Abstract
Streptomycin is used in plant agriculture for bacterial disease control, particularly against fire blight in pome fruit orchards. Concerns that this may increase environmental antibiotic resistance have led to bans or restrictions on use. Experience with antibiotic use in animal feeds raises the possible influence of formulation-delivered resistance genes. We demonstrate that agricultural streptomycin formulations do not carry producer organism resistance genes. By using an optimized extraction procedure, Streptomyces 16S rRNA genes and the streptomycin resistance gene strA were not detected in agricultural streptomycin formulations. This diminishes the likelihood for one potential factor in resistance development due to streptomycin use.
Antibiotic use in plant agriculture constitutes less than 0.5% of total antibiotic use in the United States, with the major use for controlling fire blight of pome fruits, caused by the enterobacterium Erwinia amylovora (19). Since streptomycin was first registered in the United States in 1959, it has been used up to 21 times during a particular growing season, but it is currently generally applied only during bloom and in conjunction with disease forecasting systems with zero to four applications per season in most locations (19). Resistance development in the pathogen was first reported in 1971 in California presumably due to streptomycin application selection pressure and has been found to result in efficacy loss in many areas where streptomycin has since been used for fire blight management (19). Resistance development in E. amylovora in most locations has been attributed to stable, horizontally nontransmissible chromosomal mutation in a single nucleotide of the rpsL gene (3, 17). More rarely, pathogen resistance has been attributed to acquisition of plasmid-borne strA that is transmissible (18), with other plant-associated or environmental bacteria as likely sources (11, 23, 27). The source(s) of strA acquired by E. amylovora is uncertain.
Public health and environmental concerns with antibiotic use in agriculture have lead to prohibitions and restricted use in Europe and elsewhere (4). A main concern is a hypothesized horizontal gene transfer to clinically relevant bacteria (22, 24), although such a link has never been documented (30). Antibiotic formulations used in animal feeds have been suggested as a potential delivery vehicle for resistance genes (1, 8, 21), since low-grade formulations have been shown to be contaminated with the resistance genes of producer organism used to prepare formulations (15, 28, 29). Streptomycin formulations used in plant agriculture are produced using Streptomyces griseus subsp. griseus, which carries strA for self-resistance to the antibiotic (20). Tolba et al. (25) reported that resistant environmental bacteria in an orchard with streptomycin application carried the strA resistance gene. We examined whether plant agriculture formulations of streptomycin are contaminated with streptomycin resistance genes and thus could serve as a factor in accelerated resistance development in the pathogen and/or environmental bacteria.
Relatively few firms produce streptomycin formulations that are distributed internationally. We evaluated 18 batches representative of formulations commercially used in the United States, New Zealand, and Europe from 1998 to 2008 (Table 1). Reagent-grade streptomycin sulfate salt (Fluka, Buchs, Switzerland) without carrier was included as a control. Extractions were done by suspending formulations in deionized H2O at a concentration of 200 mg ml−1 and then dividing the suspension into 100-μl portions. Detection limits were determined by spiking samples with 10 μl of fivefold dilution series of purified DNA from S. griseus subsp. griseus type strain DSMZ 40236 (ATCC 10137, obtained from an overnight LB liquid culture [Difco, Allschwil, Switzerland] using the Wizard Genomic DNA purification kit [Promega Corp., Madison, WI] and quantified using a NanoDrop ND-1000 spectrophotometer [Witec AG, Littau, Switzerland]) to obtain a formulation with a range of 0.3 ng to 30.0 μg DNA g−1. Spiked DNA was adsorbed to the matrix prior to extraction by gentle overnight shaking in aqueous suspension at 20 to 25°C, with gentle agitation; under these conditions, maximum adsorption was expected to occur within 90 min (12).
TABLE 1.
