Abstract
Some strains of Pseudomonas aeruginosa produce R-type pyocins, which are high-molecular-weight phage tail-like protein complexes that have bactericidal activity against other Pseudomonas strains. These particles recognize and bind to bacterial surface structures via tail fibers, their primary spectrum determinant. R-type pyocins kill the cell by contracting a sheath-like structure and inserting their hollow core through the cell envelope, resulting in dissipation of the cellular membrane potential. We have retargeted an R-type pyocin to Escherichia coli O157:H7 by fusing a tail spike protein from an O157-specific phage, φV10, to the pyocin tail fiber. The φV10 tail spike protein recognizes and degrades the O157 lipopolysaccharide. This engineered pyocin, termed AVR2-V10, is sensitive and specific, killing 100% of diverse E. coli O157:H7 isolates but no other serotypes tested. AVR2-V10 can kill E. coli O157:H7 on beef surfaces, making it a candidate agent for the elimination of this pathogen from food products. All rare AVR2-V10-resistant mutants isolated and examined have lost the ability to produce the O157 antigen and are expected to have compromised virulence. In addition, E. coli O157:H7 exposed to and killed by AVR2-V10 do not release Shiga toxin, as is often the case with many antibiotics, suggesting potential therapeutic applications. The demonstration that a novel R-type pyocin can be created in the laboratory by fusing a catalytic tail spike from the family Podoviridae to a tail fiber of a member of the family Myoviridae is evidence that the plasticity observed among bacteriophage tail genes can, with modern molecular techniques, be exploited to produce nonnatural, targeted antimicrobial agents.
Shiga toxin (Stx)-producing Escherichia coli (STEC) strain O157:H7 is a major cause of food-borne outbreaks of colitis, which can sometimes lead to hemolytic-uremic syndrome (19). Cattle are a major, unaffected reservoir for the pathogen, and outbreaks are often associated with contaminated (adulterated) beef. However, outbreaks are also associated with dairy and other food products, including fresh leafy vegetables and fruit juices, possibly as a result of contamination with water from manure runoff or wildlife intrusions (26). The application of a method that specifically and rapidly kills this food-borne pathogen without compromising food quality might decrease the occurrence of these outbreaks.
R-type pyocins are high-molecular-weight protein complexes, produced by some Pseudomonas aeruginosa strains, which specifically kill other strains of the same species (for a review, see reference 17). These complexes resemble the tail structures of bacteriophages of members of the family Myoviridae, particularly the P2-like phages, and consist of a hollow core surrounded by a sheath, at one end of which is a base plate with six tail fibers (Fig. 1). R-type pyocins are encoded in the bacterial genome on a single locus of 16 open reading frames, of which less than 10 probably encode structural proteins of the pyocin (18, 25). Pyocin production is induced by the SOS response, which yields about 100 to 200 particles per cell (15). The cells then lyse by a process similar to phage-mediated lysis, releasing the pyocin particles into the surroundings. The particles kill the target bacteria by first binding to a cell surface structure via their tail fibers, followed by contraction of the sheath and insertion of their hollow core through the cell envelope. The resulting channel leads to the depolarization of the cytoplasmic membrane and cell death (29). A single pyocin particle can kill a cell (6, 28, 29). In many respects, R-type pyocins can be considered defective prophages, and it is highly likely that they share a common ancestry with related phages. However, pyocins do not contain a genome and they do not infect target cells; replication occurs only as a genetic component of the producer strain chromosome (9, 10, 24). Cells carrying an R-type pyocin gene cluster are genetically resistant to their own pyocin. However, cells that express pyocin must lyse to release the particles. Thus, it seems that the selective advantage is to protect kin strains from nonkin strains, a form of altruism or kin selection.
FIG. 1.
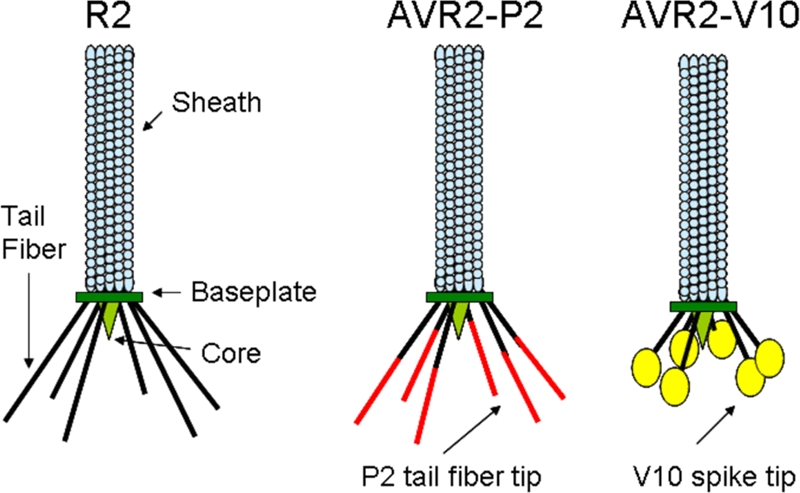
Cartoon representation of the R-type pyocin structure. R2 is the wild type and kills certain P. aeruginosa strains. AVR2-P2 has been retargeted to kill some rough strains of E. coli by using the tail fiber of phage P2. AVR2-V10 is the engineered pyocin characterized in this study, in which the phage φV10 tail spike was fused to the pyocin tail fiber and is specific for E. coli strains that produce the O157 antigen.
Five naturally occurring R-type pyocins, termed R1 to R5, each with a unique bactericidal spectrum, have been described (7). Each pyocin spectrum is related to that of the others, such that R4 encompasses the spectrum of R3, R2 encompasses the spectra of R3 and R4, and R5 has the broadest spectrum and includes the spectra of all the other pyocins, plus those of some additional strains. The spectrum of R1 is unrelated to that of R2, R3, or R4; but it is still a subset of that of R5. This places the R5 receptor at the root of a spectrum tree with two branches, one consisting of the R1 receptor and the other consisting of the receptors for R2, R4, and R3. The well-studied strain P. aeruginosa PAO1 produces R2 pyocin and is the platform R-type pyocin used in this study.
