bound nucleotide, adopt the same general conformation in all the structures in complex with a nucleotide. However, in contrast to the AMPPNP binary structure of APH(3′)-IIIa (9), an additional interaction is observed between the main chain amide of Met26 and a γ-phosphate oxygen. This observation is in line with the postulated role of Met26 in stabilizing the transfer of the phosphoryl group (49).
FIG. 1.
(A) Cartoon representation of the crystal structure of APH(3′)-IIIa in complex with AMPPNP and butirosin A. Magnesium ions are shown in black. (B) Simulated annealing Fo − Fc omit map for AMPPNP, Mg ions, and butirosin A, contoured at 2.0 σ.
Aminoglycoside binding.
Butirosin A occupies the acidic substrate-binding pocket in the same manner as neomycin B, utilizing subsites A and C to make contact with the enzyme, where subsite A holds the core 2-deoxystreptamine and the prime rings and subsite C accommodates the ribose substituted at position 5 of the central ring (18) (Fig. 1 and 2). In the presence of AMPPNP, a hydrogen bond is observed between a γ-phosphate oxygen and the 4′ OH of butirosin A. This interaction plausibly contributes to the proper alignment of the 3′ OH for catalysis. Phosphoryl transfer can occur in an entirely associative or dissociative manner. The associative mechanism is distinguished by the small reaction coordinate distance and with evidence of a trigonal bipyramidal intermediate, corroborating with a nucleophilic attack that precedes leaving group bond breakage. At the other extreme, phosphoryl transfer can take place via a dissociative mechanism, where the nucleophilic attack follows leaving group bond breakage. This is characterized by the large reaction coordinate distance, generating a full metaphosphate structure (7, 35, 36). The structure reveals that the 3′ OH of the aminoglycoside is located 3.8 Å from the γ-phosphorus atom. Although evidence so far supports a dissociative phosphoryl transfer mechanism for APH(3′)-IIIa (7), the distance of 3.8 Å observed here appears to be more consistent with a transition state of greater associative properties (46). Nonetheless, it has been noted that most enzymes are unlikely to adopt a fully associative or dissociative mechanism. The actual mechanism may lie anywhere between the two extremes (39). The crystal structure reported here could be a representation of the ground state of the enzyme, with both ligands bound, prior to any bond breakage. The distances and positions of the ligands may undergo adjustment as the reaction proceeds.
FIG. 2.
(A) Cartoon representation of the aminoglycoside-binding site of the butirosin A ternary complex of APH(3′)-IIIa. Residues interacting with butirosin A are drawn as gray sticks, and their hydrogen bond interactions with butirosin A are shown as dashed lines. The color scheme is the same as that shown in Fig. 1. (B) Schematic representation of the hydrogen bonding interactions between butirosin A and APH(3′)-IIIa. The distance between the γ-phosphate of the nucleotide and the reactive 3′-hydroxyl of the aminoglycoside substrate is also indicated. The asterisks denote the amino acid residues that underwent a conformation change in the amikacin-bound model of APH(3′)-IIIa.