Streptomycin formulations tested in this study
| Batch no. | Formulation | Sourcea | Yr | Mixture codeb |
|---|---|---|---|---|
| 1 | Agri-Mycin 17 WP | NuFarm Americas Inc. (Burr Ridge, IL, USA) | 2006 | A |
| 2 | Strepto | Globachem NV (Sint-Truiden, BE) | 2007 | B |
| 3 | Agri-Mycin 17 WP | Novartis Crop Protection Inc. (Greensboro, NC, USA) | 2003 | A |
| 4 | Agri-Mycin 17 | NuFarm Americas Inc. (Burr Ridge, IL, USA) | 2005 | A |
| 5 | Agri-Mycin 17 WP | NuFarm Americas Inc. (Burr Ridge, IL, USA) | 2007 | A |
| 6 | Plantomycin WG | W. Neudorf GmbH KG (Emmerthal, DE) | 1998 | |
| 7 | Strepto | Globachem NV (Sint-Truiden, BE) | 2005 | B |
| 8 | Plantomycin | Asepta B.V. (Delft, NL) | 2006 | |
| 9 | Agri-Mycin 17 | NuFarm Americas Inc. (Burr Ridge, IL, USA) | 2004 | |
| 10 | Agri-Mycin 17 WP | NuFarm Americas Inc. (Burr Ridge, IL, USA) | 2008 | A |
| 11 | Strepto | Globachem NV (Sint-Truiden, BE) | 2008 | B |
| 12 | Ag-Streptomycin | Leu+Gygax AG (Birmenstorf, CH) | 2008 | D |
| 13 | Strepto | Schneiter Agro AG (Seon, CH) | 2008 | D |
| 14 | KeyStrepto | Key Industries Ltd. (Auckland, NZ) | 2008 | C |
| 15 | Agri-Mycin 17 WP | NuFarm Americas Inc. (Burr Ridge, IL, USA) | 2008 | A |
| 16 | KeyStrepto | Key Industries Ltd. (Auckland, NZ) | 2006 | C |
| 17 | KeyStrepto | Key Industries Ltd. (Auckland, NZ) | 2007 | C |
| 18 | KeyStrepto | Key Industries Ltd. (Auckland, NZ) | 2007 | C |
Country abbreviations: USA, United States of America; CH, Switzerland; DE, Germany; BE, Belgium; NL, The Netherlands; NZ, New Zealand.
Detection limits were established for mixtures of formulation batches with similar textures, i.e., all formulations with the same mixture code were pooled. All formulation batches were tested individually using the final protocol (i.e., unspiked formulations supplemented with 40 mM sodium metaphosphate and magnetic bead extraction, with three independent tests per formulation batch).
Direct PCR on water suspensions of antibiotic formulations spiked with S. griseus subsp. griseus DNA was unsuccessful. Our preliminary efforts to concentrate DNA from formulation suspensions using Pierce SnakeSkin 10,000-molecular-weight-cutoff dialysis tubes (Fisher Thermo Scientific, Rockford, IL) with 1× Tris-EDTA buffer, followed by ethanol precipitation, yielded the lowest detection limit of 30 ng g−1 formulation. Dialysis was also cumbersome and time-consuming and accompanied by a cross-contamination risk when processing many samples. We then developed a high-throughput, higher-sensitivity method for DNA extraction from antibiotic formulations which yielded largely intact DNA (>500 bp) and had a detection limit under 1 to 3 ng DNA g−1 formulation. Agricultural antibiotics are formulated with water-insoluble carriers (e.g., kaolin clays) that adsorb the active ingredient. Clays render DNA resistant to degradation (12) despite retaining biological activity and also can inhibit PCR so that intact DNA may be present in a formulation but undetectable (7). To optimize detection of DNA in formulations, we first evaluated the addition of nonspecific competitor DNA to reduce adsorption of target DNA to carrier material. Subsequently, we evaluated sodium metaphosphate and extraction of DNA with magnetic beads to lower the detection limit.