Because of their specific and potent bactericidal activities, R-type pyocins are candidate antibacterial agents. Although a few studies have shown that parenterally administered R-type pyocins can rescue mice from lethal infections (4, 16, 22), interest has been limited. One of the concerns about the use of R-type pyocins as antibacterial agents is their limited spectrum, which is determined primarily by the specificity of the tail fiber binding to bacterial surface structures or receptors. While it is unlikely that an R-type pyocin could be engineered to have a very broad spectrum, we have shown that it is possible to retarget their spectra to other bacterial species and genera by making fusions between the pyocin tail fibers and the tail fibers of related bacteriophages (32). One such pyocin, AVR2-P2 was made by fusing the C-terminal portion of the phage P2 tail fiber to the N-terminal portion of the R2 tail fiber, thereby retargeting the bactericidal activity toward rough E. coli strains that serve as the host for P2, including E. coli strain C. Unfortunately, AVR2-P2 does not kill any E. coli O157:H7 isolates. Here we describe some of the properties and applications of a new O157-specific R-type pyocin created by fusing to the R2 tail fiber an O157-specific tail spike protein from an unrelated phage from the family Podoviridae.
MATERIALS AND METHODS
Construction and expression of R2-V10 fiber-spike fusions.
Portions of R2 prf15 encoding the N-terminal fragments of the tail fiber were amplified from cloned regions of the P. aeruginosa strain PAO1 genome by using primers with a BspHI site and either a HindIII or a BbsI site. R2-V10 tail fusions were constructed by methods similar to those previously described for the construction of R2-P2 fusions (32). For the R2 fragments encoding amino acids 1 to 127 and 1 to 161, the coding sequences were fused directly to the phage φV10 sequences by using engineered BbsI sites with nonpalindromic overhangs. For the R2 fragment encoding amino acids 1 to 164, codon 164 was changed from V to L, introducing a HindIII site. For the R2 fragment encoding amino acids 1 to 241, a silent mutation was introduced, and this also created a HindIII site. The φV10 tail spike-encoding fragments were amplified with primers containing overhanging HindIII and XbaI ends. These fragments were assembled and cloned into pUCPtac, a derivative of E. coli-Pseudomonas shuttle vector pUCP30T (32), or pUCPBAD, a vector created by inserting into pUCP30T the portion of plasmid pBAD24 (3) comprising the araC gene, PBAD, multiple-cloning site, and rrnB terminator. These plasmids were then transformed into a derivative of PAO1 in which the tail fiber gene was deleted (PAO1 Δprf15).
Pyocin purification and assays.
Pyocin production and purification were performed on the basis of previously described methods (22). P. aeruginosa strain PAO1 Δprf15 containing the fusion expression plasmids under the control of either PTAC or PBAD was grown at 37°C and shaken at 225 rpm to an optical density at 600 nm of 0.25 in G medium (20 g sodium glutamate, 5 g glucose, 2.23 g Na2HPO4, 100 mg MgSO4·7H2O, 250 mg KH2PO4, 500 mg yeast extract per liter) containing gentamicin (50 μg/ml). At this point, the pyocin genes were induced by adding mitomycin C to 3 μg/ml and either 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG) or 0.2% arabinose. The cultures were incubated for an additional 2.5 h or until complete lysis occurred. The culture was allowed to incubate for an additional 30 min, after which 5 μl of DNase I (1 U/μl; Invitrogen) was added per 200 ml culture. Lysates were collected, and the debris was removed from the lysate by centrifugation at 22,000 × g in a Beckman Coulter Avanti J-25I centrifuge with a JLA-16.250 rotor for 1 h. To each 100 ml of supernatant, 65 ml of saturated ammonium sulfate was slowly added at a rate of 1 ml/min with stirring on ice. The suspension was stored at 4°C overnight. The ammonium sulfate precipitate was sedimented at 22,000 × g for 1 h, and the pellet was resuspended in 10 ml of 50 mM NaCl-10 mM Tris HCl, pH 7.5 (TN50 buffer). R-type pyocin particles were then sedimented at 61,000 × g in a centrifuge with a Beckman JA-25.50 rotor for 1 h at 4°C and resuspended in 3 ml of TN50 buffer.
Quantitative pyocin assays were performed by the method of Kageyama et al. (9), as modified previously (22). In a typical assay, fivefold serial dilutions of pyocins in TN50 buffer were incubated for 40 min at 37°C in a microwell with a known quantity of target bacteria (E. coli O157:H7 strain EDL933 in the case of AVR2-V10), for example, 1 × 108 CFU/100 μl, in Trypticase soy broth (TSB). The contents of the microwells were then serially diluted and aliquots were spotted on Trypticase soy agar plates to count the surviving bacteria. A bactericidal event was the death of a bacterium as a result of contact with a pyocin particle(s). The number of bactericidal events was related to the fraction of bacterial survivors in a Poisson distribution, m = −lnS, where m is the average number of bactericidal events per bacterial cell, and S is the fraction of survivors. The total number of bactericidal events per ml equals m times the number of bacterial cells per ml. A microwell that has an average of one bactericidal event per bacterium will yield, at or near equilibrium, 37% survivors, and a well with an average of 2.3 bactericidal events per bacterium will yield 10% survivors. It was within this survival range that the m values were calculated.
Semiquantitative assays and determinations of the bactericidal spectra of the R-type pyocins were performed by a spot method. Aliquots of 100 μl of overnight cultures were added to 5 ml of soft agar (0.5% agar with LB medium) and poured onto a 1.5% LB agar plate. After the agar overlay set, 5-μl aliquots of pyocins serially diluted 1:4 in TN50 buffer were spotted onto the plate and allowed to dry, and the plates were incubated overnight. Killing was noted as a sharp-bordered circular clearing of the bacteria corresponding to the placement of the 5-μl aliquot.
LPS analysis.