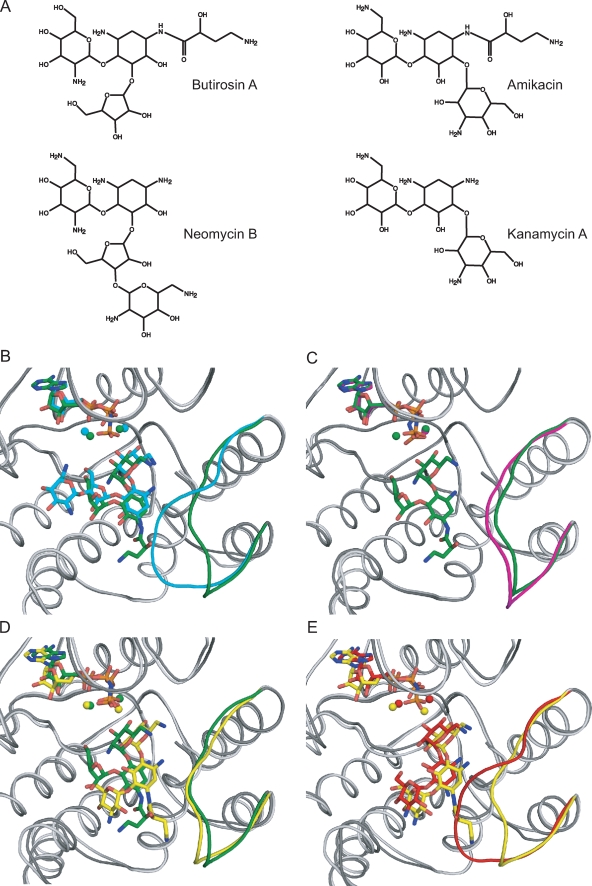
Although the binding properties of butirosin and neomycin are largely analogous, the enzyme's antibiotic-binding loop adopts dramatically different conformations. The conformation of this loop differs, depending on the presence or absence as well as the structure of the antibiotic substrates. The antibiotic-binding loop located between α-helices A and B has been shown to be flexible in the absence of a bound aminoglycoside substrate (9). However, upon the binding of kanamycin A and neomycin B, the loop undergoes a significant conformation change (18). This segment folds over toward the antibiotic substrate, and several residues in this section form interactions with the substrate, thereby completing the aminoglycoside-binding pocket. However, in the presence of butirosin A, the loop assumes an open conformation, akin to that observed in the binary enzyme structures, in order to create room for the AHB group (Fig. 3B and C). Consequently, the antibiotic-binding pocket cannot be completed in the same manner, and some stabilizing interactions are forfeited. The open loop conformation in the presence of an N1-substituted aminoglycoside was not anticipated, based on the information obtained from our previous studies. This structure illustrates the importance of nonspecific electrostatic and van der Waals interactions for aminoglycoside binding to APH(3′)-IIIa.
FIG. 3.
(A) Chemical structure of several aminoglycoside antibiotics. (B to E) Superpositions of APH(3′)-IIIa, shown in ribbon representation, bound with various ligands, drawn as sticks. The nucleotide, magnesium ions, aminoglycoside, and antibiotic-binding loops are highlighted in color. (B) APH(3′)-IIIa bound with butirosin A shown in dark gray/green is superposed on the neomycin B complex colored in light gray/cyan. (C) The butirosin A ternary complex is shown in dark gray/green, and the AMPPNP binary structure is shown in light gray/magenta. (D) The butirosin A-bound structure shown in dark gray/green is overlaid with the amikacin-bound model shown in light gray/yellow. (E) The kanamycin A ternary complex is shown in dark gray/red, and the APH(3′)-IIIa model bound with amikacin is shown in light gray/yellow. The RMSD value of the α-carbon atoms for residues 150 to 165 of the butirosin A ternary complex and the binary structure is 1.63 Å, whereas this value is 4.23 Å between the APH(3′)-IIIa-AMPPNP-butirosin A and APH(3′)-IIIa-ADP-kanamycin A or -neomycin B structures.
The difference in the conformation of the antibiotic-binding loop between the butirosin A ternary complex and the kanamycin A as well as the neomycin B ternary complexes is reflected in their root mean square deviation (RMSD) values in this region (Fig. 3). Moreover, the temperature factors in this region of the butirosin A-bound complex are comparable to those in the nucleotide-bound enzyme structures, which are noticeably higher than those in the kanamycin A and neomycin B ternary structures. The average B factor in the antibiotic-binding loop for the ADP-bound and butirosin A-bound enzymes is 1.4 standard deviations above the mean, whereas the corresponding values for kanamycin A- and neomycin B-bound complexes are 0.4 and 1.0, respectively. The open loop conformation can be ascribed to the AHB at N1 of the central ring, which hinders the loop from approaching and forming as many interactions with the aminoglycoside.