To estimate the detection limit, antibiotic formulations with similar characteristics were pooled in equal proportions prior to extraction (mixtures A to D [Table 1]), and all pooled formulations were extracted in parallel with spiked controls. Formulations with the same mixture letter as those in Table 1 were pooled. For each antibiotic sample, 10-ml aliquots of extraction suspension (200 mg formulation ml−1) were added, and the mixture was diluted in 90 μl of deionized H2O. These were used directly for PCR amplification of S. griseus subsp. griseus 16S rRNA genes and the strA streptomycin resistance gene (Table 2) for comparison with optimization approaches described below. All PCRs were carried out in 10-μl reaction mixtures using either 1 μl of 10-fold diluted formulation suspension (without extraction) or 1 μl of purified DNA solution plus 0.4 mM each primer in a final concentration of 1× master mix of the HotStar-Taq MasterMix kit (Qiagen, Basel, Switzerland). Cycling conditions consisted of the following steps: (i) an initial denaturation and activation of the HotStar Taq enzyme for 15 min at 95°C; (ii) 40 cycles of PCR, with 1 cycle consisting of 30 s of denaturation at 95°C, 30 s of annealing at the appropriate temperature for each primer set (Table 2), and 1 min of elongation at 72°C; and (iii) a final elongation step of 10 min at 72°C. Amplicons were observed on a 1.2% agarose gel containing ethidium bromide.
TABLE 2.
Primers used in this study
| Primer | Target | Sequence (5′-3′) | Product size (bp) | TAa (°C) | Reference |
|---|---|---|---|---|---|
| strA-F | strA | GCGGCTGCTCGACCACGAC | 598 | 63 | 18 |
| strA-R | strA | CCGTCCTCGATGTCCCACAGGG | |||
| 16Str-F | 16S rRNA gene | TTCGGGTGTTACCGACTTTC | 898 | 47 | This work |
| 16Str-R | 16S rRNA gene | CAAGCGTTGTCCGGAATTAT |
TA, primer annealing temperature used in the PCR.
Optimization approaches were evaluated using 220-μl samples of aqueous antibiotic suspensions as described above. In the first set of samples, competitor DNA of Pantoea agglomerans (20 μl), prepared as described above for S. griseus subsp. griseus DNA, was added to antibiotic suspensions to give a concentration of 50 μg nontarget DNA g−1 formulation. In the second set of samples, sodium metaphosphate (SMP) (Sigma-Aldrich, Buchs, Switzerland) was added to antibiotic suspensions at six concentrations (0 to 180 nM). Positive controls were prepared by spiking double-distilled H2O (no antibiotic formulations) to the fivefold dilution series of purified DNA from S. griseus subsp. griseus DNA. The final volume of each sample was brought to 230 μl, and the samples were incubated for 60 h with gentle shaking at room temperature. Incubated samples were then heated to 95°C for 10 min just prior to DNA extraction (or direct use in PCR). A further optimization approach incorporated the Qiagen BioSprint96 magnetic beads extraction kit (Qiagen AG, Hombrechtikon, Switzerland) following the manufacturer's protocol for plant samples. Automatic extraction using BioSprint96 was done after adding 200 μl isopropanol and 20 μl magnetic beads to aqueous suspensions of antibiotic formulation or to samples amended with competitor P. agglomerans DNA and/or SMP. Purified and concentrated DNA from this magnetic bead separation was then used as the PCR template. After the extraction procedure was optimized, all antibiotic formulations were individually tested without the addition of spiked S. griseus subsp. griseus DNA.
Direct PCR on water suspensions of streptomycin formulations failed to yield any amplicons using either strA or 16S rRNA gene primers when spiked with producer bacterium DNA (30 μg g−1 formulation) (Fig. 1A). The addition of competitor DNA slightly improved PCR detection, lowering the detection limit to 5 μg g−1 of formulation (Fig. 1B), probably by competing with S. griseus subsp. griseus DNA for binding sites on the carrier matrix. As expected, nontarget competitor DNA by itself did not yield any amplicons with strA primers, but it produced a clear uniform amplification with 16S rRNA gene primers, irrespective of the amount of spiked target DNA present, thus rendering this primer set useless for the detection of residual S. griseus subsp. griseus DNA (data not shown). Better detection was obtained with the addition of SMP. Pretreatment of the aqueous formulation suspension with SMP yielded a faint but positive amplification down to 1 μg DNA g−1 formulation. The optimal SMP concentration was assessed by performing the procedure described above on formulation batch 12 (Leu+Gygax AG, Birmenstorf, Switzerland) spiked with 1 μg S. griseus subsp. griseus DNA g−1 formulation and pretreated with six SMP concentrations. For both primer pairs, the best amplification was obtained when samples were pretreated with 20 to 40 mM SMP (Fig. 1 and 2). This supports prior observations of SMP-mediated recovery of bacterial DNA from clay substrates (2).