Bacterial lipopolysaccharide (LPS) was extracted by using a kit from iNtRON Biotech. Examination of LPS degradation was done by incubating approximately 2 μg of LPS with 108 purified pyocin particles in 10 μl at 37°C for 2 h. Each entire sample was electrophoresed on 4 to 20% sodium dodecyl sulfate (SDS)-polyacrylamide gels from Invitrogen and silver stained (Owl Scientific). Immunoblots were performed by transferring the electrophoresed LPS samples onto nitrocellulose with an Invitrogen X Cell II blot module. The filters were incubated overnight in Tris-buffered saline (TBS)-Tween (0.05%) with 5% nonfat dried milk. The samples were then washed three times for 10 min in TBS-Tween. The blot was subsequently incubated with anti-O157 serum (Becton Dickinson) at a 1:2,000 dilution in TBS-Tween with 5% nonfat dried milk for 1 h, followed by three more washes in TBS-Tween. The secondary antibody was an antirabbit antibody-horseradish peroxidase conjugate (Promega), which was also diluted 1:2,000 in TBS-Tween and incubated for 1 h. After three final washes, the blot was developed by using the Novex horseradish peroxidase colorimetric reagent.
AVR2-V10-resistant mutants.
For N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) mutagenesis, strain EDL933 was grown to log phase in LB medium at 37°C. Cells were then harvested by centrifugation at 5,000 × g and resuspended in a solution of 5 μg of MNNG per ml of 10 mM Tris HCl, pH 7.5-50 mM NaCl, and the mixture was incubated for 1 h at 37°C. The bacteria were washed three times in LB medium by centrifugation and resuspension. After the final resuspension, the cells were grown at 37°C to log phase, diluted to approximately 106 cells in 1 ml, and incubated with 1010 AVR2-V10 particles overnight. The cells were harvested and plated on LB agar plates. Individual surviving colonies were picked, grown in 5 ml LB broth, and tested by spot assay for sensitivity to AVR2-V10. For transposon mutagenesis, we used the EZ-Tn5 system from Epicentre Biotechnologies and followed the manufacturer's procedure. After mutagenesis, the cells were amplified and exposed to AVR2-V10 as described above for MNNG mutagenesis, and the survivors were picked and screened for resistance.
Beef decontamination.
Strain EDL933 was transformed with pUCP30T to confer gentamicin resistance. The bacteria were grown to stationary phase, which was an optical density at 600 nm of approximately 6.0, in LB medium containing gentamicin (15 μg/ml); and the culture was incubated at 4°C overnight. Groups of four slices (25 cm2 each) of beef steak were contaminated by spreading 2 × 104 CFU of bacteria directly onto the surface. This was allowed to dry at ambient temperature, and the beef samples were stored at 4°C for 48 h. Chilled pyocin samples were sprayed onto the contaminated surface of the meat such that complete moistening could be seen (approximately 150 μl liquid). This was then allowed to dry, and the meat was placed at 4°C for 2 h. EDL933 bacteria were recovered by placing the samples in 20 ml of LB medium and vortexing for 30 s. Aliquots were then plated on LB plates containing gentamicin (15 μg/ml) to determine the numbers of CFU. In samples that were treated with 109 AVR2-V10, the plating of 1-ml aliquots (of the 20-ml recovery volume) yielded no colonies. For these samples, gentamicin was added to each remaining 19 ml, and the samples were incubated overnight to enrich for the bacteria present at less than 1 CFU/ml. Aliquots (1 ml) of these enriched cultures were then spread onto LB agar plates to detect gentamicin-resistant EDL933 colonies. The data shown in Fig. 6 are representative of those from three separate experiments. Similar analyses were also conducted by dropping inoculated and treated pieces of meat in 20 ml of 50 mM sodium acetate buffer at pH 3.8 for 1 min. The liquid was subsequently vortexed, serially diluted, and plated as described above to count the surviving gentamicin-resistant EDL933 bacteria. The AVR2-V10 pyocins were completely inactivated by such brief acid exposure (data not shown), while EDL933 bacteria could be recovered without loss.
FIG. 6.
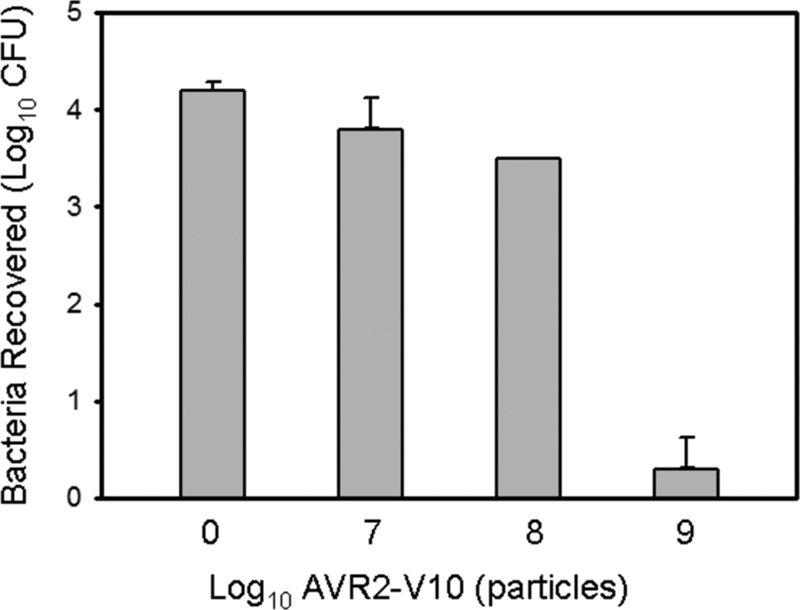
AVR2-V10 can reduce E. coli O157 from the surface of beef. Beef slices were contaminated with EDL933 and treated with various concentrations of AVR2-V10 (see Materials and Methods). The error bars indicate standard deviations. The data shown are representative of three independent experiments.
Stx activity in culture supernatants of E. coli O157:H7 grown in the presence of AVR2-V10.
Stx2 levels were measured by the Verotoxin-producing E. coli reverse passive latex agglutination assay (VTEC-RPLA; Oxoid), according to the manufacturer's instructions. E. coli O157:H7 strain RM5202 (Stx2+) (2) was grown to 5 × 107 CFU/ml and incubated with AVR2-V10 at the following concentrations: 109/ml (20:1 pyocin/bacteria), 2.5 × 108/ml (5:1 pyocin/bacteria), and 6 × 107 (1.2:1 pyocin/bacteria). The cells were also incubated at 37°C with various amounts of ciprofloxacin at concentrations of 25 ng/ml, 6 ng/ml, and 2 ng/ml. Untreated cells were used as a control for basal-level toxin production. The samples were incubated for 6 h at 37°C. The supernatants were then serially diluted twofold and assayed for toxin. The presence of toxin was visually determined by agglutination and was scored from − to +++, according to the manufacturer's directions.