Most of the hydrogen bond interactions made between APH(3′)-IIIa and butirosin A, kanamycin A, or neomycin B, notably those bonds made with the core moiety (the central 2-deoxystreptamine and the prime rings) of the aminoglycosides, are conserved. However, since the antibiotic-binding loop adopts an open conformation in order to accommodate the AHB, differences in the patterns of hydrogen bond interactions are observed (Fig. 2). For example, the side chain of Glu157 no longer makes contacts with the core moiety of butirosin A but is now involved in hydrogen bonding of the hydroxyl of the butyryl group. In addition, the higher level of flexibility in the aminoglycoside-binding loop, as indicated by elevated temperature factor values, may result in less-stable interactions with the aminoglycoside substrate. This may account for the lower binding affinity (higher Km) of butirosin A relative to those of kanamycin A and neomycin B. The APH(3′)-IIIa-AMPPNP-butirosin A structure illustrates the importance of the pliable aminoglycoside-binding loop in the recognition and binding of structurally diverse aminoglycoside substrates.
Model of amikacin binding to APH(3′)-IIIa.
Amikacin is the first semisynthetic aminoglycoside with an N1-substituted AHB. It differs from butirosin A in that it is a derivative of kanamycin A, a 4,6-disubstituted aminoglycoside. In contrast to butirosin A, which lacks good pharmaceutical properties, amikacin has had great success and remains an effective treatment for serious infections, especially those caused by pathogens resistant to other aminoglycosides such as kanamycin, tobramycin, and gentamicin (2). Although amikacin can be inactivated by APH(3′)-IIIa in vitro, only a low level of resistance is detected in vivo. Nevertheless, synergy of amikacin with a cell wall-active antibiotic is eliminated in S. aureus and enterococcal isolates expressing APH(3′)-IIIa (55). Attempts were made to crystallize APH(3′)-IIIa with amikacin without success. In the absence of a crystal structure, we have modeled amikacin into the antibiotic-binding pocket of APH(3′)-IIIa, based on our knowledge of butirosin A binding, in order to speculate how a 4,6-disubstituted aminoglycoside with an AHB substitution at N1 is accommodated in the enzyme.
The conformation of amikacin was taken from the crystal structure of an RNA fragment solved with a bound amikacin (28). When the amikacin is overlaid with kanamycin A and butirosin A from the APH(3′)-IIIa ternary structures, it is evident that their conformations are remarkably similar. This similarity has previously been illustrated by the comparison of APH(3′)-IIIa-bound kanamycin A and neomycin B (18), ribosome-bound paromomycin I, RNA fragment-bound tobramycin (53), and APH(3′)-IIa-bound kanamycin A (43). Based on this knowledge, it can be deduced that the ring structures of amikacin bound to APH(3′)-IIIa should not deviate significantly from those found in the RNA fragment-bound structure. Therefore, the conformation of the ring structures of amikacin was maintained by imposing restraints on the dihedral angles during the course of molecular dynamics. The resulting model shows that AMPPNP, magnesium ions, and amikacin remain in their respective binding sites, adopting positions and orientations similar to their equivalents found in the other aminoglycoside-bound ternary structures (Fig. 3D and E). Although the AHB group of amikacin is in a different conformation than that of butirosin A, the modeled aminoglycoside is able to fit in the binding site without any significant movement in the antibiotic-binding loop and the rest of the enzyme compared to the butirosin A-bound structure. The overall and the main chain RMSD between the butirosin-bound APH(3′)-IIIa and the amikacin-bound model of the enzyme are 1.2 Å and 0.8 Å, respectively. These values are in the same range as those between the kanamycin A- and neomycin B-bound ternary structures of APH(3′)-IIIa (1.2 Å and 0.4 Å, respectively). This result is not unexpected, since similar types and magnitudes of movements in the protein would be expected of binding a 4,5-disubstituted and a 4,6-disubstituted aminoglycoside. However, the side chains of Glu157 and Glu262, which make direct contact with the AHB group of butirosin, adopt different rotamers in order to accommodate the AHB group of amikacin. Based on this model, it can be inferred that isepamicin would bind to APH(3′)-IIIa in the same manner as amikacin. Since isepamicin is substituted at N1 with a 4-amino-2-hydroxypropionyl group containing one less carbon than the butyryl group in amikacin, isepamicin should be able to be accommodated by the enzyme without any difficulty. Although the other semisynthetic N1-substituted aminoglycosides arbekacin and netilmicin are not substrates of APH(3′)-IIIa since they lack the 3′-hydroxyl functional group, they are competitive inhibitors of the enzyme (38). It can also be extrapolated that arbekacin and netilmicin, which are substituted with an AHB group and an ethyl group at N1, respectively, would also be able to bind to APH(3′)-IIIa.