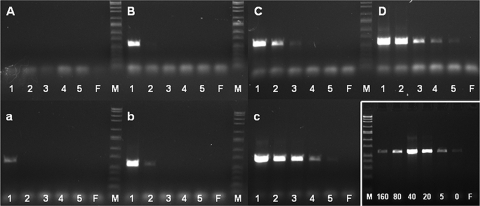
FIG. 1.
PCR amplification of a 570-bp fragment from the strA gene of S. griseus subsp. griseus from agricultural streptomycin formulation batch 12 (Table 1) spiked with different amounts of S. griseus subsp. griseus DNA. PCR was performed directly on a suspension of the formulation (A to C) or after DNA extraction using the BioSprint96 DNA plant kit (a to c). Sample pretreatments were as follows: none (A and a), 50 μg nontarget (P. agglomerans) DNA g−1 formulation added (B and b), 40 mM sodium metaphosphate added to formulation suspension (C and c), and S. griseus subsp. griseus DNA only (formulation absent) (D). The amounts of spiked S. griseus subsp. griseus DNA g−1 formulation in the different lanes are as follows: 30 μg (lanes 1), 5 μg (lanes 2), 1 μg (lanes 3), 0.2 μg (lanes 4), 0.04 μg (lanes 5), and 0 μg (formulation only) (lanes F). (Bottom right panel) Effect of pretreatment with different concentrations of sodium metaphosphate (0 to 160 mM) on DNA extraction and strA amplification efficiency on a sample of formulation batch 12 containing 1 μg of spiked S. griseus subsp. griseus DNA g−1 formulation. The optimal concentration of sodium metaphosphate is about 40 mM. F lanes are negative controls and contain formulation with no spiked S. griseus subsp. griseus DNA and 40 mM sodium metaphosphate. Lanes M, size markers (base pair ladder from Sigma). Positive bands in the gels are shown in white boxes.
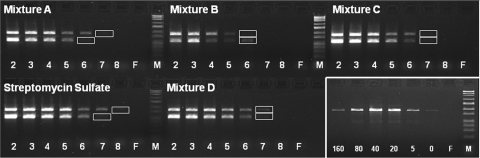
FIG. 2.
Detection limit of 16S rRNA genes (top band in gels) and strA (bottom band in gels) of S. griseus subsp. griseus in different agricultural streptomycin formulation mixtures after sample pretreatment with 40 mM sodium metaphosphate and DNA extraction using the BioSprint96 DNA plant kit. PCR of 16S rRNA genes and PCR of strA were performed as separate reactions, and then the products were combined before loading on the gel. Formulation mixtures consisted in equal parts of different batches of the same formulation manufactured by the same producer, with the exception of mixture D (Table 1), where different formulations with similar textures were used. Spiked S. griseus subsp. griseus DNA was added at the following rates (g−1 formulation): 5 μg (lanes 2), 1 μg (lanes 3), 200 ng (lanes 4), 40 ng (lanes 5), 8 ng (lanes 6), 1.6 ng (lanes 7), 0.3 ng (lanes 8), and 0 ng (formulation only) (lanes F). (Bottom right panel) Effect of pretreatment with different concentrations of sodium metaphosphate (0 to 160 mM) on DNA extraction and 16S rRNA gene amplification on a sample of formulation batch 12 amended with 1 μg of spiked S. griseus subsp. griseus DNA g−1 formulation. The optimal concentration of sodium metaphosphate was about 40 mM. F lanes are negative controls and contain formulation with no spiked S. griseus subsp. griseus DNA and 40 mM sodium metaphosphate. Lanes M, size markers (base pair ladder from Sigma). The observable detection limits on the gels are indicated by the white boxes.