RESULTS
Construction an E. coli O157-specific R-type pyocin.
Bacteriophage φV10 is a phage of the family Podoviridae that specifically infects E. coli O157 strains (30). φV10 is similar to P22-like phages, particularly phage ɛ15 (8, 12). Such phages typically do not encode tail fibers but instead encode tail spikes that recognize and often degrade surface polysaccharide structures (for a review, see reference 21). φV10 encodes an 875-amino-acid open reading which we anticipated might be a tail spike responsible for the recognition and the degradation of the O157 antigen.
On the basis of these assumptions, we designed fusions between N-terminal portions of the R2 pyocin tail fiber and various C-terminal portions of the φV10 tail spike (see Table S1 in the supplemental material). Previous work showed that amino acids 1 to 164 and 1 to 241 of the R2 tail fiber were good acceptors for tail fiber fusions, at least when the donor tail fiber came from a related phage of the family Myoviridae or other R-type pyocin (32). We fused these two regions of the R2 tail fiber gene (prf15) to DNA encoding various C-terminal portions of the φV10 tail spike. These gene fusions were expressed individually in trans from a plasmid under the control of either PTAC or PBAD in a P. aeruginosa PAO1 mutant from which the wild-type tail fiber gene was deleted (PAO1 Δprf15 [32]). Thus, the tail fiber fusion genes might complement the missing wild-type tail fiber gene, prf15. Pyocin particles were produced upon mitomycin C induction of the remaining chromosomal pyocin genes simultaneously with the induction of the tail fiber fusion gene on the plasmid with either arabinose for the PBAD constructs or IPTG for the PTAC constructs. Pyocin particles from the resulting lysates were then purified and analyzed for bactericidal activity (see Table S1 in the supplemental material).
Seven of the tail fiber fusions resulted in the production of functional pyocin particles that had bactericidal activity against E. coli O157:H7 strain EDL933 (see Table S1 in the supplemental material). Only those fused to amino acids 1 to 164 of the R2 tail fiber produced functional pyocin particles. It is noteworthy that the portion of the R2 tail fiber from amino acids 1 to 241 was unable to form functional fusions with any of the φV10 tail spike truncations, yet this region worked well for the creation of fusions with the P2 tail fiber (32). Functional pyocins were also produced only with φV10 tail spike fusions starting at amino acids 204 to 217. This region may represent a junction between functional domains. The construct formed with amino acids 1 to 164 of the R2 tail fiber and amino acids 217 to 875 of the φV10 tail spike gave the best yield of pyocin. We chose to further characterize pyocins that incorporated this tail fiber fusion, hereafter termed AVR2-V10. SDS-polyacrylamide gel electrophoresis (PAGE) analysis of purified AVR2-V10 confirmed that the tail fiber-spike fusion is incorporated into the active pyocin particle (Fig. 2). Fusion constructs that did not yield bactericidal activity against O157 strains produced very small quantities of particles that had detectable pyocin sheath and core structures but no detectable protein at the molecular weight of the fused tail fibers expected (data not shown).
FIG. 2.
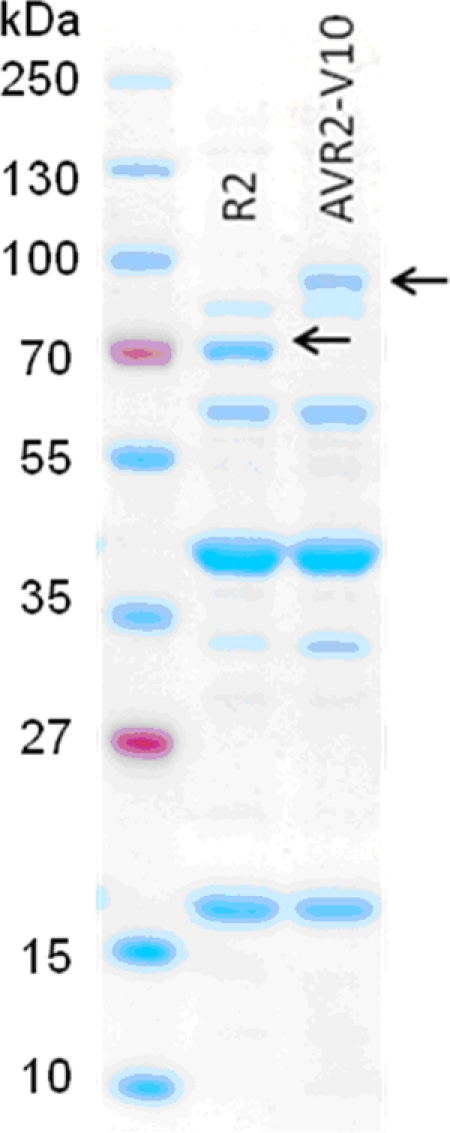
Coomassie blue-stained SDS-polyacrylamide gel of purified wild-type R2 pyocin and AVR2-V10. The arrows indicate the positions of the stained bands consistent with the molecular weights of the tail fiber of R2 and the tail fiber-tail spike R2-V10 fusion. The first (unmarked) lane contains molecular weight markers.
AVR2-V10 bactericidal spectrum.
The spectrum of activity of AVR2-V10 was determined by spotting serial dilutions of pyocin preparations on lawns of 49 diverse isolates of E. coli O157:H7 from multiple geographic areas and diverse sources, e.g., animals, plants, water, and outbreaks (U.S. Department of Agriculture, Western Regional Research Center collection). All were sensitive to AVR2-V10, but of 15 different non-O157 E. coli isolates, including isolates of many other O serotypes, none was sensitive to the AVR2-V10 pyocin (see Table S2 in the supplemental material). Several rough laboratory E. coli strains were also tested, including K-12 (MG1655), E. coli B (BL21), and E. coli C; none was sensitive to AVR2-V10. The AVR2-V10 pyocin killed EDL933 but it did not kill TEA026, a mutant of EDL933 that lacks the O157 polysaccharide O antigen (5) (Fig. 3). On the contrary, the rough strain-specific AVR2-P2 killed mutant strain TEAO26 but not wild-type EDL933.