Implications for future semisynthetic N1-substituted aminoglycosides.
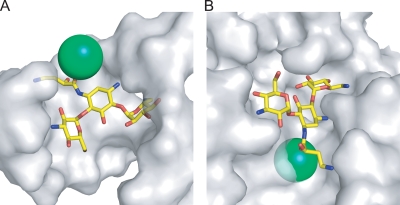
The clinical success of amikacin, isepamicin, and arbekacin have validated the efficacy of aminoglycosides derivated at N1 of the 2-deoxystreptamine ring. Nonetheless, incidents of resistance to these semisynthetic N1-substituted aminoglycosides are slowly rising, including those to arbekacin, the most recent addition to the aminoglycoside antibiotic arsenal (3). Previously, the crystal structures of various aminoglycoside-bound A site and kanamycin A- or neomycin B-bound ternary complexes of APH(3′)-IIIa (18) have contributed to the design of several promising paromomycin derivatives (19-21). The additional information provided by the butirosin A crystal structure and the amikacin-bound model of APH(3′)-IIIa reported here could be implemented to further improve the prospect of new aminoglycoside derivatives. Our analysis suggests that even though enzyme-mediated resistance to the existing N1-substituted aminoglycosides has developed, further derivation of the AHB tail may still be a viable option for generating antibiotics with dramatically reduced susceptibility to resistance mechanisms (Fig. 4). In both the butirosin-bound structure and amikacin-bound model of APH(3′)-IIIa, the carbonyl points toward the center of the enzyme. In contrast, the same group in the amikacin bound to an RNA fragment (28) points to an open end of the groove and is not involved in any interaction with the A site. Therefore, it can be deduced that a bulky group substituting the oxygen at this position of the AHB tail would generate an aminoglycoside capable of evading resistance mediated by APH(3′)-IIIa, while retaining its ability to bind to the A site and precipitate bacterial cell death.
FIG. 4.
Molecular surfaces of the ribosomal A site (A) (28) and APH(3′)-IIIa around the amikacin-binding site (B). Amikacin is shown in yellow sticks, and the green sphere represents the bulky group that could be added to replace the carbonyl of the AHB tail. An appropriate substitution at this location should not interfere with the binding of the aminoglycoside derivative to the A site. However, such an addition would not be accommodated by the APH(3′)-IIIa resistance enzyme.
Concluding remarks.
The significance of the aminoglycoside-binding loop in accepting and positioning structurally diverse aminoglycosides in the binding pocket of APH(3′)-IIIa is emphasized by the structure of the butirosin ternary complex. While the presence of AHB at N1 of aminoglycosides results in a reduction in binding affinity toward many resistance factors, butirosin, amikacin, and isepamicin are unable to evade the recognition and inactivation by APH(3′)-IIIa. The structure of the butirosin A ternary complex illustrates that the substrate-binding pocket of APH(3′)-IIIa is highly malleable, due to the flexible substrate-binding loop. The loop accommodates butirosin A by receding from the core of the aminoglycoside to make space for the AHB group at the central ring. Modeling of the amikacin-bound enzyme suggests that this conformational adaptation is the basis for conferring resistance to other clinically relevant N1-substituted aminoglycosides. Thus, these studies provide structural insights into APH(3′)-IIIa-mediated resistance to AHB-containing aminoglycoside antibiotics. These insights can be exploited for the development of aminoglycoside derivatives with reduced sensitivity to antibiotic resistance.
Supplementary Material
Acknowledgments
We thank past and present members of the Berghuis lab for their assistance and suggestions.
This work was supported by grant FRN13107 (A.M.B.) from the Canadian Institutes of Health Research. A.M.B. holds a Canada Research Chair in Structural Biology.
Footnotes
Published ahead of print on 11 May 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Allinger, N. L. 1977. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 99:8127-8134. [Google Scholar]
- 2.Arya, D. P. 2007. Aminoglycoside antibiotics: from chemical biology to drug discovery. Wiley-Interscience, Hoboken, NJ.