A procedure combining SMP pretreatment with magnetic bead extraction of DNA provided the lowest detection limit (Fig. 1). Using magnetic beads on aqueous formulations spiked with S. griseus subsp. griseus DNA gave a detection limit around 30 μg DNA g−1 formulation (Fig. 1a) and 5 μg g−1 formulation in samples augmented with nontarget DNA (Fig. 1b). Samples treated with 40 mM SMP prior to DNA extraction lowered the detection limit to 40 ng DNA g−1 formulation (Fig. 1c). Our approach combining SMP pretreatment followed by magnetic DNA extraction enhanced the detection of resistance genes in streptomycin formulations.
The influence of formulation on the limit of detection of introduced strA and 16S rRNA genes by PCR was evaluated in different streptomycin formulation mixtures pretreated with 40 mM SMP and then extracted with magnetic beads. The mixtures (Table 1) consisted of equal amounts of different batches of formulation manufactured by the same producer, with the exception of mixture D (which had different formulations of similar textures) and reagent-grade streptomycin sulfate. The detection limit for each mixture was at least 8 ng DNA g−1 formulation (mixture Β), and in some cases, it was as low as 1.6 ng DNA g−1 formulation (mixtures C and D) (Fig. 2). This lower detection limit (compared to that from Fig. 1) was obtained because on the basis of the results from the first set of trials, we included lower concentrations of spiked DNA; it does not indicate different limits of detection per se. Among formulations, the limit of detection of the strA gene was 1.6 to 8.0 ng DNA g−1 formulation, which was similar to the detection limit of 16S rRNA genes (0.3 to 8.0 ng DNA g−1 formulation) (Fig. 2). For extensive screenings of formulations, focusing detection methods on amplification of target resistance genes should be adequate because a positive response would indicate the presence of the genes of concern, whereas amplification of 16S rRNA genes would require additional testing to verify that the DNA was due to the presence of the producer organism harboring resistance genes and not a contaminant.
We developed an optimized procedure to detect antibiotic resistance genes in formulations applied to plants. The combination of SMP pretreatment and magnetic beads to extract DNA mitigated the challenges of amplifying target DNA in a suspension of kaolin, the carrier for many antibiotic formulations intended for plants. Earlier, Lu et al. (15) detected 30 μg DNA g−1 chicken feed amended with avoparcin. The highest detection limit for our optimized procedure (8 ng DNA g−1 formulation) is several orders of magnitude lower than that needed to detect DNA contamination in antibiotic formulations used in animal production.
In three repeated tests, none of the 18 streptomycin formulation batches analyzed individually showed a positive amplification of either strA or 16S rRNA genes with the optimized procedure when the formulations were not spiked with producer DNA. This demonstrates that commercial streptomycin formulations from the United States, New Zealand, Belgium, Germany, and Switzerland used to control bacterial plant disease worldwide lack detectable intact S. griseus subsp. griseus 16S rRNA genes or more importantly intact strA genes which could be horizontally transferred in the environment. Although theoretically, a single copy of a resistance gene would be sufficient to result in acquired resistance, the main epidemiological concern is with inundation of resistance gene copies that influence resistance in populations to a degree beyond what may occur via natural sources of resistance genes in the environment (25). Little information is available relating gene copy number and probability of horizontal transfer. Kloos et al. (13) found approximately 2.1 × 10−6 transformants per μg DNA, or approximately one transformation event per 106 bacteria. With the 1.6-ng DNA detection limit in our study, corresponding to approximately 105 S. griseus subsp. griseus genome equivalents, this translates to one transformation event per 109 bacteria. In terms of exposure rates across an orchard, approximately 600 g formulation ha−1 are applied following product label guidelines to approximately 106 flowers per ha (the primary exposure site), and each flower may be colonized by 106 bacteria (10). Assuming a formulation contained DNA that was just below our detection limit of 1.6 ng g−1, this means that the maximum exposure would be 0.6 mg per flower or 0.00094 ng DNA applied to a population of 106 bacteria per flower. Thus, standard application rates for streptomycin in orchards represent an insignificant introduction of resistance genes from formulations.