FIG. 3.
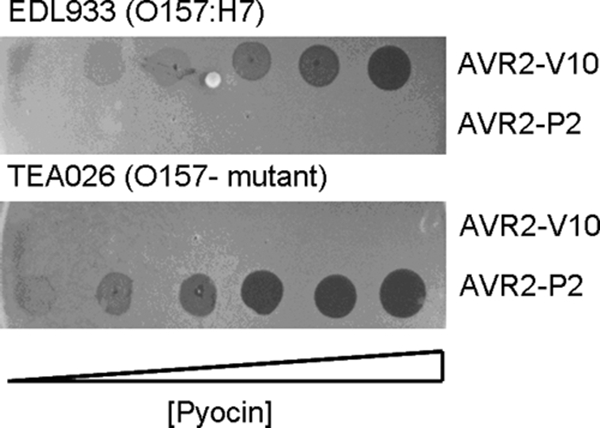
Semiquantitative spot tests of engineered pyocins AVR2-V10 and AVR2-P2 on lawns of strains EDL933 and TEA026, a mutant of EDL933 that cannot produce the O157 antigen. Pyocin samples (highest concentration, 1012 particles per ml) were serially diluted 1:4, and 5 μl of each dilution was spotted onto lawns seeded with the target bacteria (the highest concentrations are to the right). The plates were incubated at 37°C. Zones of clearing indicate bactericidal activity.
AVR2-V10 degrades the O157 LPS.
On the basis of the mechanism of infection by related phages, we predicted that the φV10 tail spike protein possesses catalytic activity that degrades the O157 polysaccharide structure. If the φV10 tail spike functions as predicted, then we would expect that AVR2-V10 would also degrade the O157 antigen. LPS was extracted from O157 strain EDL933 and incubated with purified AVR2-V10. The resulting LPS preparations were analyzed on SDS-polyacrylamide gels and silver stained for the O antigen. Incubation of the EDL933 LPS with AVR2-V10 resulted in the degradation of the high-molecular-weight polysaccharide (Fig. 4). The wild-type R2 pyocin had no detectable effect on the LPS.
FIG. 4.
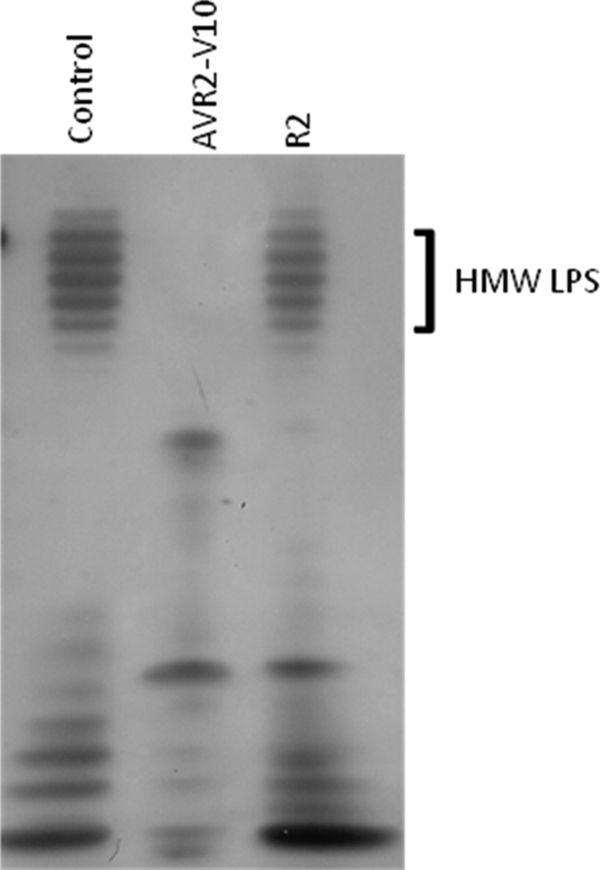
AVR2-V10 degrades O157 LPS. A silver-stained SDS-polyacrylamide gel of purified O157 LPS that was incubated with either AVR2-V10 or wild-type R2 pyocin is shown. The control is LPS alone (see Materials and Methods). HMW, high molecular weight.
E. coli mutants selected for resistance to AVR2-V10 have reduced O antigen.
A concern with any antibacterial agent is the emergence of resistant target bacteria. We isolated spontaneous AVR2-V10-resistant mutants of strain EDL933 at a frequency of less than 10−8. We also generated AVR2-V10-resistant mutants by treatment of EDL933 with MNNG or by transposon insertional mutagenesis. Of more than 20 such AVR2-V10-resistant EDL933 isolates, all displayed a phenotype in which O-antigen expression was either greatly reduced or completely eliminated. Figure 5 shows the results of analysis of the LPS from one such representative mutant, EDL933-AVr-7, obtained by MNNG mutagenesis.
FIG. 5.
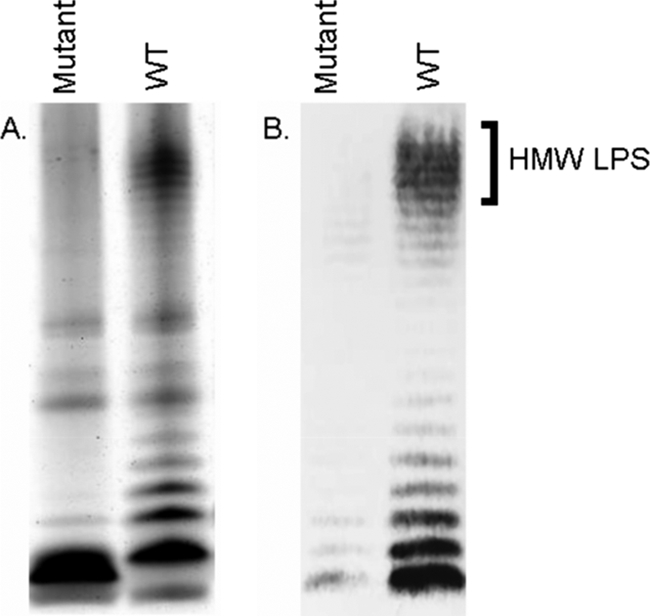
AVR2-V10-resistant mutants lack the O157 antigen. EDL933-AVr-7 (obtained by MNNG mutagenesis) is a representative AVR2-V10-resistant mutant of EDL933. (A) Silver-stained SDS-polyacrylamide gel of LPS isolated from EDL933-AVr-7 (mutant) and EDL933 (wild type [WT]). (B) Western blot of a parallel SDS-polyacrylamide gel run with an anti-O157 monoclonal antibody (see Materials and Methods). HMW, high molecular weight.