- 3.Barada, K., H. Hanaki, S. Ikeda, Y. Yamaguchi, H. Akama, T. Nakae, T. Inamatsu, and K. Sunakawa. 2007. Trends in the gentamicin and arbekacin susceptibility of methicillin-resistant Staphylococcus aureus and the genes encoding aminoglycoside-modifying enzymes. J. Infect. Chemother. 13:74-78. [DOI] [PubMed] [Google Scholar]
- 4.Berendsen, H. J. C., J. P. M. Postma, W. F. Vangunsteren, A. Dinola, and J. R. Haak. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684-3690. [Google Scholar]
- 5.Berendsen, H. J. C., D. Vanderspoel, and R. Vandrunen. 1995. Gromacs—a message-passing parallel molecular-dynamics implementation. Comput. Phys. Commun. 91:43-56. [Google Scholar]
- 6.Berman, J. D., and L. Fleckenstein. 1991. Pharmacokinetic justification of antiprotozoal therapy. A US perspective. Clin. Pharmacokinet. 21:479-493. [DOI] [PubMed] [Google Scholar]
- 7.Boehr, D. D., P. R. Thompson, and G. D. Wright. 2001. Molecular mechanism of aminoglycoside antibiotic kinase APH(3′)-IIIa: roles of conserved active site residues. J. Biol. Chem. 276:23929-23936. [DOI] [PubMed] [Google Scholar]
- 8.Brünger, A., P. Adams, G. Clore, W. DeLano, P. Gros, R. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. Pannu, R. Read, L. Rice, T. Simonson, and G. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 9.Burk, D. L., W. C. Hon, A. K.-W. Leung, and A. M. Berghuis. 2001. Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase. Biochemistry 40:8756-8764. [DOI] [PubMed] [Google Scholar]
- 10.Burkert, U., and N. L. Allinger. 1982. Molecular mechanics. American Chemical Society, Washington, DC.
- 11.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 12.Cox, J. R., D. R. Ekman, E. L. DiGiammarino, A. Akal-Strader, and E. H. Serpersu. 2000. Aminoglycoside antibiotics bound to aminoglycoside-detoxifying enzymes and RNA adopt similar conformations. Cell Biochem. Biophys. 33:297-308. [DOI] [PubMed] [Google Scholar]
- 13.Cox, J. R., G. A. McKay, G. D. Wright, and E. H. Serpesu. 1996. Arrangement of substrates at the active site of an aminoglycoside antibiotic 3′-phosphotransferase as determined by NMR. J. Am. Chem. Soc. 118:1295-1301. [Google Scholar]
- 14.Cox, J. R., and E. H. Serpersu. 1997. Biologically important conformations of aminoglycoside antibiotics bound to an aminoglycoside 3′-phosphotransferase as determined by transferred nuclear Overhauser effect spectroscopy. Biochemistry 36:2353-2359. [DOI] [PubMed] [Google Scholar]
- 15.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald—an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089-10092. [Google Scholar]
- 16.Davies, J. E. 1991. Aminoglycoside-aminocyclitol antibiotics and their modifying enzymes, p. 691-713. In V. Lorian (ed.), Antibiotics in laboratory medicine. Williams & Wilkins, Baltimore, MD.