Our finding removes one of the potential risk factors for antibiotic resistance development that has been overlooked in previous studies of the impact of environmental applications of antibiotics as a fire blight management option. Antibiotic formulations applied to plants are highly unlikely to serve as a direct conduit for introduction of genes encoding streptomycin resistance from the producer organism into the orchard microbial communities or exposed agricultural workers and subsequently to contribute to horizontal transfer of streptomycin resistance. Our results are not intended to decide the wider debate either for or against the agricultural use of streptomycin. However, by removing agricultural formulation-introduced genes as an important potential resistance gene acquisition factor, our results facilitate focus on other more likely factors involved in environmental antibiotic resistance development, including the contribution of natural resistance reservoirs (e.g., native Streptomyces [6, 22, 25, 27]), as well as agricultural applications of animal waste (5, 14), livestock transmission (26), insect vectors (16), or use strategies in clinical therapy (9).
Acknowledgments
We thank E. Holliger, J. Vanneste, C. Scherr, and E. Moltmann for assistance in obtaining commercial antibiotic formulations and J. E. Frey and B. Frey for technical advice.
Financial support was provided by the Swiss Federal Department for Agriculture (BLW Fire Blight Research-Control Project), and the U.S. Department of Agriculture (USDA-CSREES grant 2007-51100-03852). This study was conducted within the European Science Foundation-funded research network COST Action 864 (www.cost864.eu).
Footnotes
Published ahead of print on 4 May 2009.
REFERENCES
- 1.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, A. J., M. Khanna, G. A. Toranzos, and G. Stotzky. 1998. Amplification of DNA bound on clay minerals. Mol. Ecol. 7:775-778. [Google Scholar]
- 3.Chiou, C.-S., and A. L. Jones. 1995. Molecular analysis of high-level streptomycin resistance in Erwinia amylovora. Phytopathology 85:324-328. [Google Scholar]
- 4.Duffy, B., H.-J. Schärer, M. Bünter, A. Klay, and E. Holliger. 2005. Regulatory measures against Erwinia amylovora in Switzerland. EPPO Bull. 35:239-244. [Google Scholar]
- 5.Duriez, P., and E. Topp. 2007. Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial swine farm. Appl. Environ. Microbiol. 73:5486-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egan, S., P. Wiener, D. Kallifidas, and E. M. H. Wellington. 1998. Transfer of streptomycin biosynthesis gene clusters within streptomycetes isolated from soil. Appl. Environ. Microbiol. 64:5061-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallori, E., M. Bazzicalupo, L. Dal Canto, R. Fani, P. Nannipieri, C. Vettori, and G. Stotzky. 1994. Transformation of Bacillus subtilis by DNA bound on clay in non-sterile soil. FEMS Microbiol. Ecol. 15:119-126. [Google Scholar]
- 8.Goldman, E. 2004. Antibiotic abuse in animal agriculture: exacerbating drug resistance in human pathogens. Hum. Ecol. Risk Assess. 10:121-134. [Google Scholar]
- 9.Gould, I. M. 1999. A review of the role of antibiotic policies in the control of antibiotic resistance. J. Antimicrob. Chemother. 43:459-465. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, K. B., and V. O. Stockwell. 1998. Management of fire blight: a case study in microbial ecology. Annu. Rev. Phytopathol. 36:227-248. [DOI] [PubMed] [Google Scholar]
- 11.Jones, A. L., and E. L. Schnabel. 2000. The development of streptomycin-resistant strains of Erwinia amylovora, p. 235-251. In Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, Wallingford, United Kingdom.