AVR2-V10 can eliminate E. coli O157:H7 from beef surfaces.
One potential application for AVR2-V10 is to kill O157:H7 cells on the surface of contaminated beef. Slices (25 cm2) of beef were contaminated with 2 × 104 CFU of gentamicin-resistant strain EDL933 previously grown to stationary phase. The samples were incubated at 4°C for 2 days, sprayed with AVR2-V10, incubated for 2 h, and washed vigorously by vortexing to release the bacteria; and then the bacteria were collected and plated on gentamicin-containing plates for enumeration. Greater than 90% of the gentamicin-resistant EDL933 could be recovered from untreated samples. Samples that were treated by spraying 109 pyocin particles per 25 cm2 resulted in a decrease in the numbers of bacteria below the countable level (<20 CFU) (Fig. 6). Similar results were seen in a control experiment in which the bacterial recovery buffer was at pH 3.8 (data not shown; see Materials and Methods). This inactivates AVR2-V10 but allows the recovery of the bacteria, negating the chance that the actual killing could be occurring during the plating process.
Stx production from E. coli O157:H7 is not increased by the presence of AVR2-V10.
The treatment of human E. coli O157:H7 infections with certain antibiotics is contraindicated due to the concern that antibiotics can induce Stx production in vitro. Stx is the most important virulence factor in STEC isolates, and there is clinical evidence that antibiotic therapy of STEC-induced colitis may lead to an increased risk of hemolytic-uremic syndrome (33). Therefore, an antimicrobial agent considered for use as a therapeutic agent to eliminate E. coli O157:H7 must do so without enhancing Stx production. Using a latex agglutination assay, we analyzed the amount of Stx2 present in supernatants from cultures of E. coli O157:H7 strain RM5202 (Stx2+) grown in the presence and the absence of AVR2-V10. Inocula of 5 × 107/ml E. coli O157:H7 were incubated with AVR2-V10 at ratios of pyocin to bacteria ranging from 20:1 to 1.2:1 (see Materials and Methods). At the highest pyocin/bacteria ratio, greater than 99.9% of the cells were killed rapidly; at the lowest ratio, less than 80% of the cells were killed. This range represents a potential therapeutic concentration down to a concentration that would likely be subtherapeutic. Compared to the negative control cells in TSB alone, no increase in Stx production was evident at any of the AVR2-V10 concentrations (Table 1). In contrast, treatment of the bacteria with ciprofloxacin resulted in a marked increase in the amount of Stx2. For example, even a sublethal concentration of ciprofloxacin (2 ng/ml) that resulted in partial cell lysis and death induced an increase in Stx production above that of cells exposed to 109 AVR2-V10 particles per ml; the amount of Stx detected in a 256-fold dilution of supernatant from cells exposed to ciprofloxacin at 2 ng/ml was comparable to the amount of Stx detected in a two- to fourfold dilution of supernatant from bacteria exposed to 109 AVR2-V10 particles per ml (Table 1).
TABLE 1.
Effects of lethal and sublethal concentrations of ciprofloxacin and AVR2-V10 on Stx2 production in culture supernatants of E. coli O157:H7 strain RM5202a
| Reciprocal dilution | Agglutination with:
|
||||||
|---|---|---|---|---|---|---|---|
| Ciprofloxacin concn (ng/ml)
|
No. of AVR2-V10 particles/ml
|
Control | |||||
| 25 | 6 | 2 | 1 × 109 | 2.5 × 108 | 6 × 107 | ||
| 2 | ++ | ++ | ++ | + | ± | ± | + |
| 4 | ++ | ++ | ++ | + | ± | ± | + |
| 8 | ++ | ++ | ++ | ± | ± | ± | + |
| 16 | ++ | ++ | + | − | − | − | ± |
| 32 | ++ | ++ | + | − | − | − | − |
| 64 | ++ | ++ | + | − | − | − | − |
| 128 | ++ | ++ | + | − | − | − | − |
| 256 | ++ | ++ | + | − | − | − | − |
E. coli O157:H7 strain RM5202 is stx2 positive and was grown for 6 h in TSB. Agglutination was scored as ++, +, ±, or − according to the manufacturer's instructions. The control consisted of bacteria grown in the absence of ciprofloxacin or AVR2-V10.
DISCUSSION
We have previously shown that it is possible to retarget R2 pyocins to kill rough strains of E. coli by fusing the C-terminal portion of the closely related bacteriophage P2 tail fiber to the N-terminal portion of the R2 tail fiber (32). This nonnatural pyocin, termed AVR2-P2, has bactericidal activity against E. coli C and K-12 but does not kill O157:H7 strains. This failure to kill the latter strains is likely due to the relatively thick O157 antigen itself, which blocks the adsorption of the pyocin to the cell surface (5). A similar phenomenon was reported for phage P1, the adsorption of which is blocked by the O157 antigen. A mutant of EDL933 defective in the production of O157 antigen polysaccharide, designated TEA026, however, was susceptible to P1 phage (5). Similarly, TEA026 was susceptible to AVR2-P2, but its wild-type O157:H7 parent was not. On the basis of these observations, we reasoned that a phage that recognizes the O157 antigen specifically as a receptor might be a source of a tail fiber (or tail spike) donor. Since R-type pyocins are related to the tail structures of P2-like phages and P2 tail fibers can successfully be fused with pyocin tail fibers to create pyocins active against P2 hosts, we tried to identify an O157-specific P2-like phage to serve as the donor of the receptor-binding domain. However, no such phage has been described in the literature, and our own efforts to isolate one from the environment have been unsuccessful to date (unpublished data). For these reasons, we chose to use the tail spike of O157-specific φV10, even though its virion is structurally unrelated to R-type pyocins. Despite this dissimilarity, the construction of functional fusions between the R2 tail fiber and the φV10 tail spike was successful, although it appears that fusion sites are limited to a few specific regions on both tail proteins. The bactericidal spectra of AVR2-V10, the fact that it can degrade the O157 high-molecular-weight LPS, and the observation that AVR2-V10 mutants have lost the O antigen all support our success in generating a pyocin that specifically targets the O antigen itself.