- 17.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577-8593. [Google Scholar]
- 18.Fong, D. H., and A. M. Berghuis. 2002. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 21:2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanessian, S., S. Adhikari, J. Szychowski, K. Pachamuthu, X. Wang, M. T. Migawa, R. H. Griffey, and E. E. Swayze. 2007. Probing the ribosomal RNA A-site with functionally diverse analogues of paromomycin-synthesis of ring I mimetics. Tetrahedron 63:827-846. [Google Scholar]
- 20.Hanessian, S., J. Szychowski, S. S. Adhikari, G. Vasquez, P. Kandasamy, E. E. Swayze, M. T. Migawa, R. Ranken, B. Francois, J. Wirmer-Bartoschek, J. Kondo, and E. Westhof. 2007. Structure-based design, synthesis, and A-site rRNA cocrystal complexes of functionally novel aminoglycoside antibiotics: C2″ ether analogues of paromomycin. J. Med. Chem. 50:2352-2369. [DOI] [PubMed] [Google Scholar]
- 21.Hanessian, S., J. Szychowski, N. B. Campos-Reales Pineda, A. Furtos, and J. W. Keillor. 2007. 6-Hydroxy to 6‴-amino tethered ring-to-ring macrocyclic aminoglycosides as probes for APH(3′)-IIIa kinase. Bioorg. Med. Chem. Lett. 17:3221-3225. [DOI] [PubMed] [Google Scholar]
- 22.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463-1472. [Google Scholar]
- 23.Holm, S. E., B. Hill, A. Lowestad, R. Maller, and T. Vikerfors. 1983. A prospective, randomized study of amikacin and gentamicin in serious infections with focus on efficacy, toxicity and duration of serum levels above the MIC. J. Antimicrob. Chemother. 12:393-402. [DOI] [PubMed] [Google Scholar]
- 24.Hon, W. C., G. A. McKay, P. R. Thompson, R. M. Sweet, D. S. Yang, G. D. Wright, and A. M. Berghuis. 1997. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89:887-895. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 26.Kawaguchi, H., T. Naito, S. Nakagawa, and K. I. Fujisawa. 1972. BB-K 8, a new semisynthetic aminoglycoside antibiotic. J. Antibiot. (Tokyo) 25:695-708. [DOI] [PubMed] [Google Scholar]
- 27.Kohl, A., P. Amstutz, P. Parizek, H. K. Binz, C. Briand, G. Capitani, P. Forrer, A. Pluckthun, and M. G. Grutter. 2005. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure 13:1131-1141. [DOI] [PubMed] [Google Scholar]
- 28.Kondo, J., B. Francois, R. J. Russell, J. B. Murray, and E. Westhof. 2006. Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the gamma-amino-alpha-hydroxybutyryl (haba) group. Biochimie 88:1027-1031. [DOI] [PubMed] [Google Scholar]
- 29.Kondo, S., and K. Hotta. 1999. Semisynthetic aminoglycoside antibiotics: development and enzymatic modifications. J. Infect. Chemother. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 30.Kondo, S., K. Iinuma, H. Yamamoto, Y. Ikeda, and K. Maeda. 1973. Letter: synthesis of (S)-4-amino-2-hydroxybutyryl derivatives of 3′,4′-dideoxykanamycin B and their antibacterial activities. J. Antibiot. (Tokyo) 26:705-707. [DOI] [PubMed] [Google Scholar]
- 31.Kondo, S., K. Iinuma, H. Yamamoto, K. Maeda, and H. Umezawa. 1973. Letter: syntheses of 1-n-(S)-4-amino-2-hydroxybutyryl)-kanamycin B and -3′, 4′-dideoxykanamycin B active against kanamycin-resistant bacteria. J. Antibiot. (Tokyo) 26:412-415. [DOI] [PubMed] [Google Scholar]
- 32.Lindahl, E., B. Hess, and D. Van Der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306-317. [Google Scholar]
- 33.Litovchick, A., A. Lapidot, M. Eisenstein, A. Kalinkovich, and G. Borkow. 2001. Neomycin B-arginine conjugate, a novel HIV-1 Tat antagonist: synthesis and anti-HIV activities. Biochemistry 40:15612-15623. [DOI] [PubMed] [Google Scholar]
- 34.Llano-Sotelo, B., E. F. Azucena, Jr., L. P. Kotra, S. Mobashery, and C. S. Chow. 2002. Aminoglycosides modified by resistance enzymes display diminished binding to the bacterial ribosomal aminoacyl-tRNA site. Chem. Biol. 9:455-463. [DOI] [PubMed] [Google Scholar]