- 12.Khanna, M., and G. Stotzky. 1992. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect on DNase on the transforming ability of bound DNA. Appl. Environ. Microbiol. 58:1930-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloos, D. U., M. Stratz, A. Guttler, R. J. Steffan, and K. N. Timmis. 1994. Inducible cell lysis system for the study of natural transformation and environmental fate of DNA released by cell death. J. Bacteriol. 176:7352-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kümmerer, K. 2004. Resistance in the environment. J. Antimicrob. Chemother. 54:311-320. [DOI] [PubMed] [Google Scholar]
- 15.Lu, K., R. Asano, and J. Davies. 2004. Antimicrobial resistance gene delivery in animal feeds. Emerg. Infect. Dis. 10:679-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macovei, L., and L. Zurek. 2006. Ecology of antibiotic resistance genes: characterization of enterococci from houseflies collected in food settings. Appl. Environ. Microbiol. 72:4028-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manulis, S., F. Kleitman, D. Shtienberg, H. Shwartz, D. Oppenheim, M. Zilberstaine, and E. Shabi. 2003. Changes in the sensitivity of Erwinia amylovora populations to streptomycin and oxolinic acid in Israel. Plant Dis. 87:650-654. [DOI] [PubMed] [Google Scholar]
- 18.McManus, P. S., and A. L. Jones. 1994. Epidemiological and genetic analysis of streptomycin-resistant Erwinia amylovora from Michigan and evaluation of oxytetracycline for control. Phytopathology 84:627-633. [Google Scholar]
- 19.McManus, P. S., V. O. Stockwell, G. W. Sundin, and A. L. Jones. 2002. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40:443-465. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi, Y., J. Ishikawa, H. Hara, H. Suzuki, M. Ikenoya, H. Ikeda, A. Yamashita, M. Hattori, and S. Horinouchi. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pezzella, C., A. Ricci, E. DiGiannatale, I. Luzzi, and A. Carattoli. 2004. Tetracycline and streptomycin resistance genes, transposons, and plasmids in Salmonella enterica isolates from animals in Italy. Antimicrob. Agents Chemother. 48:903-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seveno, N. A., D. Kallifidas, K. Smalla, J. D. van Elsas, J.-M. Collard, A. D. Karagouni, and E. M. H. Wellington. 2002. Occurrence and reservoirs of antibiotic resistance genes in the environment. Rev. Med. Microbiol. 13:15-27. [Google Scholar]
- 23.Sundin, G. W. 2002. Distinct recent lineages of the strA-strB streptomycin-resistance genes in clinical and environmental bacteria. Curr. Microbiol. 45:63-69. [DOI] [PubMed] [Google Scholar]
- 24.Szczepanowski, R., I. Krahn, B. Linke, A. Goesmann, A. Puhler, and A. Schluter. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613-3630. [DOI] [PubMed] [Google Scholar]
- 25.Tolba, S., S. Egan, D. Kallifidas, and E. M. H. Wellington. 2002. Distribution of streptomycin resistance and biosynthesis genes in streptomycetes recovered from different soil sites. FEMS Microbiol. Ecol. 42:269-276. [DOI] [PubMed] [Google Scholar]
- 26.van den Bogaard, A., and E. E. Stobberingh. 2000. Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14:327-335. [DOI] [PubMed] [Google Scholar]
- 27.van Overbeek, L. S., E. M. H. Wellington, S. Egan, K. Smalla, H. Heuer, J.-M. Collard, G. Guillaume, A. D. Karagouni, T. L. Nikolakopoulou, and J. D. van Elsas. 2002. Prevalence of streptomycin-resistance genes in bacterial populations in European habitats. FEMS Microbiol. Ecol. 42:277-288. [DOI] [PubMed] [Google Scholar]
- 28.Webb, V., and J. Davies. 1993. Antibiotic preparations contain DNA: a source of drug resistance genes? Antimicrob. Agents Chemother. 37:2379-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegener, H. C. 2003. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 6:439-445. [DOI] [PubMed] [Google Scholar]
- 30.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]