Podoviridae phage tail spikes (and tail fibers, in the case of T7) have been found to be homotrimers, including those of ɛ15, a close relative of φV10 (8, 13, 23, 27). We therefore suspect that the φV10 tail spike is also a homotrimer, and because it can form functional fusions with the R2 tail fiber, we believe that the R2 tail fiber polypeptide forms trimers as well.
While we have previously shown that it is possible to change the specificity of pyocin with chimeric tail fibers from the closely related P2-like phages, the fact that we can now change the specificity of pyocin using tail spikes of phages of the family Podoviridae greatly expands the potential target bacteria, particularly to strains that produce thick surface polysaccharide structures. Of particular note is the fact that the AVR2-V10 chimeric pyocin retains the catalytic O-antigen-degrading activity of the donor phage tail spike, a feature not known in any natural phage tail-like bacteriocin.
In nature, bacteriophages have clearly evolved chimeric tail structures based on polysaccharide-degrading tail spikes. For example, a similar tail spike protein, the K1 endosialidase, has been identified as a natural component of diverse phages, including a P22-like phage of the family Podoviridae (11), a T7-like phage of the family Podoviridae (20), and a phage of the family Siphoviridae (14); in each case, the tail spikes are fused to a different head-binding (or base plate-binding) portion of a tail protein. A similar organization has been reported with the P22-like endorhamnosidase; and in the case of this enzyme, the tail spike is also part of a Myoviridae tail structure (31). The adaptability of these tail spikes to unrelated and different phage virion structures provides an obvious evolutionary advantage, especially in light of the fact that phages readily exchange genetic information. The mechanism by which phages naturally acquire tail spike genes and adapt the proteins to the virion structure is unclear, but it could simply involve some combination of illegitimate recombination to acquire the gene followed by gene rearrangements and/or small mutations that result in the formation of functional tail fibers and spikes. In some cases, a more specific mechanism might be involved. For example, in SP6-like phages, the sequences for all the tail spikes are encoded in a conserved cassette near the end of the genome, suggesting that tail spikes might be readily swapped; but this is yet to be proven (21). Whatever the natural course of events, we have shown, using modern molecular techniques, that this structural flexibility of phage tail fibers and spikes can be exploited to generate novel phage tail-like bacteriocins. We expect to be able to further exploit this information to expand the range of target bacteria using the tail spikes of other diverse phages.
Although phage φV10 has been shown to be specific to E. coli O157, it does not plaque on all O157 strains (30). On the other hand, AVR2-V10 killed all 49 O157 isolates tested and may therefore provide complete coverage within the serotype. This difference in spectrum may be explained by the mechanism of R-type pyocin action. To be bactericidal, AVR2-V10 likely needs only to attach to the surface, insert its core, and disrupt the membrane potential. On the other hand, the bactericidal action of phages involves many more functions of both the phage and the host bacterium, particularly the ability to defeat DNA restriction systems, to replicate, to assemble within the target organism, and ultimately, to kill the target organism while escaping.
The O157 antigen itself is a virulence factor, the loss of which compromises the bacterial colonization of the mammalian gut (5). We showed that the resistance of E. coli O157:H7 to AVR2-V10 resulted in the loss of expression of that O antigen. Thus, the rare AVR2-V10-resistant mutants would be expected to be less pathogenic and the emergence of resistance of less concern. This is potentially an advantageous property for a bactericidal agent.
E. coli O157:H7 continues to be a major cause of food-borne illness. Because patients presenting with signs and symptoms of STEC-caused colitis probably have a Shiga toxemia at the time of diagnosis (NIH Workshop on Prevention of Hemolytic Uremic Syndrome Caused by STEC, 19 June 2002, Bethesda, MD), a major focus has been on eliminating the pathogen from foodstuffs rather than therapeutic intervention after infection. The clinical intervention with antibiotics is also contraindicated because of the concern that antibiotics may enhance toxin production by STEC. A highly specific, cost-effective agent that kills this human pathogen in or on food without affecting the quality of food products could theoretically reduce the incidence of illness due to this particularly virulent strain. Engineered R-type pyocins are good candidates for such an application. They consist of proteins, have no known toxicities, and are bactericidal at low concentrations. In addition, because AVR2-V10 does not enhance the production or cause the release of Stx from STEC, it is also a potential candidate therapeutic agent both for the treatment of infections in humans and possibly for the minimization of human-to-human transmission by reducing the shedding of bacteria from infected patients (1).
ADDENDUM IN PROOF
Relevant to Fig. 6, the further binding of unbound AvR2-V10 to gentamicin-resistant EDL933 bacteria upon or after their recovery from meat can be effectively quenched by vortexing the meat slices in selective medium containing 104 excess of gentamicin-sensitive (wild-type) EDL933 bacteria. We also found that the recovery of gentamicin-resistant E. coli O157:H7 CFUs from exposure to pH 3.8 is greater and complete from 50 mM citrate/citric acid buffer than from acetate/acetic acid buffer when 1 ml is diluted into 3 ml of soft agar medium.
Supplementary Material
Acknowledgments
We thank Richard Calendar for helpful discussions.
Part of this work was supported by USDA-ARS CRIS project 5325-42000-045-00D and grants from USDA-CSREES section 32.1 (projects 2006-01240 and 2007-02029).