- 35.Madhusudan, P. Akamine, N. H. Xuong, and S. S. Taylor. 2002. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat. Struct. Biol. 9:273-277. [DOI] [PubMed] [Google Scholar]
- 36.Matte, A., L. W. Tari, and L. T. Delbaere. 1998. How do kinases transfer phosphoryl groups? Structure 6:413-419. [DOI] [PubMed] [Google Scholar]
- 37.McKay, G., P. Thompson, and G. Wright. 1994. Broad spectrum aminoglycoside phosphotransferase type III from Enterococcus: overexpression, purification, and substrate specificity. Biochemistry 33:6936-6944. [DOI] [PubMed] [Google Scholar]
- 38.McKay, G. A., and G. D. Wright. 1995. Kinetic mechanism of aminoglycoside phosphotransferase type IIIa. Evidence for a Theorell-Chance mechanism. J. Biol. Chem. 270:24686-24692. [DOI] [PubMed] [Google Scholar]
- 39.Mildvan, A. S. 1997. Mechanisms of signaling and related enzymes. Proteins 29:401-416. [PubMed] [Google Scholar]
- 40.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature (London) 327:389-394. [DOI] [PubMed] [Google Scholar]
- 41.Nagabhushan, T. L., A. B. Cooper, H. Tsai, P. J. Daniels, and G. H. Miller. 1978. The syntheses and biological properties of 1-N-(S-4-amino-2-hydroxybutyryl)-gentamicin B and 1-N-(S-3-amino-2-hydroxypropionyl)-gentamicin B. J. Antibiot. (Tokyo) 31:681-687. [DOI] [PubMed] [Google Scholar]
- 42.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 43.Nurizzo, D., S. C. Shewry, M. H. Perlin, S. A. Brown, J. N. Dholakia, R. L. Fuchs, T. Deva, E. N. Baker, and C. A. Smith. 2003. The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327:491-506. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 45.Pape, T., W. Wintermeyer, and M. V. Rodnina. 2000. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 7:104-107. [DOI] [PubMed] [Google Scholar]
- 46.Parang, K., J. H. Till, A. J. Ablooglu, R. A. Kohanski, S. R. Hubbard, and P. A. Cole. 2001. Mechanism-based design of a protein kinase inhibitor. Nat. Struct. Biol. 8:37-41. [DOI] [PubMed] [Google Scholar]
- 47.Schroeder, R., C. Waldsich, and H. Wank. 2000. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 19:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serpersu, E. H., J. R. Cox, E. L. DiGiammarino, M. L. Mohler, D. R. Ekman, A. Akal-Strader, and M. Owston. 2000. Conformations of antibiotics in active sites of aminoglycoside-detoxifying enzymes. Cell Biochem. Biophys. 33:309-321. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, P. R., D. D. Boehr, A. M. Berghuis, and G. D. Wright. 2002. Mechanism of aminoglycoside antibiotic kinase APH(3′)-IIIa: role of the nucleotide positioning loop. Biochemistry 41:7001-7007. [DOI] [PubMed] [Google Scholar]
- 50.Tok, J. B., L. J. Dunn, and R. C. Des Jean. 2001. Binding of dimeric aminoglycosides to the HIV-1 rev responsive element (RRE) RNA construct. Bioorg. Med. Chem. Lett. 11:1127-1131. [DOI] [PubMed] [Google Scholar]
- 51.Tsukiura, H., K. Saito, S. Kobaru, M. Konishi, and H. Kawaguchi. 1973. Aminoglycoside antibiotics. IV. BU-1709 E1 and E2, new aminoglycoside antibiotics related to the butirosins. J. Antibiot. (Tokyo) 26:386-388. [PubMed] [Google Scholar]
- 52.van Aalten, D. M. F., R. Bywater, J. B. C. Findlay, M. Hendlich, R. W. W. Hooft, and G. Vriend. 1996. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 10:255-262. [DOI] [PubMed] [Google Scholar]
- 53.Vicens, Q., and E. Westhof. 2002. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 9:747-755. [DOI] [PubMed] [Google Scholar]
- 54.Wright, J. J. 1976. Synthesis of 1-N-ethylsisomicin: a broad-spectrum semisynthetic aminoglycoside antibiotic. J. Chem. Soc. Chem. Commun. 6:206-208. [Google Scholar]
- 55.Zarrilli, R., M. F. Tripodi, A. Di Popolo, R. Fortunato, M. Bagattini, M. Crispino, A. Florio, M. Triassi, and R. Utili. 2005. Molecular epidemiology of high-level aminoglycoside-resistant enterococci isolated from patients in a university hospital in southern Italy. J. Antimicrob. Chemother. 56:827-835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.