Footnotes
Published ahead of print on 6 April 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ahn, C. K., E. Klein, and P. I. Tarr. 2008. Isolation of patients acutely infected with Escherichia coli O157:H7: low tech, highly effective prevention of hemolytic uremic syndrome. Clin. Infect. Dis. 46:1197-1199. [DOI] [PubMed] [Google Scholar]
- 2.Cooley, M., D. Carychao, L. Crawford-Miksza, M. T. Jay, C. Myers, C. Rose, C. Keys, J. Farrar, and R. E. Mandrell. 2008. Incidence and tracking of E. coli O157:H7 in a major produce production region in California. PLoS One 2:e1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas, H., T. Sacks, and N. Saltz. 1974. Protective effect of pyocin against lethal Pseudomonas aeruginosa infections in mice. J. Infect. Dis. 129:470-472. [DOI] [PubMed] [Google Scholar]
- 5.Ho, T. D., and M. K. Waldor. 2007. Enterohemorrhagic Escherichia coli O157:H7 gal mutants are sensitive to bacteriophage P1 and defective in intestinal colonization. Infect. Immun. 75:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishii, S., Y. Nishi, and F. Egami. 1965. The fine structure of a pyocin. J. Mol. Biol. 13:428-431. [DOI] [PubMed] [Google Scholar]
- 7.Ito, S., M. Kageyama, and F. Egami. 1970. Isolation and characterization of pyocins from several strains of Pseudomonas aeruginosa. J. Gen. Appl. Microbiol. 16:205-214. [Google Scholar]
- 8.Jiang, W., J. Chang, J. Jakana, P. Weigele, J. King, and W. Chiu. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kageyama, M. 1964. Studies of a pyocin. I. Physical and chemical properties. J. Biochem. 55:49-53. [DOI] [PubMed] [Google Scholar]
- 10.Kageyama, M., K. Ikeda, and F. Egami. 1964. Studies of a pyocin. III. Biological properties of the pyocin. J. Biochem. 55:59-64. [DOI] [PubMed] [Google Scholar]
- 11.King, M. R., R. P. Vimr, S. M. Steenbergen, L. Spanjaard, G. Plunkett III, F. R. Blattner, and E. R. Vimr. 2007. Escherichia coli K1-specific bacteriophage CUS-3 distribution and function in phase-variable capsular polysialic acid O acetylation. J. Bacteriol. 189:6447-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kropinski, A. M., I. V. Kovalyova, S. J. Billington, A. N. Patrick, B. D. Butts, J. A. Guichard, T. J. Pitcher, C. C. Guthrie, A. D. Sydlaske, L. M. Barnhill, K. A. Havens, K. R. Day, D. R. Falk, and M. R. McConnell. 2007. The genome of epsilon15, a serotype-converting, group E1 Salmonella enterica-specific bacteriophage. Virology 369:234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long, G. S., J. M. Bryant, P. W. Taylor, and J. P. Luzio. 1995. Complete nucleotide sequence of the gene encoding bacteriophage E endosialidase: implications for K1E endosialidase structure and function. Biochem. J. 309:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida, Y., K. Miyake, K. Hattori, S. Yamamoto, M. Kawase, and S. Iijima. 2000. Structure and function of a novel coliphage-associated sialidase. FEMS Microbiol. Lett. 182:333-337. [DOI] [PubMed] [Google Scholar]
- 15.Matsui, H., Y. Sano, H. Ishihara, and T. Shinomiya. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J. Bacteriol. 175:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merrikin, D. J., and C. S. Terry. 1972. Use of pyocin 78-C2 in the treatment of Pseudomonas aeruginosa infection in mice. Appl. Microbiol. 23:164-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel-Briand, Y., and C. Baysse. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499-510. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama, K., K. Takashima, H. Ishihara, T. Shinomiya, M. Kageyama, S. Kanaya, M. Ohnishi, T. Murata, H. Mori, and T. Hayashi. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213-231. [DOI] [PubMed] [Google Scholar]
- 19.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of E. coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholl, D., and C. Merril. 2005. The genome sequence of bacteriophage K1F, a T7-like phage that has acquired the ability to infect K1 strains of E. coli. J. Bacteriol. 187:8499-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholl, D., and C. Merril. 2005. Polysaccharide-degrading phages, p. 400-414. In Phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC.
- 22.Scholl, D., and D. W. Martin, Jr. 2008. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a mouse peritonitis model. Antimicrob. Agents Chemother. 52:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seckler, R. 1998. Folding and function of repetitive structure in the homotrimeric phage P22 tailspike protein. J. Struct. Biol. 122:216-222. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu, Y., T. Kamazaki, and S. I. Ishii. 1982. Specific cleavage at fibers of a bacteriophage-tail-like bacteriocin, pyocin R1 by successive treatment with organomercurial compounds and trypsin. J. Virol. 44:692-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinomiya, T., S. Shiga, and M. Kageyama. 1983. Genetic determinant of pyocin R2 in Pseudomonas aeruginosa PAO. I. Localization of the pyocin R2 gene cluster between the trpCD and trpE genes. Mol. Gen. Genet. 189:375-381. [DOI] [PubMed] [Google Scholar]
- 26.Sivapalasingam, S., C. R. Friedman., L. Cohen, and R. V. Tauxe. 2004. Fresh produce: a rowing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67:2342-2353. [DOI] [PubMed] [Google Scholar]
- 27.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. D. Parry. J. S. Wall, J. F. Hainfield, and W. F. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 28.Strauch, E., H. Kaspar, C. Schaudinn, P. Dersch, K. Madela, C. Gewinner, S. Hertwig, J. O. Wecke, and B. Appel. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl. Environ. Microbiol. 67:5634-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uratani, Y., and T. Hoshino. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157:632-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waddell, T. E., and C. Poppe. 2000. Construction of mini-Tn10luxABcam/Ptac-ATS and its use for developing a bacteriophage that transduces bioluminescence to Escherichia coli O157:H7. FEMS Microbiol. Lett. 182:285-289. [DOI] [PubMed] [Google Scholar]
- 31.Walter, M., C. Fiedler, R. Grassl, M. Biebl, R. Rachel, X. L. Hermo-Parrado, A. L. Llamas-Saiz, R. Seckler, S. Miller, and M. J. van Raaij. 2008. Structure of the receptor-binding protein of bacteriophage det7: a podoviral tail spike in a myovirus. J. Virol. 82:2265-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, S., D. Gebhart, D. W. Martin, and D. Scholl. 2008. Re-targeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74:3868-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


